Abstract
Posttranscriptional and translational controls mediated by microRNAs (miRNA) regulate diverse biologic processes. We dissected regulatory effects of miRNAs relevant to megakaryocytopoiesis and platelet biology by analyzing expression patterns in 79 subjects with thrombocytosis and controls, and integrated data with transcriptomic and proteomic platforms. We validated a unique 21-miRNA genetic fingerprint associated with thrombocytosis, and demonstrated that a 3-member subset defines essential thrombocythemia (ET). The genetic signature includes functional guide and passenger strands of the previously uncharacterized miR 490 (5p and 3p), which displayed restricted, low-level expression in megakaryocytes/platelets (compared with leukocytes), and aberrant expression during thrombocytosis, most profound in ET. Overexpression of miR 490 in a bilineage differentiation model of megakaryocyte/erythroid progenitor formation was insufficient for hematopoietic colony differentiation and/or lineage specification. Integration of transcriptomic and mass spectrometric datasets with functional reporter assays identified dishevelled associated activator of morphogenesis 1 (DAAM1) as a miR 490 5p protein target demonstrating decreased expression in ET platelets, putatively by translational control (and not by mRNA target degradation). Our data define a dysregulated miRNA fingerprint in thrombocytosis and support a developmentally restricted function of miR 490 (and its putative DAAM1 target) to conditions associated with exaggerated megakaryocytopoiesis and/or proplatelet formation.
Introduction
Recent data have demonstrated that both megakaryocytes and platelets retain an abundant and diverse array of microRNAs (miRNAs). Largely studied within the context of mammalian development and lineage specification, miRNAs are a class of noncoding 21- to 24-bp species that primarily regulate protein translation by posttranscriptional targeting of 3′-untranslated regions (UTRs).1 Emerging evidence has implicated miRNAs in the control of megakaryocytopoiesis2 and in progenitor fate during the megakaryocyte-erythroid transition,3 presumably by modulating expression of key transcriptional regulators.2,3 Furthermore, distinct patterns of miRNA expression have been seen in differentiated hematopoietic cells4 and in subsets of patients with myeloproliferative neoplasms,5,6 further implicating discrete miRNAs in lineage commitment during normal or dysregulated hematopoiesis.
Anucleate platelets retain megakaryocyte-derived mRNAs7 and have evolved unique adaptive signals for maintenance of genetic and protein diversity.8,9 Quiescent platelets generally display minimal translational activity, although maximally activated platelets retain the capacity for protein synthesis, with implications for modulating arthritis-associated inflammation,10 or the production of platelet progeny in vivo.11 Interestingly, platelets also retain a competent miRNA pathway capable of converting precursor miRNAs through functional Dicer/Argonaute 2 (Ago2) complexes,12 with evidence that Ago2-miRNA-223 complexes specifically regulate expression of the functionally important platelet purinergic P2Y12 adenosine-diphosphate (ADP) receptor. Additional evidence suggests that miRNAs (miR 28) can modulate expression of the c-mpl thrombopoietin (Tpo) platelet receptor5 and that miR 96–mediated regulation of endobrevin/VAMP8 (vesicle-associated membrane protein 8) affects human platelet functional responsiveness.13 The potential importance of a functionally competent miRNA pathway is further highlighted by the enrichment of platelet miRNAs compared with other hematopoietic cells, such as granulocytes and megakaryocytes.12
We have previously shown that distinct platelet phenotypes can be genetically classified using a restricted set of mRNA biomarkers.14 We have now expanded these observations to further investigate the impact of miRNAs in thrombocytosis, and we have identified a unique fingerprint associated with enhanced megakaryocytopoiesis and/or proplatelet formation. Systematic analyses of miRNA with transcriptomic and proteomic studies using an integrated computational framework identified miR 490 5p and its putative target dishevelled associated activator of morphogenesis 1 (DAAM1) as reciprocally regulated pairs in thrombocythemic platelets, presumably by translational control and not by mRNA target degradation. These data suggest a unique mechanism for modifying the platelet proteome in times of exaggerated platelet production.
Methods
Human subjects
A total of 79 platelet RNAs were randomly chosen from a State University of New York at Stony Brook institutional review board–approved study of thrombocytosis subjects and healthy controls14 ; samples were assigned to 3 cohorts based on previous phenotypic characterization: essential thrombocythemia (ET; N = 27), reactive thrombocytosis (RT; N = 22), and healthy controls (N = 30; patient characteristics are provided in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Leukocytes were isolated by density-gradient centrifugation from sodium citrate-treated blood (0.4% volume/volume final concentration), whereas platelet-rich plasma served as the source of platelet mRNA. Leukocyte contamination of platelet-rich plasma was < 1 × 10−5, and the isolation, quantification, and quality control of both leukocyte and platelet RNAs were established using an 2100 Bioanalyzer (Agilent Technologies) as previously described.14 JAK2V617F mutation genotyping (exon 12, 1849G→T transversion) was completed on all subjects.15 This study was conducted in accordance with the Declaration of Helsinki.
miRNA profiling and data analysis
Platelet RNA (100 ng/sample) was labeled with Cy3 amine reactive dyes using a miRNA labeling kit (Ambion), and samples were hybridized to an Agilent G4470C human miRNA gene chip that incorporates 866 human and 89 viral miRNAs (miRBase database Version 12.0). Images were acquired using a GenePix 4000B scanner (Molecular Devices), followed by quantification and processing using Agilent Feature Extraction software (Version 10.7). Within-group coefficient of variation was determined to evaluate intra-array reproducibility, and samples with values below the default (coefficient of variation < 15%) were re-arrayed until they fulfilled the acceptable requirement. The microarray data were analyzed using Bioconductor package on R (Version 2.11) and deposited in the public GEO database (GEO accession no. GSE39046). A moderate filtering step was adopted to specifically include miRNAs with > 70% present cells in any of the 3 groups (ET, normal, and RT), followed by quantile normalization with K-nearest neighbor imputation (K = 10) for missing values16 ; the same standard was applied to each sample to ensure data integrity on both dimensions. Two-way unsupervised hierarchical clustering was performed to generate dendrograms using median-centered miRNA profiles (1-Spearman correlation). Significance analysis of microarrays was performed to identify differentially expressed genes among cohorts (ie, ET, RT, and normal), using fold change ≥ 2 and false discovery rate ≤ 0.05 as cutoffs.
A comprehensive discriminant analysis was used to identify an initial biomarker subset that separated class on the basis of microarray data, restricted to 2-4 miRNAs to prevent over-fitting. The fidelity of the genetic biomarker subsets as class prediction tools for both microarray and quantitative RT-PCR was established using linear discriminant analysis with a leave-one-out cross-validation analysis.17 The posterior classification probability for each subject was derived, and the binary decision for group assignment was based on individual highest probability; the same biomarker set using the microarray data was applied to the quantitative RT-PCR data.
Quantitative RT-PCR
Confirmatory miRNA expression (ET, normal, or CD34+-differentiated) was determined using 10-ng RNA as template and TaqMan miRNA stem-loop primers (Applied Biosystems). Expression values for individual miRNAs were obtained from triplicate wells run in parallel, calculated in arbitrary units as 2[-ΔCt], where ΔCt represents the Ct values of internal standards miR 22 and miR 720 minus that of individual sample wells. The group means of miR 22 and miR 720 were specifically chosen as internal standards because they displayed the lowest coefficient of variations across gene arrays. Cross-platform (ie, Agilent microarray and quantitative RT-PCR) expression interrelatedness was established for individual miRNAs using the Pearson correlation of data uniformly scaled and standardized to mean = 0 and SD = 1. The miR 490 threshold sensitivity was established by serial dilution and linear regression modeling of CD34+ cells expressing lentivirus-transduced miR 490 or empty vector control. Platelet DAAM1 mRNA was quantified using 125 ng RNA and fluorescence-based SYBR Green 1 standardized to β-actin (oligonucleotide primers are available in supplemental Table 2).7,14
Hematopoietic assays and lentiviral transduction
Lentiviruses expressing miR 490 (LV 490) or empty virus control (LV 000) driven by the elongation factor 1 α (EF1α) promoter and containing the puromycin-resistant cassette were obtained from Biosettia. Vesicular stomatitis virus-pseudotyped lentiviruses were generated in 293T cells, and concentrated stocks were titered in NIH-3T3 cells.18 Transduction efficiencies in 293T cells for both viruses were nearly identical with infectious units titers as follows: LV000 (1.8 × 107/mL) and LV490 (2.0 × 107/mL). Cryopreserved, peripheral blood-mobilized human CD34+ hematopoietic stem cells were obtained from the NHLBI Program of Excellence in Gene Therapy at the Fred Hutchinson Cancer Center (Seattle, WA) and propagated using well-defined and specified reagents from Stem Cell Technologies. CD34+ hematopoietic stem cells were cultured in SFEM expansion medium, and 24 hours after thaw were lentivirus-transduced using multiplicity of infection of 1 by spin-transduction (1000g) in the presence of 4 μg/mL polybrene for 2 hours at 25°C. Cells were washed free of lentivirus/polybrene and selected with 2 μg/mL puromycin 24 hours after infection; noninfected controls in the presence of puromycin displayed 0% cell viability by 48 hours. Colony assays were completed in methylcellulose cultures using MethoCult (H4034 Optimum) for quantification of hematopoietic progenitors or MegaCult collagen-based semi-solid media (no. 04901) supplemented with 50 ng/mL thrombopoietin, 10 ng/mL IL-6, and 10 ng/mL IL-3 for Mk progenitor assays. Hematopoietic progenitors (500 cells/plate) were morphologically enumerated at day 14, whereas Mk colony formation was quantified from cells (1 × 105/mL) fixed and stained at day 10. Liquid cultures (for 21-day time course and bilineage expansion) were maintained in puromycin-selected cells using SFEM supplemented with CC100 expansion cocktail, 50 ng/mL thrombopoietin, and 2 U/mL erythropoietin. Cells were fixed at day 7 using 1% formalin and flow cytometric quantification using FITC-conjugated anti-CD41 MAb (integrin αIIb, Mk marker) or anti-glycophorin A (erythroid) for determination of lineage specificity; alternatively, cells were pelleted at distinct time points for RNA isolation.7
Reporter gene activity assays
Human embryonic kidney (HEK) 293 cells at passage 30-34 were diluted to 2 × 105/mL and plated in 96-well microtiter plates to reach ∼ 70% confluency by the time of transfection (usually at 16-24 hours). Transfections were completed using DharmaFect Duo (Thermo Fisher Scientific) and 30 ng/μL pLightSwitch plasmid (SwitchGear Genomics) in which the terminal 975-bp DAAM1 3′-UTR was inserted downstream of Renilla luciferase (RLuc). Cotransfections were completed using chemically optimized double-stranded miR 490 3p or miR 490 5p mimics that act as functional equivalents to endogenous human miRNAs (SwitchGear Genomics); a nontargeting Caenorhabditis elegans miR 239b stem-loop primer at identical concentrations was used as control (supplemental Table 2). At 24 hours after transfection, cells were lysed by freezing at −80°C, followed by luminescence detection after 30-minute incubation at 25°C in 100 μL of the RLuc-optimzed assay substrate, using a SpectraMax L luminometer (Molecular Devices); in all situations, cellular toxic effects of individual miRNAs were excluded by MTT assays completed in parallel wells using identical conditions.
Platelet mass spectrometric and protein analyses
Platelet-rich plasma from 3 healthy controls and 3 ET subjects was incubated with 20μM acetylsalicylic acid, 0.1μM prostaglandin E1, and 13mM EDTA at 37°C for 20 minutes to ensure quiescence, followed by isolation of gel-filtered platelets over a Sepharose 2B column as previously described.7 Solubilized proteins obtained from ∼ 3 × 108 platelets were reduced using 4mM DTT, alkylated in 8.4mM iodoacetamide, digested for 16 hours at 37°C using sequencing-grade trypsin (mass ratio 1:40), and immediately frozen at −80°C. Samples were pressure bomb-loaded onto a column constructed of 250-μm ID fused silica tubing packed with 3 cm of strong cation exchanger (5 μm) and 3 cm of C18 matrix (5 μm) for multidimensional protein identification technology (MudPIT) using microcapillary liquid chromatography and tandem mass spectrometry (μLC-MS/MS).19,20 HPLC was completed using a 12-step fully automated separation interfaced with an LTQ linear ion trap mass spectrometer for full mass spectra determination over a 400-2000 m/z range; 5 tandem MS/MS events were recorded for optimal peptide identification linked to a human IPI (International Protein Index) database, retaining only peptides with a P value < 0.01 and proteins identified by 2 distinct peptides for further analysis. Spectral counts were quantile-normalized to ensure cross-experimental comparisons, and the overall concordance between technical replicates was excellent (r = 0.987). For select proteins, multiple reaction monitoring was used as a highly sensitive validation assay for peptide quantification21 ; in brief, samples were rerun on the LTQ mass spectrometer, and a protein-specific peptide(s) and its corresponding fragment ions were monitored (and quantified) over the chromatographic elution time; the specific pairs of m/z transitions corresponding to precursor and fragment ions allow precise spectral identification and quantification as a validation of semiquantitative MudPIT analyses. Immunoblot analysis was completed using 4%-15% SDS-PAGE and anti-DAAM1 (Abcam; 1:100) or antiactin (1:1000) mAbs.7
miRNA target prediction algorithms linking miRNA/mRNA/proteomic datasets
An integrated workflow model was developed to identify miR 490 protein targets, specifically applying 2 sequential filters designed to (1) pool all potential targets derived from the union of computational prediction databases using R bioconductor package (Version 2.6) RmiR.Hs.miRNA (referred to as our “knowledge base”), followed by (2) a second (“data-driven base”) for comparative integration of mRNA and proteomic data. Microarray mRNA profiles included our previously described 6 ET subjects and 5 healthy controls (GEO accession no. GPL1716)22 ; a filtering step was applied to retain genes expressed in at least 6 samples, and genes represented by duplicate probes were combined and averaged before quantile normalization. For proteomics data, quantile-normalized MS spectral counts were averaged by cohort, restricting the analyses to proteins displaying spectral counts > 10 to bypass the inherent limitations of protein quantification at low spectral counts. Samples were specifically filtered to identify mRNA/protein pairs with counts present in one group and absent in another, or were analyzed by significance analysis of microarrays when expressed in a minimum of 4 of 6 samples. In the first iteration, an inverse correlation between miRNA/mRNA abundance was built to reflect miRNA-mediated target degradation mechanism, whereas in the second iteration we applied statistically unchanged mRNA expression levels (group mean folds within the range 0.5 < group mean folds < 2.0 with insignificant P values > .05). For all statistical algorithms, P values were corrected by Benjamini-Hochberg multiple testing correction.
Results
Dysregulated miRNA expression patterns among normal and thrombocytotic cohorts
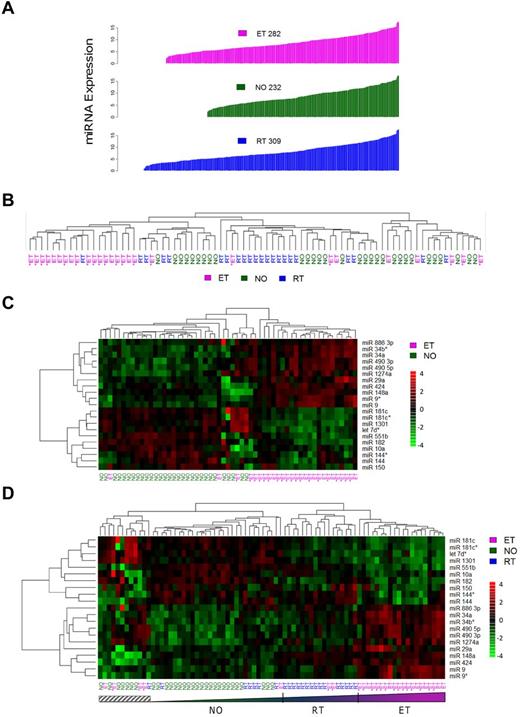
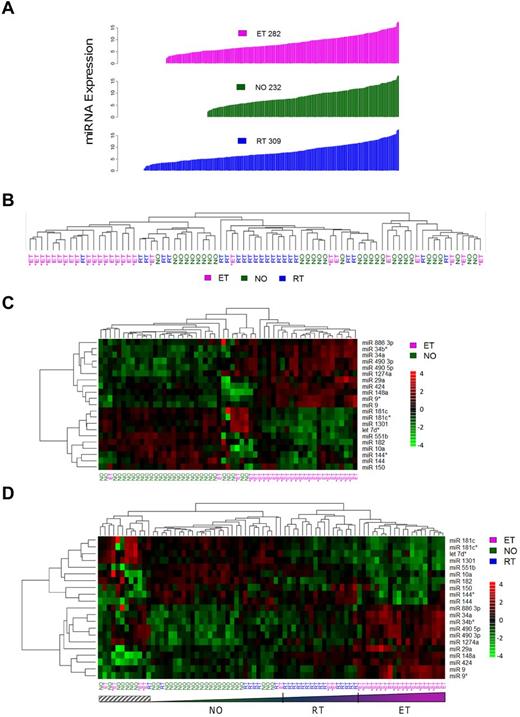
To gain insight into platelet miRNA function and expression patterns in normal and pathologic conditions (thrombocytosis), we microarrayed 79 human platelet samples from a previously described cohort,14 composed of normal controls (NO; n = 30), patients with RT (n = 22), or patients with ET (n = 27). Absolute platelet miRNA expression patterns ranged from N = 232 (healthy controls) to N = 309 (RT; Figure 1A; supplemental Table 3). These aggregate data considerably expand on the previously described platelet miRNA expression repertoire12 ; more importantly, they establish the dynamic nature of the megakaryocytic/platelet transcriptome during situations of megakaryoctyopoiesis and/or enhanced proplatelet formation.
General miRNA expression patterns across normal and thrombocytotic cohorts. (A) Aggregate expression patterns are displayed for platelet miRNAs displaying > 70% present calls by cohort, in order of increasing fluorescence intensities. Expression data for each cohort represent normalized means from healthy controls (n = 30), ET (n = 27), or RT (n = 22). (B-D) Unsupervised hierarchical clustering was completed based on correlation proximity; miRNA expression data are standardized to mean = 0, SD = 1 before clustering, and displayed as a continuous data range from −4 to +4. ET cohorts are substratified by the presence (ET+) or absence (ET−) of JAK2V617F. (B) The clustering dendrograms using the aggregate 392-member miRNAs, which demonstrates relative segregation of ET from RT and NO cohorts, whereas (C) ET and NO phenotypes alone and (D) all 3 phenotypes delineate the phenotypic segregation using the 21-member miRNA list. Note the relative continuum of phenotypes in panel D as defined by the 21-member miRNAs, exclusive of a small genetically heterogeneous subset (hatched box).
General miRNA expression patterns across normal and thrombocytotic cohorts. (A) Aggregate expression patterns are displayed for platelet miRNAs displaying > 70% present calls by cohort, in order of increasing fluorescence intensities. Expression data for each cohort represent normalized means from healthy controls (n = 30), ET (n = 27), or RT (n = 22). (B-D) Unsupervised hierarchical clustering was completed based on correlation proximity; miRNA expression data are standardized to mean = 0, SD = 1 before clustering, and displayed as a continuous data range from −4 to +4. ET cohorts are substratified by the presence (ET+) or absence (ET−) of JAK2V617F. (B) The clustering dendrograms using the aggregate 392-member miRNAs, which demonstrates relative segregation of ET from RT and NO cohorts, whereas (C) ET and NO phenotypes alone and (D) all 3 phenotypes delineate the phenotypic segregation using the 21-member miRNA list. Note the relative continuum of phenotypes in panel D as defined by the 21-member miRNAs, exclusive of a small genetically heterogeneous subset (hatched box).
Identification of differentially expressed miRNAs
Initial unsupervised hierarchical clustering of 392 aggregate miRNAs revealed an expression pattern that genetically segregated the groups by phenotype (Figure 1B). Coclustering of RT with normal platelets provided genetic segregation distinct from ET. Direct visual quantification confirmed that 18 of 27 ET samples (67%) were assigned to the same cluster, whereas RT and normal platelets were randomly grouped within a larger cluster. Further ET substratification by the JAK2V617F mutation demonstrated genetic segregation from the RT/normal cluster irrespective of JAK2 genotype. Based on the unsupervised clustering results, 13 of 19 (68%) JAK2V617F and 5 of 8 (63%) JAK2 wild-type ET samples were assigned to the ET-enriched cluster, results consistent with previous genetic classification data using aggregate and condensed mRNA expression profiles.14
Significance analysis of microarrays identified 21 miRNAs that were most differentially expressed among the cohorts and most pronounced comparing ET to normal (Figure 1C; Table 1). Aggregate group-wise comparisons demonstrated less pronounced differences between NO and RT, results that were consistent with the phenotypic cosegregation using unsupervised hierarchical clustering. We repeated the unsupervised hierarchical clustering among the 3 groups using the 21-member miRNAs (Figure 1D), results that demonstrated clear cosegregation by phenotype. Furthermore, this 21-miRNA fingerprint defined clustered phenotypic progression whereby ET subjects displayed the most exaggerated expression patterns whereas those with RT represented an intermediary subset flanked by normal and ET cohorts. These data strongly suggested that normal and exaggerated thrombopoiesis represent genetically defined continua along a convergent pathway, rather than qualitatively distinct genetic entities.
The majority of differentially expressed miRNAs (19 of 21; 90.5%) displayed concordant expression patterns in thrombocytosis (ET:NO or RT:NO), suggesting that the final common pathway(s) controlling megakaryocytopoiesis and/or proplatelet formation are regulated by comparable developmental networks. Eight of the 21 miRNAs (miR 9/9*, miR 144/144*, miR 181c/181c*, and miR 490 3p/490 5p) represented complementary sequences derived from single pre-miRNAs23 and demonstrated parallel fold-changes of the 5′- and 3′-strands. Of the differentially expressed miRNAs, 2 (miR 34a and miR 182) were previously described as displaying aberrant expression patterns in polycythemia vera granulocytes,6 and 3 others (miR 10a, miR 150, and miR 144) have been implicated in regulation of cell-lineage fate during megakaryocytopoiesis,2 megakaryocyte-erythroid progenitors,3 or erythroid development,24 respectively. Both miR 10a and miR 150 were down-regulated in ET and RT platelets, and the miR 144/miR144* pair was singularly noted to display paradoxical responses in ET compared with RT.
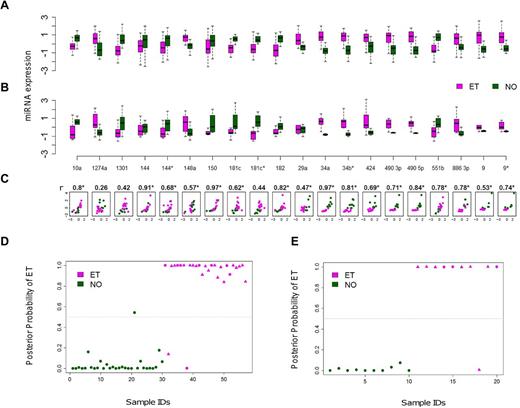
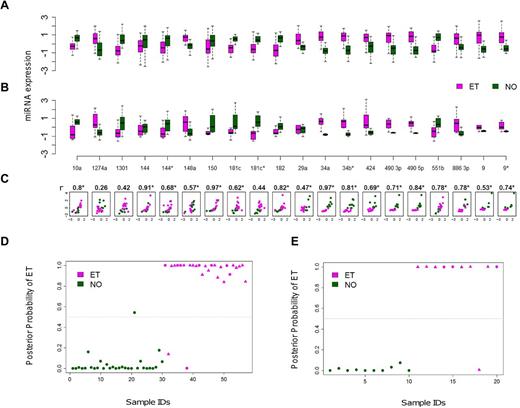
A subset of 20 of the 21 miRNAs was reanalyzed using quantitative RT-PCR as a second platform to validate the microarray expression differences between ET and NO (the pri-miRNA let 7d was excluded). A high correlation coefficient was observed between the 2 methods in aggregate (r = 0.69, P = .02), or individually where 19 of 20 miRNAs exhibited concordant group medians (Figure 2A-C). The consistency of miRNA expression profiles using 2 independent platforms verified the fidelity of the 20 biomarkers and prompted more focused studies.
Cross-platform validation and class prediction models. (A-B) Normalized miRNA expression data using paired microarray (A) or quantitative RT-PCR (B) are displayed for a randomly selected subset of ET (N = 10) and control (N = 10) platelets (each group included 5 males and 5 females). For both microarray and quantitative RT-PCR, boxes represent the within-group interquartile range encompassing 50% of the values, whereas the 95% confidence intervals and outliers are depicted; the horizontal bar within each box represents the group median. (C) Scatter plots demonstrate between-platform concordance for individual miRNAs by phenotype; the x- and y-axes represent the microarray and quantitative RT-PCR data, respectively, using a unified scale standardized to mean = 0 and SD = 1; 19 of 20 miRNAs (except for miR 1274a) exhibit concordant group medians, validating the expression data using independent platforms. *Statistically significant Spearman correlation coefficients (r) with P < .05. (D-E) Linear discriminant analysis plots display the posterior classification probability of each subject using a 3-biomarker subset (miR 10a, miR 148a, and miR 490 5p) based on microarray (D; N = 57) or quantitative RT-PCR (E; N = 20). Triangles represent samples containing the JAK2V617F mutation (either homozygous or heterozygous).
Cross-platform validation and class prediction models. (A-B) Normalized miRNA expression data using paired microarray (A) or quantitative RT-PCR (B) are displayed for a randomly selected subset of ET (N = 10) and control (N = 10) platelets (each group included 5 males and 5 females). For both microarray and quantitative RT-PCR, boxes represent the within-group interquartile range encompassing 50% of the values, whereas the 95% confidence intervals and outliers are depicted; the horizontal bar within each box represents the group median. (C) Scatter plots demonstrate between-platform concordance for individual miRNAs by phenotype; the x- and y-axes represent the microarray and quantitative RT-PCR data, respectively, using a unified scale standardized to mean = 0 and SD = 1; 19 of 20 miRNAs (except for miR 1274a) exhibit concordant group medians, validating the expression data using independent platforms. *Statistically significant Spearman correlation coefficients (r) with P < .05. (D-E) Linear discriminant analysis plots display the posterior classification probability of each subject using a 3-biomarker subset (miR 10a, miR 148a, and miR 490 5p) based on microarray (D; N = 57) or quantitative RT-PCR (E; N = 20). Triangles represent samples containing the JAK2V617F mutation (either homozygous or heterozygous).
A discrete 3-miRNA subset discriminates normal from ET platelets
We applied various statistical classifiers to define a limited miRNA subset capable of discriminating normal and thrombocythemic phenotypes. The initial searching algorithm identified 19 distinct combinations of 3 miRNAs demonstrating class prediction accuracy > 90%, although repeat analysis using leave-one-out cross-validation refined this list to 3 miRNAs (miR 10a, miR 148a, and miR 490 5p). The application of this miRNA subset to both the microarray and quantitative RT-PCR data was confirmed using 3 distinct statistical classifiers (Figure 2D-E; Table 2), providing both statistical and cross-platform validation. The identical 3 miRNA subset performed less robustly in discriminating RT from NO (supplemental Table 4), and we were unable to define a more robust RT/NO discriminatory subset using the 21-miRNA subset and de novo computational algorithms. This is entirely consistent with the previously identified limited differences between RT and NO miRNA profiles.
miR 490 displays lineage (Mk/platelet)–restricted expression and is aberrantly expressed during thrombocytosis
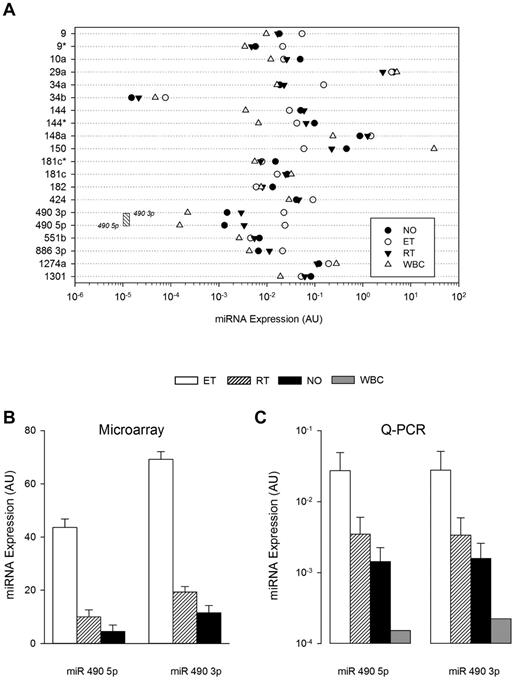
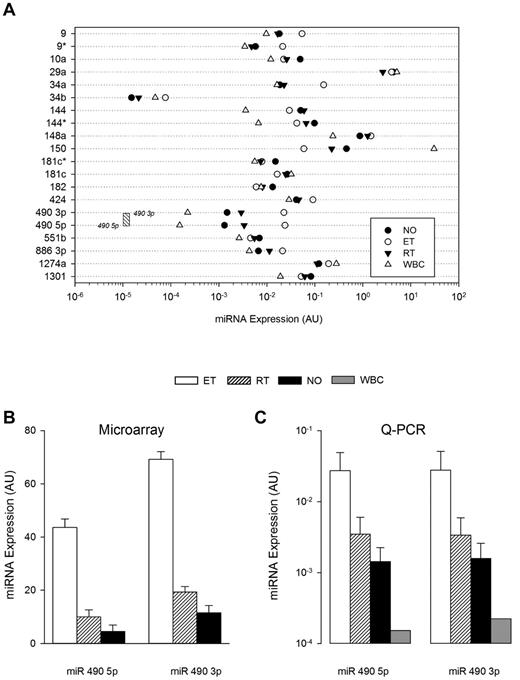
The abundance and lineage specificity of the miRNAs were established by simultaneously comparing expression patterns in leukocytes and platelets (both normal and thrombocytosis; Figure 3). Only miR 490 (both 5p and 3p) demonstrated relative lineage specificity with low-level leukocyte expression that was minimally 50-fold less than any of the other miRNAs studied (except for miR 34b); consistent with previous observations, miR 150 displayed the highest leukocyte expression.25 Among the 21-member miRNA list, miR 490 demonstrated the lowest expression in normal platelets, displaying ∼ 10-fold less abundance than other miRNAs. Comparative sequence alignments of miR 490 demonstrated stringent sequence homology across mammalian species (supplemental Figure 1), evolutionarily conserved in critical seed regions known to be important for assembly of the RNA-inducing silencing complex.26 Given its lineage-focused expression and evolutionarily conserved sequence—coupled with lack of genetic/functional information on miR 490—we specifically studied its expression patterns using ex vivo expanded CD34+ hematopoietic stem cells in a bilineage model of megakaryocyte-erythroid progenitor lineage specification.3 As shown (Figure 4A), both miR 490 5p and miR 490 3p demonstrated coordinate low-level expression beyond D7, patterns that contrasted sharply with the megakaryocyte-erythrocyte progenitor marker miR 1503 whose expression was readily evident at D6 and generally > 2-logs higher throughout the 21-day time course. Furthermore, miR 490 abundance at D21 was remarkably similar to that found in normal subjects (Figure 3A-B below). These studies were entirely consistent with the presence of low-level miR 490 abundance in normal platelets, and aberrant expression restricted to states of exaggerated Mk expansion and/or proplatelet formation.
Cellular and cohort-specific miRNA expression. (A) Quantitative RT-PCR was used to quantify miRNA expression patterns in leukocytes (WBC), normal platelets, or thrombocytosis (ET or RT), displayed by group mean (N = 5 samples/group). The threshold sensitivity of miR 490 3p and 5p assays is delineated by boundaries of the stippled box. (B-C) Normalized aggregate expression data for miR 490 3p and miR 490 5p are displayed for microarray (B; N = 79) or quantitative RT-PCR data (C; N = 10/group). Data are presented as the mean ± SEM.
Cellular and cohort-specific miRNA expression. (A) Quantitative RT-PCR was used to quantify miRNA expression patterns in leukocytes (WBC), normal platelets, or thrombocytosis (ET or RT), displayed by group mean (N = 5 samples/group). The threshold sensitivity of miR 490 3p and 5p assays is delineated by boundaries of the stippled box. (B-C) Normalized aggregate expression data for miR 490 3p and miR 490 5p are displayed for microarray (B; N = 79) or quantitative RT-PCR data (C; N = 10/group). Data are presented as the mean ± SEM.
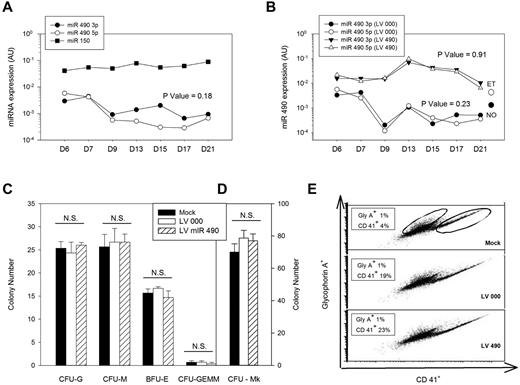
Genetic and functional characterization of miR 490. (A) Endogenous miR 490 expression patterns were monitored by quantitative RT-PCR using CD34+ cells differentiated in vitro over 21 days using a bilineage culture system; miR 150 expression is shown for comparison. (B-E) CD34+ cells transduced with lentiviruses LV 000 or LV490 were puromycin-selected (or not for mock-treated controls) and used for monitoring miR 490 expression patterns over a 21-day period (B); expression values for normal (NO) and ET platelets are shown for comparison. Hematopoietic progenitor assays were completed at day 14 after lentivirus infection (C), megakaryocyte colony assays on day 10 after infection (D), and megakaryocyte/erythroid lineage specification was analyzed at day 7 after infection by flow cytometry using anti-CD41 (Mk marker) or anti-glycophorin A (erythroid) antibodies for quantification (displayed in boxes; E). All results are from a single representative experiment repeated on one occasion. (A-B) P values were calculated by paired t test comparing aggregate miR 490 3p and miR 490 5p time series data.
Genetic and functional characterization of miR 490. (A) Endogenous miR 490 expression patterns were monitored by quantitative RT-PCR using CD34+ cells differentiated in vitro over 21 days using a bilineage culture system; miR 150 expression is shown for comparison. (B-E) CD34+ cells transduced with lentiviruses LV 000 or LV490 were puromycin-selected (or not for mock-treated controls) and used for monitoring miR 490 expression patterns over a 21-day period (B); expression values for normal (NO) and ET platelets are shown for comparison. Hematopoietic progenitor assays were completed at day 14 after lentivirus infection (C), megakaryocyte colony assays on day 10 after infection (D), and megakaryocyte/erythroid lineage specification was analyzed at day 7 after infection by flow cytometry using anti-CD41 (Mk marker) or anti-glycophorin A (erythroid) antibodies for quantification (displayed in boxes; E). All results are from a single representative experiment repeated on one occasion. (A-B) P values were calculated by paired t test comparing aggregate miR 490 3p and miR 490 5p time series data.
Functional effects of miR 490 in hematopoietic stem cells
Human CD34+ cells were genetically modified using lentiviruses expressing miR 490 (LV 490) or empty virus control (LV 000) to assess effects on colony formation or megakaryocyte/erythroid lineage specification.3 Transduction using LV 490 resulted in coordinate overexpression of both miR 490 5p and miR 490 3p at levels comparable with those of both RT and ET platelets, whereas LV 000–transduced cells demonstrated low-level miR 490 expression as identified in human blood platelets and during situations of normal hematopoietic differentiation (Figure 4B). Furthermore, the level of miR 490 expression using LV 490 was not dissimilar to that of miR 150, which was used as a standard to assess expression levels of a physiologically relevant Mk miRNA.3 LV 490-transduced CD34+ cells failed to enhance CFU-Mk formation, nor was there any effect on lineage-specification as assayed by colony formation of granulocytes, macrophage, erythroid cells, granulocyte-erythroid-macrophage-megakaryocyte, or megakaryocytes (Figure 4C-D). Given the interrelatedness between the vertebrate erythroid and Mk developmental repertoire,3 we applied the bilineage cell culture system to quantify effects on Mk/erythroid lineage specification. As assessed using flow cytometry for GlyA (erythroid) or CD41 (Mk) expression 7 days after transduction, we saw no evidence that miR 490 shifted the Mk/erythroid progenitor balance (Figure 4E). Thus, although miR 490 displayed lineage restrictions and ectopic expression associated with the thrombocytotic phenotype, these collective data demonstrate that its expression is not sufficient for enhanced Mk expansion or lineage specification during hematopoiesis.
miR 490 target identification using integrated datasets
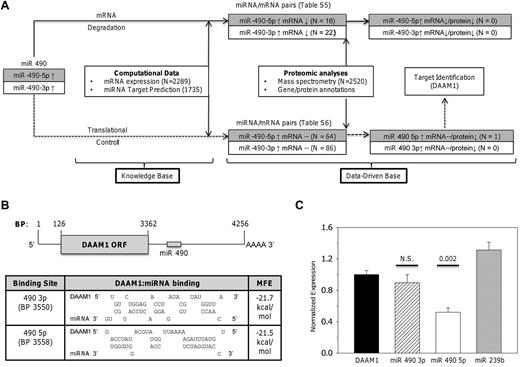
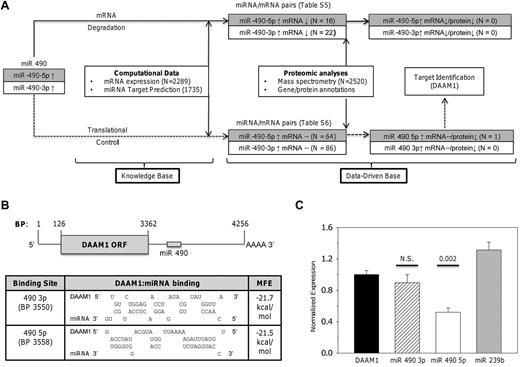
We developed a heuristic algorithm to identify putative miR 490 targets whose expression could be modulated during thrombocytosis. Given the limitations of miRNA target identification using computational databases,27 we systematically dissected the effects of dysregulated miRNA 490 expressions by incorporating a workflow model linking miRNA, mRNA, and proteomic expression patterns in normal and thrombocytotic states (Figure 5A). We used previously described mRNA genetic profiles from 6 ET patients and 5 healthy controls,22 and spectral (peptide) counts generated by multidimensional protein identification technology (MudPIT)20 as a semiquantitative means of platelet protein estimation, using samples from 3 healthy controls and 3 ET subjects. If miRNA 490 modulates protein expression, 2 distinct patterns would be expected: (1) inverse relationship of the miRNA and mRNA/protein pair (ie, a mRNA degradation pathway), or (2) inverse relationship of miRNA and protein without affecting mRNA (ie, translation repression). Using the 2-step target identification algorithm, we identified 38 mRNAs predicted to be directly repressed by transcriptional repression (supplemental Table 5) and 150 mRNAs putatively regulated by translational control (supplemental Table 6). Detailed integration of the protein profiles with these mRNA/miRNA candidates narrowed the candidate target list from 188 genes (none of which was represented in the degradation pathway) to a single mRNA/protein pair identified as DAAM1 (dishevelled-associated activator of morphogenesis 1). Furthermore, DAAM1 expression appeared as a putative target restricted to translation repression (ie, diminished ET protein expression unrelated to platelet DAAM1 mRNA levels).
Identification and characterization of miR 490 targets. (A) Schema delineating the workflow model for target identification delineates 2 distinct pathways based on mRNA degradation or translational control. Supplemental Tables 5 and 6 detail miRNA/mRNA pairs for either pathway. (B) Schema of DAAM1 mRNA (RefSeq accession no. NM_014992) with open-reading frame (ORF) and miR 490 binding sites is shown (top); topology of the DAAM1/miR 490 binding and calculated minimum free energy (MFE) are displayed as predicted using RNA hybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid). (C) Luciferase reporter assay quantification was completed 24 hours after transfection of HEK 293 cells using the 975-bp DAAM1 3′-UTR inserted downstream of Renilla luciferase (DAAM1) or cotransfected with a fixed concentration (10nM) of miR 490 3p or miR 490 5p chemical mimetics (or nontargeting Caenorhabditis elegans miR 239b stem-loop primer as control). Functional luciferase data from individual wells are expressed as the normalized mean ± SEM relative to DAAM1 expression (N = 6 wells/group representing aggregate data from 2 independent experiments). P values are calculated by paired t test.
Identification and characterization of miR 490 targets. (A) Schema delineating the workflow model for target identification delineates 2 distinct pathways based on mRNA degradation or translational control. Supplemental Tables 5 and 6 detail miRNA/mRNA pairs for either pathway. (B) Schema of DAAM1 mRNA (RefSeq accession no. NM_014992) with open-reading frame (ORF) and miR 490 binding sites is shown (top); topology of the DAAM1/miR 490 binding and calculated minimum free energy (MFE) are displayed as predicted using RNA hybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid). (C) Luciferase reporter assay quantification was completed 24 hours after transfection of HEK 293 cells using the 975-bp DAAM1 3′-UTR inserted downstream of Renilla luciferase (DAAM1) or cotransfected with a fixed concentration (10nM) of miR 490 3p or miR 490 5p chemical mimetics (or nontargeting Caenorhabditis elegans miR 239b stem-loop primer as control). Functional luciferase data from individual wells are expressed as the normalized mean ± SEM relative to DAAM1 expression (N = 6 wells/group representing aggregate data from 2 independent experiments). P values are calculated by paired t test.
DAAM1 characterization in normal and thrombocythemic platelets
The formin DAAM1 is required for directional cell migration and polarity during development, regulatory effects acting indirectly or directly via nucleating properties of actin.28 Limited information is available for platelet DAAM1, although recent data suggest that it functions as an activation-dependent intermediary linking Rho GTPase to actin assembly29 ; furthermore, its inhibitory effects on dishevelled may modulate noncanonical Wnt signaling pathways in platelets.28,30 We confirmed DAAM1 target identification using a reporter construct encompassing the 3′-UTR and synthetic double-stranded RNAs specific for either miR 490 5p or miR 490 3p, thereby providing further dissection of the relative roles of either of the strands in modulating DAAM1 protein expression. As shown in Figure 5B and C, inhibition of DAAM1 translation efficiency was restricted to miR 490 5p, with no inhibition using either miR 490 3p or the nonrelevant C elegans miR 239b stem-loop primer. These data are entirely consistent with our workflow model, which specifically identified miR 490 5p as the DAAM1 inhibitory miRNA.
Analysis of DAAM1 mRNA expression patterns in hematopoietic cells using a previously described database established a megakaryocyte-restricted pattern of expression31 (supplemental Figure 2), prompting us to compare expression patterns in ET and normal platelets. As shown in Figure 6A, DAAM1 mRNA was readily identified (and comparable) in both cohorts with no evidence that diminished ET DAAM1 was explained by decreased mRNA levels; these data were entirely consistent with and predicted by our computational algorithm. Detailed review of the MS chromatograms confirmed that the DAAM1 spectral counts in ET were 52% of those in normal platelets (Figure 6B), results that were validated by specifically identifying and comparing elution profiles of 2 peptides expressed across all platelet samples. Comparative analysis using DAAM1-specific peptides (DAAM1 I974-K995 and DAAM1 F162-K176) paralleled the aggregate spectral determinations, consistently demonstrating ET DAAM1 levels lower than those found in normal platelets (DAAM1974-994; 22% of normal; DAAM1162-176 ∼ 35% of normal; Figure 6C). These MS data were entirely consistent with immunoblot analysis, although there was heterogeneity of DAAM1 expression across the cohorts (Figure 6D). Given the limiting resolution of immunoblots to quantitative differences at the low dynamic range found in ET, we applied multiple reaction monitoring to reliably characterize and quantify DAAM1 peptide abundance free of coeluting peptides.21 We specifically tracked the m/z 1215.8 transition to its doubly charged Y19 ion from an individual ET and normal platelet sample, specifically quantifying ET DAAM1 abundance ∼ 40% of normal. The specificity of the elution peptide was established by following the m/z 1094.6 transition (Figure 6E-F). Collectively, these studies using immunoblot analysis, DAAM1-specific peptides quantification, and multiple reaction monitoring consistently established ET DAAM1 abundance at ∼ 40% of normal platelets.
Characterization of disheveled-associated activator of morphogenesis 1 in platelets. (A) DAAM1 quantitative RT-PCR was completed using ET or normal platelets (N = 5/group). (B) Normalized DAAM1-specific spectral counts from MudPit analyses are expressed as the mean ± SEM from 3 normal and 3 ET subjects. (C) DAAM1 peptide abundance was quantified between cohorts for each of 2 peptides and expressed as the mean ± SEM (N = 3/cohort). (D) Immunoblot analysis was completed using 4%-15% SDS-PAGE for each of 6 platelet lysates. (E) Quantitative multiple reaction monitoring from one normal and one ET platelet sample is displayed as the area under the curve (in parentheses) of the m/z 1215.8 transition to its doubly charged Y19 ion. (F) MS/MS spectra of the DAAM1 peptide IQPDEFFGIFDQFLQAVSEAK specifies the fragmentation and Y19 ion(s) used for abundance quantification.
Characterization of disheveled-associated activator of morphogenesis 1 in platelets. (A) DAAM1 quantitative RT-PCR was completed using ET or normal platelets (N = 5/group). (B) Normalized DAAM1-specific spectral counts from MudPit analyses are expressed as the mean ± SEM from 3 normal and 3 ET subjects. (C) DAAM1 peptide abundance was quantified between cohorts for each of 2 peptides and expressed as the mean ± SEM (N = 3/cohort). (D) Immunoblot analysis was completed using 4%-15% SDS-PAGE for each of 6 platelet lysates. (E) Quantitative multiple reaction monitoring from one normal and one ET platelet sample is displayed as the area under the curve (in parentheses) of the m/z 1215.8 transition to its doubly charged Y19 ion. (F) MS/MS spectra of the DAAM1 peptide IQPDEFFGIFDQFLQAVSEAK specifies the fragmentation and Y19 ion(s) used for abundance quantification.
Discussion
We have identified a unique 21-miRNA signature occurring in the setting of exaggerated platelet production, either associated with the myeloproliferative neoplasm (MPN) ET or self-limiting situations of enhanced platelet production (RT). The majority of the differentially expressed miRNAs displayed congruent directional behavior irrespective of the thrombocytosis etiology, supporting a molecular model whereby normal and exaggerated thrombopoiesis represents genetically defined continua along a convergent pathway, rather than qualitatively distinct genetic entities. Thus, subjects with ET display the most exaggerated expression patterns, whereas those with RT represent an intermediary subset, collectively defined by quantitatively distinct expression patterns of the 21-miRNA signature. This concept of quantitative genetics and phenotypic progression is well exemplified in MPN whereby the “strength” (or allelic burden) of the JAK2V617F signal may represent one determinant of the MPN phenotype.32 Similarly, data using various murine models also support the concept that the level of JAK2V617F can initiate and sustain differential MPN phenotypes.33 Our data reinforce and extend to miRNAs the paradigm that quantitative differences involving the genetic fingerprint may be associated with (or confer) distinct phenotypes (ie, a self-limiting phenotype seen in RT compared with the sustained phenotype associated with ET).
Given the convergence of c-mpl/Tpo and Jak/Stat pathways in normal and MPN-associated platelet production, it is not unexpected that the 21-miRNA thrombocytosis expression signatures remained concordant, the notable exception being miR 144/144*, which was specifically up-regulated in RT and down-regulated in ET. Interestingly, miR 144 is known to be transcribed with miR 451 on a single precursor RNA (pri-miRNA),24 and both are lineage (erythroid)–enriched in hematopoiesis models using zebrafish (Danio rerio). The miR 144/451 locus is a direct GATA1 transcriptional target, although effects on erythropoiesis appear to be restricted to miR 451, with no evidence that either miRNA has an effect on thrombopoiesis.24 Although the orthologous miR 144/451 locus is conserved on human chromosome 17, we saw no evidence for dysregulated miR 451 during thrombocytosis, further supporting the concept that these contiguous genes appear to have divergent effects on the megakaryocyte/erythroid transition. Furthermore, these data imply that distinct pathways of thrombopoiesis may be defined by the lineage-restricted expression patterns (or cognizant transcriptional regulators) of miR 144.
What exactly is the functional significance of miRNA 490 overex-pression in thrombocytosis? We identified miRNA 490 (5p and 3p) as an evolutionarily conserved miRNA demonstrating low-level expression during normal thrombopoiesis and exaggerated levels during thrombocytosis, although most profound in ET. Of the 21-member list, only miR 490 displayed a generally restricted pattern of expression in Mk/platelets (relative to leukocytes), with no prior description of this miRNA in MPN studies,5,6,34 and/or models of hematopoiesis4,24 or megakaryocytic differentiation.2,35,36 Given its aberrant expression during thrombocytosis, our data suggest that miR 490 represents a unique biomarker of exaggerated megakaryocytopoiesis and/or proplatelet formation, although its physiologic function(s) remain to be precisely elucidated. An in vitro differentiation model demonstrated progressive miR 490 down-regulations, results entirely consistent with the low-level expression seen in healthy controls during normal thrombopoiesis. Likewise, miR 490 overexpression was not sufficient for Mk proliferation and/or differentiation in CD34+ hematopoietic stem cells. These results collectively support a developmentally restricted function across species, apparently restricted to conditions associated with exaggerated megakaryocytopoiesis and/or proplatelet formation.
We developed an integrated proteomic/genetic approach to identify putative miR 490 targets by studying the ET proteome, and validated the specificity of miR 490 5p/DAAM1 inhibition. What are the implications for diminished DAAM1 expression on platelet and/or megakaryocyte functions in ET? Characteristic features of formins are the forming homology (FH) domains that are essential for mediating linear actin polymerization, functions distinct from proteins such as Arp2/3, which initiate and elongate branched actin filaments. DAAM1 was initially identified as a disheveled interacting protein mediating the Wnt/planar cell polarity pathway,37 and recent data have established both the functional capacity of platelet DAAM1 in actin assembly,29 and the relevance of the canonical Wnt signaling pathway as a negative modulator of platelet responsiveness.30 The latter is especially relevant given known platelet functional abnormalities and thrombohemorrhagic consequences in ET.14 In contrast, no information is available for DAAM1 function in Mk despite the requisite role of actin-based forces on proplatelet formation. Recent data using Daam1-knockout mice have established a crucial role in heart morphogenesis by regulating filamentous actin assembly and cytoskeletal organization.38 The associated embryonic lethality precluded detailed analysis of the hematopoietic system, although DAAM1's megakaryocyte-enriched pattern of expression presumably connotes functional properties relevant to megakaryocytopoiesis and/or proplatelet formation.
Further studies are required to establish whether miR 490 or DAAM1 has requisite functions in exaggerated thrombopoiesis occurring in the background of MPNs or RT. Given its low-level expression during Mk differentiation, the functional effects of miR 490 inhibition using specific miRNA inhibitors (or other approaches) are best evaluated using in vivo models associated with miR 490 overexpression (such as enhanced platelet formation). We are currently in the process of evaluating and testing murine models of thrombocytosis (along with evaluation of their Mks that presumably will demonstrate altered miR 490 and/or DAAM1 expression) as a comprehensive approach to precisely define these inter-relationships relevant to exaggerated proplatelet formation. Such an approach is especially relevant given the promiscuity of miRNAs for their cognate targets (ie, 1 miRNA typically has multiple targets), coupled with the converse regulation of mRNAs by a multiplicity of miRNAs.1 Similarly, although our data define altered DAAM1 expression in ET platelets, miR 490/DAAM1 functional consequences in RT remain incomplete. In preliminary data using a small RT cohort, we noted that DAAM1 levels appeared comparable with those found in healthy controls (not unexpected given the ∼ 2-fold miR 490 increase in RT), although intersubject variability precludes definitive comparisons and awaits ongoing, integrated studies.
Our integrated workflow model to identify putative miR 490 targets (both 490 5p and 490 3p) suggests that miR 490 5p-loaded RNA-inducing silencing complex modulates DAAM1 protein expression by suppressing translation independent of an effect on DAAM1 mRNA stability. Although the disparate miRNA regulatory pathways (ie, degradation vs translation arrest) are well described,1 a recent study suggests that miRNAs retain dual capacity for enhanced mRNA translation, depending on the proliferative phase of the cell cycle.39 This physiologic switch is controlled by altered protein composition of target mRNA 3′-UTRs, either through stimulatory FXR1 or inhibitory GW12 recruitments to the miRNA/Ago complex.39,40 Although the cell cycle is largely irrelevant to functions of circulating platelets, recent data suggest that platelets produce functional progeny, which involves a form of cell division not requiring a nucleus.11 Furthermore, extracellular signals reconfigure the platelet proteome, converting generally quiescent platelets into active translational states8,41 with clinically relevant implications for inflammatory disorders.10 It remains unknown whether platelets retain comparable FXR1/GW12 proteins capable of regulating miRNA functions with dichotomous effects on protein expression. If so, it would provide an additional dimension for activation-dependent modulation of platelet functions, with functional implications for ET platelets harboring aberrant miRNA expression patterns.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Antonius Koller (University Proteomics Center) for detailed analysis and helpful discussions and Jean Wainer for technical assistance.
This work was supported by the New York State Stem Cell (board grant C024317, W.F.B.; NYSTEM grant C026716; Stony Brook Stem Cell Center) and the National Institutes of Health (grant HL086376, W.F.B.; and grant RR032112, D.V.G.).
National Institutes of Health
Authorship
Contribution: I.S.H., D.V.G., and D.W.M. performed relevant experiments; X.X. and W.Z. analyzed results and generated the figures; and W.F.B., J.J., and X.X. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wadie F. Bahou, Department of Medicine, Health Sciences Center T15-040, Stony Brook University School of Medicine, Stony Brook, NY 11794-8151; e-mail: wadie.bahou@sbumed.org.