Abstract
Although chronic myelogenous leukemia (CML) is effectively controlled by Bcr-Abl kinase inhibitors, resistance to inhibitors, progressive disease, and incomplete eradication of Bcr-Abl–expressing cells are concerns for the long-term control and suppression of this disease. We describe a novel approach to targeting key proteins in CML cells with a ubiquitin-cycle inhibitor, WP1130. Bcr-Abl is rapidly modified with K63-linked ubiquitin polymers in WP1130-treated CML cells, resulting in its accumulation in aggresomes, where is it unable to conduct signal transduction. Induction of apoptosis because of aggresomal compartmentalization of Bcr-Abl was observed in both imatinib-sensitive and -resistant cells. WP1130, but not Bcr-Abl kinase inhibitors, directly inhibits Usp9x deubiquitinase activity, resulting in the down-regulation of the prosurvival protein Mcl-1 and facilitating apoptosis. These results demonstrate that ubiquitin-cycle inhibition represents a novel and effective approach to blocking Bcr-Abl kinase signaling and reducing Mcl-1 levels to engage CML cell apoptosis. This approach may be a therapeutic option for kinase inhibitor–resistant CML patients.
Introduction
Chronic myelogenous leukemia (CML) is associated with a chromosomal abnormality in the hematopoietic stem cell that results in the expression of Bcr-Abl with unregulated tyrosine kinase activity.1 These observations supported the development and clinical testing of the first Bcr-Abl kinase inhibitor, imatinib, which demonstrated remarkable clinical efficacy in CML patients. Imatinib is the frontline therapy for CML and other Bcr-Abl–expressing leukemias, and most patients treated with imatinib in the chronic phase achieve a complete cytogenetic response.2,3 However, molecular studies of imatinib-treated patients in remission demonstrated that Bcr-Abl expression is still detectable in most cases, and discontinuation of imatinib therapy often results in disease relapse.4,5 Limited duration of imatinib response is also common in advanced CML patients, and imatinib resistance can occur at any stage of the disease.5,6 Acquired imatinib resistance and disease progression are frequently characterized by Bcr-Abl mutations and posttranslational modifications that affect imatinib binding and kinase inhibition.5,6 Some of the molecular changes in imatinib-resistant disease can be overcome with second-generation tyrosine kinase inhibitors, which bind Bcr-Abl with higher affinity or inhibit imatinib-insensitive kinases associated with resistance.7-11 However, the activity of these inhibitors can also be limited by mutations and other mechanisms.12-14 These observations suggest that additional approaches and targeting strategies may be needed to provide long-term effective treatment for CML patients.
Kinase inhibition by small molecules that bind the ATP or the switch pocket region of Bcr-Abl are effective inhibitors but require continuous treatment because Bcr-Abl protein levels themselves are not directly regulated through kinase inhibition. Some evidence suggests that Bcr-Abl can function as a protein scaffold to organize signaling complexes that are not fully dependent on kinase activity.15,16 These observations suggest that compounds that modulate Bcr-Abl protein levels may be more effective and appropriate for CML therapy in some settings. In light of that possibility and as a novel approach to overcoming kinase inhibitor resistance, several compounds have been described that affect Bcr-Abl function through mechanisms other than direct kinase inhibition. These include heat shock protein 90 (Hsp90) inhibitors, arsenic trioxide, homoharringtonine, histone deacetylase inhibitors, proteasome inhibitors, PP2A activators, and others.5,10 All are reported to lead to CML cell death through down-regulation of Bcr-Abl expression or a loss of Bcr-Abl stability. However, most of these compounds reduce Bcr-Abl levels only after extended incubation intervals and display only limited selectivity for Bcr-Abl, which may increase their toxicity and decrease their clinical utility.17-19 Additional compounds that induce rapid changes in Bcr-Abl levels with limited impact on other proteins are still needed.
In a previous study, we described the discovery and antileukemic activity of WP1130, a small molecule with an unknown mechanism of Bcr-Abl down-regulation.20 In the present study, we demonstrate that WP1130 rapidly induces ubiquitination of Bcr-Abl, resulting in its relocalization from the cytoplasm into compact, intracellular protein complexes called aggresomes. This modification results in the loss of downstream Bcr-Abl oncogenic signaling. We further demonstrate that WP1130 directly inhibits Usp9x, a deubiquitinase (DUB) recently reported to regulate the stability of Mcl-1,21 an antiapoptotic protein expressed in many tumors, including hematologic malignancies.22,23 Mcl-1 is associated with drug resistance and survival in hematopoietic malignancies.24 WP1130-mediated Usp9x inhibition is associated with reduced Mcl-1 levels, and together with blocked Bcr-Abl kinase signaling, results in the rapid onset of apoptosis. These results suggest that targeting specific ubiquitin-cycle regulators may emerge as a novel therapeutic approach to inhibiting oncoprotein signaling and reducing elevated apoptotic thresholds.
Methods
Compounds, chemical reagents, and affinity matrices
WP1130, imatinib, and dasatinib were synthesized and purified by William Bornmann (M. D. Anderson Cancer Center, Houston, TX). TG101209 was synthesized and provided by Dr Hollis Showalter (University of Michigan, Ann Arbor). ABT-263 was purchased from Accela ChemBio. All compounds were made up as 20mM stock solutions (in dimethylsulfoxide [DMSO]), stored frozen at −20°C, and diluted into aqueous medium just before use. Other reagents used in this study were: bortezomib (Millennium Pharmaceuticals); Mini-Complete and PhosSTOP inhibitory cocktails (Roche Applied Science); ubiquitin C-terminal 7-amido-4-methylcoumarin (Ub-AMC), and hemagglutinin-tagged ubiquitin vinyl methyl sulfone (HA-UbVs), Suc-LLVY-AMC, Boc-LRR-AMC, MG-132, lactacystin, and 20S human proteasome (Boston Biochem). Affinity matrices of Rap80-agarose beads, ataxin-agarose beads, and purified poly-ubiquitin chains (K48/K63-linked) were also obtained from Boston Biochem. 17-Allylamino-17-demethoxygeldanamycin (17-AAG) was purchased from LC Laboratories, made up and stored frozen as a 20mM stock solution.
Cell lines and patient samples
K562, K562R, BV-173, BV-173R, and WDT-2 cell lines were grown and maintained as described previously.7,20,25 BaF3 cells were maintained in the same medium supplemented with 1 ng/mL of interleukin-3 (IL-3; PeproTech). BaF3 cells were also transformed with unmutated or mutant Bcr-Abl, as described previously, or transformed with an upstream enhanced green fluorescent protein (eGFP) coding sequence inserted in-frame into the Bcr-Abl expression vector.20 All cells were cultured and maintained at 37°C in a humidified atmosphere.
Specimens were from CML patients who had signed an informed consent document approved by the University of Michigan institutional review board and in accordance with the Declaration of Helsinki. Samples were derived from patients when imatinib therapy failed to continuously control their disease.26,27 Mononuclear cells were isolated from blood samples by density centrifugation (Ficoll-Hypaque), washed with phosphate-buffered saline (PBS), resuspended in cell-culture medium (RPMI 1640, 10% fetal bovine serum), and incubated overnight at 37°C in a 5% CO2 incubator before treatment with inhibitors. For some experiments, CD34+ cells from CML (Philadelphia chromosome–positive) patients or normal bone marrow were isolated by magnetic bead separation using a MACS cell-separation column, as recommended by the manufacturer (Miltenyi Biotec). Cells were cultured in serum-free medium (StemCell Technologies) containing cytokines (IL-3, 20 ng/mL; IL-6, 20 ng/mL; IL-7, 20 ng/mL; granulocyte colony-stimulating factor, 20 ng/mL; Flt-3, 50 ng/mL; and SCF, 50 ng/mL; PeproTech) and 40 μg/mL of human lipoprotein (Sigma-Aldrich) in the presence or absence of the indicated concentration of WP1130 for 24 hours. Apoptosis was assessed by annexin/propidium iodide staining and flow cytometry as recommended by the manufacturer (Southern Biotechnology).
Plasmids and electroporation
The eGFP-coding region was cloned from pLEGFPc by polymerase chain reaction using a 5′ primer with an EcoR1 restriction site, GAATTCCGCCACCATGGTGAGCAAGGGCG, and a 3′ primer with an EcoRV restriction site, GATATCGACTTGTACAGCTCGTCCATGCCGAGAGTG. The pBSsk+ vector containing p210Bcr/Abl cDNA (derived from pSG5; originally provided by Dr Ralph Arlinghaus, Department of Molecular Pathology, M. D. Anderson Cancer Center) was digested with ClaI and EcoRI and the EcoR1 site was blunted. The eGFP fragment was digested with ClaI and EcoRV and ligated into pBSsk+/p210Bcr/Abl to generate the 5′-tagged eGFPp210Bcr/Abl. The pMX/eGFPp210Bcr/Abl was constructed by ligation of the eGFPp210Bcr/Abl into the pMXpuro vector with the deletion of the internal ribosome entry site eGFP using EcoRI and NotI. BaF3 cells were electroporated20,27 with pMX/eGFPp210Bcr/Abl or pMX/eGFPp120Bcr/Abl-T315I (constructed by subcloning from the pSG5-Bcr/Abl/T315I vector using Kpn1/BsrG1 sites). All mutations and vector inserts were confirmed by sequencing. Cells were selected for puromycin resistance (2 μg/mL; 2 weeks) and enriched for Bcr-Abl expression by fluorescence-activated cell sorting of eGFP-positive cells (FACSCanto II; BD Biosciences). Bcr-Abl expression and cytokine independence were confirmed by immunoblotting and apoptotic induction by imatinib incubation but not IL-3 withdrawal.
shRNA and Usp9x silencing
BV-173 cells were infected with the lentiviral expression system for short hairpin RNA (shRNA), LVX-shRNA1, and LVX-shRNA2-Usp9x, kindly provided by Dr Dzwokai Ma (University of California, Santa Barbara). In brief, HEK293T cells were transfected with the lentiviral packaging vectors pMD2.G and psPax2 (Addgene) together with the LVX-shRNA vectors to produce virus using PolyFect as described by the manufacturer (QIAGEN). The medium was changed to RPMI 1640 with 20% fetal bovine serum and 10mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.4, the following day, and the viral supernatant was collected by centrifugation on day 2 after transfection. BV-173 cells were spin infected for 2 hours with 1.5 mL of viral supernatant containing 8 μg/mL of Polybrene (Sigma-Aldrich) at 2000g at 32°C. Two days after infection, the medium was changed and 1 μg/mL of puromycin was added. After puromycin selection (5 days), viable cells were recovered by Ficoll gradient separation, and Usp9x levels were examined by immunoblotting. Those with stable reduction of Usp9x were used to assess Mcl-1 levels and to analyze apoptotic sensitivity to imatinib and ABT-263.
Lysate preparation, antibodies, and Western blotting
Total cell lysates were prepared by boiling and sonicating cell pellets in 1× Laemmli reducing sample buffer. To prepare detergent-soluble and -insoluble fractions, cells were lysed in cold isotonic lysis buffer (10mM Tris-HCl, pH 7.5, 0.5% Triton X-100, 150mM NaCl, along with Mini-Complete and PhosSTOP) for 15 minutes on ice and centrifuged for 10 minutes at 20 000g. The clarified supernatant was used as the detergent-soluble cell fraction. The residual pellet was washed and extracted in Laemmli reducing sample buffer and briefly sonicated to derive the detergent-insoluble fraction. Equal volumes of cellular lysate or equal protein amounts were electrophoresed on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membranes (Whatmann). Proteins were detected by immunoblotting, as described previously.7,27
Antibodies used in this study were: anti–pY-Stat5, pY-CrkL, anti–poly(adenosine diphosphate)-ribose polymerase (anti-PARP), anti–caspase 3, and anti–Mcl-1 (Cell Signaling Technology); anti-actin (Sigma-Aldrich); polyclonal anti-ABL (K12), monoclonal anti-ABL (SH2 domain; 8E9), anti-ubiquitin clone P4D1, anti-Hsp90, anti-Hsp70, anti-Jak2, anti-CrkL, anti–α-tubulin, and horseradish peroxidase–conjugated goat anti–rabbit/mouse/rat immunoglobulin G (IgG; Santa Cruz Biotechnology); anti-HA clone 3F10 (Roche Applied Science); and anti-Usp9x (Bethyl Laboratories). Caspase 9 antibody was from Calbiochem.
Proteasome activity assay
The fluorogenic substrate Suc-LLVY-AMC was used to assay for the chymotryptic-like activity of the 20S proteasome. To assay for in vivo proteasome inhibition, cells were treated with WP1130 (5μM) or MG132 (5μM) for 2 hours and lysed in ice-cold lysis buffer (50mM HEPES, pH 7.5, 5mM EDTA [ethylenediaminetetraacetic acid], 150mM NaCl, 1% Triton X-100). Lysates were clarified by centrifugation at 20 000g for 10 minutes, and equal amounts of protein from each sample were incubated at 37°C with 100μM fluorogenic substrate. To assay for direct inhibition of the 20S proteasome in vitro, purified 20S human proteasome (200 ng) was incubated with WP1130 (5μM), MG132 (5μM), or lactacystin (5μM) for 30 minutes at 37°C before addition of the substrates. Fluorescence intensity was measured using a spectrophotometer at an excitation of 360 nm and an emission of 460 nm. Assays were performed in triplicate, and statistical significance was determined with a paired Student t test.
ROS assay
Leukemic cells (1 × 106) were treated with DMSO, WP1130 (5μM), or a positive (0.5mM H2O2) or negative (1mM dithiothreitol) effector of reactive oxygen species (ROS) content for 2 hours at 37°C. Cells were washed and resuspended in PBS containing 10μM 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA) and further incubated for an additional 15 minutes in the dark. After being washed in PBS, cells were transferred to individual wells of a 96-well plate and fluorescence intensity was measured using a spectrophotometer at an excitation of 492 nm and an emission of 520 nm. Assays were performed in triplicate, and statistical significance was determined with a paired Student t test.
Confocal microscopy
Control and treated eGFP-Bcr/Abl–transformed BaF3 cells were washed twice in PBS, followed by fixation using 4% formaldehyde for 15 minutes. The cells were spun onto poly-lysine–coated slides using a Cytopro centrifuge (Wescor) and permeabilized in 0.5% Triton X-100 for 5 minutes. Slides were then incubated in blocking solution (5% goat serum) for 1 hour at room temperature. Incubation with the primary antibodies (1:100) was carried out for 4 hours at room temperature or overnight at 4°C, and the slides were washed 3 times with 0.2% Triton X-100/PBS buffer. Alexa Fluor anti–mouse and Alexa Fluor anti–rabbit immunoglobulin antibodies were used as secondary antibodies. The slides were washed 3 times and the nucleus was stained with Hoechst 33342. Images were acquired using an Olympus confocal microscope FV-500.
In vitro deubiquitination assays
Ub-AMC protease assay.
Cells were lysed in ice-cold DUB buffer containing 50mM Tris-HCl, pH 7.5, 0.5% NP-40, 5mM MgCl2, 150mM NaCl, and 1mM phenylmethylsulfonylfluoride. Briefly, 5 μg of clarified lysate from untreated or treated cells was incubated with 500nM Ub-AMC in a 100-μL reaction volume at 37°C, and the release of AMC fluorescence was recorded at an excitation of 380 nm and an emission of 480 nm using a spectrofluorometer.
USP9x was immunoprecipitated from 500 μg of cell lysate and incubated in DUB buffer containing N-ethylmaleimide (NEM), WP1130, or DMSO in a 100-μL reaction volume for 30 minutes. The reaction was initiated by the addition of 500nM Ub-AMC, and the release of AMC fluorescence was recorded.
Ubiquitin chain disassembly.
In vitro disassembly of purified polyubiquitin chains (K48/K63 linked) was performed as described previously.28 Lysate (5 μg) from untreated or WP1130-treated K562 cells was prepared in DUB buffer and incubated with K48– or K63–linked chains (1 μg) for 10 minutes at 37°C. The extent of chain disassembly was assessed by ubiquitin immunoblotting.
DUB-labeling assays
To assay for changes in the activity of cellular DUB enzymes, leukemic cells were lysed in DUB buffer (50mM Tris pH 7.4, 5mM MgCl2, 150mM NaCl) for 10 minutes at 4°C. The lysates were centrifuged at 20 000g for 10 minutes, and the supernatant was used for DUB labeling. Equal amounts of lysate were incubated with 500 ng of HA-UbVs for 1 hour at room temperature, followed by boiling in reducing sample buffer and resolving by SDS-PAGE. HA immunoblotting was used to detect DUB labeling.
Results
WP1130 induces protein ubiquitination and Bcr-Abl protein redistribution in both imatinib-sensitive and -resistant cells
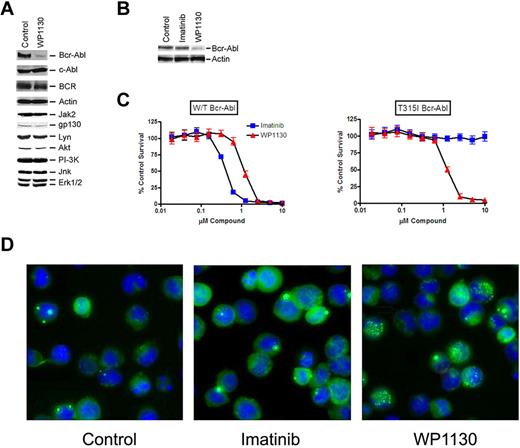
We previously reported that WP1130 induced down-regulation of Bcr-Abl in imatinib- and dasatinib-sensitive and -resistant CML cells.20 To determine the selectivity for Bcr-Abl, several kinases expressed in CML cells were assessed for WP1130-mediated down-regulation. Only Bcr-Abl protein levels were reduced after WP1130 treatment, suggesting specificity for this chimeric protein (Figure 1A). We expressed eGFP-tagged Bcr-Abl (W/T) in BaF3 cells and observed rapid down-regulation of the ectopically expressed Bcr-Abl after WP1130 treatment (Figure 1B). We further established stable BaF3 cell lines expressing W/T and T315I variants of Bcr-Abl and observed high sensitivity to WP1130-mediated apoptosis in both cases. In contrast, BaF3 cells expressing T315I eGFP-Bcr-Abl were imatinib resistant, suggesting that the eGFP tag did not influence Bcr-Abl–mediated cellular transformation or imatinib sensitivity (Figure 1C). We further investigated the effects of WP1130 on eGFP-Bcr-Abl by fluorescent microscopy and observed a rapid accumulation of eGFP-Bcr-Abl into compact, high-density Bcr-Abl clusters after WP1130 incubation (Figure 1D).
Specificity and activity of WP1130 in Bcr-Abl–expressing cells. (A) K562 cells were treated with 5μM WP1130 or DMSO (control) for 2 hours before equal amounts of detergent-soluble cell lysates were immunoblotted for the protein indicated. (B) eGFP-Bcr-Abl–transformed BaF3 cells were left untreated or treated with 5μM imatinib or WP1130 for 2 hours before analyzing equal protein cell lysates for Bcr-Abl and actin by immunoblotting. (C) BaF3 cells transformed by eGFP-Bcr-Abl without (W/T) or with the T315I Bcr-Abl mutation were treated with imatinib or WP1130 at the indicated concentration for 72 hours before assessing cell growth and survival by 3-(4,5-dimethylthiazol-2-yl)-2,5,-diphenyl tetrazolium bromide staining.7,20 The results represent the average ± SD of 4 replicates. Similar results were obtained in 2 additional independent studies. (D) BaF3 cells transformed with eGFP-Bcr-Abl were treated with 5μM imatinib or WP1130 for 2 hours before cells were fixed with paraformaldehyde, cytospun onto slides, and stained with Hoechst. Digital images were captured on an Olympus fluorescent microscope at 100× magnification (Olympus IX71, Olympus DP71). Bcr-Abl appears in compact clusters in WP1130-treated cells.
Specificity and activity of WP1130 in Bcr-Abl–expressing cells. (A) K562 cells were treated with 5μM WP1130 or DMSO (control) for 2 hours before equal amounts of detergent-soluble cell lysates were immunoblotted for the protein indicated. (B) eGFP-Bcr-Abl–transformed BaF3 cells were left untreated or treated with 5μM imatinib or WP1130 for 2 hours before analyzing equal protein cell lysates for Bcr-Abl and actin by immunoblotting. (C) BaF3 cells transformed by eGFP-Bcr-Abl without (W/T) or with the T315I Bcr-Abl mutation were treated with imatinib or WP1130 at the indicated concentration for 72 hours before assessing cell growth and survival by 3-(4,5-dimethylthiazol-2-yl)-2,5,-diphenyl tetrazolium bromide staining.7,20 The results represent the average ± SD of 4 replicates. Similar results were obtained in 2 additional independent studies. (D) BaF3 cells transformed with eGFP-Bcr-Abl were treated with 5μM imatinib or WP1130 for 2 hours before cells were fixed with paraformaldehyde, cytospun onto slides, and stained with Hoechst. Digital images were captured on an Olympus fluorescent microscope at 100× magnification (Olympus IX71, Olympus DP71). Bcr-Abl appears in compact clusters in WP1130-treated cells.
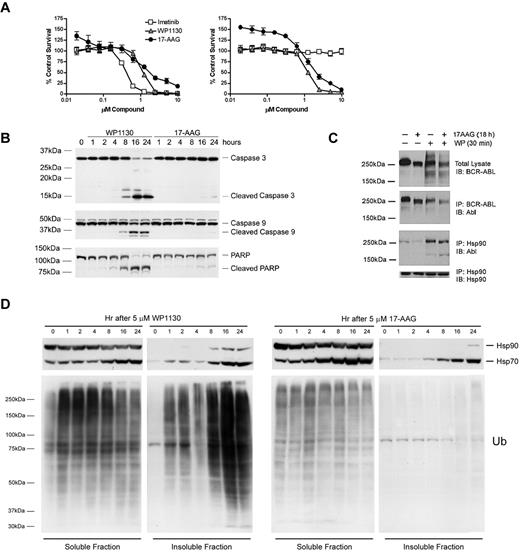
Hsp90 inhibition by geldanamycin and its analogs has been reported to affect Bcr-Abl stability and cellular distribution. We compared the antiproliferative effects of Hsp90 inhibitor (17-AAG) with those of WP1130. Both WP1130 and 17-AAG displayed antiproliferative effects in CML lines expressing either W/T Bcr-Abl (BV-173, Figure 2A left) or T315I mutant Bcr-Abl (BV-173R, Figure 2A right), but distinctions in the activation of caspases and onset of apoptosis were observed (Figure 2B). We noted a reduction in Hsp90 and Bcr-Abl association in 17-AAG–treated cells, which can lead to Bcr-Abl down-regulation through ubiquitination, as reported previously.18 However, treatment with WP1130 resulted in an increased association between Bcr-Abl and Hsp90 (Figure 2C), suggesting a distinct mechanism for Bcr-Abl down-regulation. We further assessed the impact of WP1130 and 17-AAG on cellular ubiquitinated protein and Hsp70 protein levels, because Hsp90 inhibition results in Hsp70 induction.29 17-AAG had no effect on ubiquitinated protein levels, but led to the induction of Hsp70 (Figure 2D). WP1130 induced a rapid increase in ubiquitinated proteins in the detergent-soluble cell fraction, with substantial accumulation in the detergent-insoluble fraction at later time points. Hsp70 levels were also increased in WP1130-treated cells, but based on the enhanced association of Hsp90 with Bcr-Abl in WP1130-treated cells, Hsp90 does not appear to be inhibited, and therefore is not directly associated with Hsp70 induction (Figure 2C).
WP1130 induces rapid ubiquitination and apoptosis in imatinib-sensitive and -resistant CML cells. (A) BV-173 (left) and BV-173R (right) cells were treated with the indicated compound for 72 hours before cell viability and proliferation was estimated by 3-(4,5-dimethylthiazol-2-yl)-2,5,-diphenyl tetrazolium bromide assays. The results represent the average ± SD of 4 replicates. Similar results were obtained in 2 additional independent studies. (B) BV-173R cells were treated with the indicated compound for the interval noted before cell lysates were immunoblotted for caspase 3, caspase 9, and PARP. The major cleaved forms of these proteins are also shown. (C) K562 cells were treated as noted (cells treated with the combination of agents were treated with 17-AAG followed by WP1130) before detergent-soluble cell lysates were prepared for direct Bcr-Abl blotting (top). Lysates were also subjected to Bcr-Abl or Hsp90 immunoprecipitation, followed by immunoblotting as noted. (D) K562 cells were treated as noted before analyzing the Hsp70, Hsp90, and ubiquitin content in the detergent-soluble and -insoluble fractions (fractions were resolved as described in lysate preparation section) by immunoblotting. Equal volumes of detergent-soluble and -insoluble extract were loaded in each lane. Hsp90 blotting served as a protein-loading control for the detergent-soluble fraction.
WP1130 induces rapid ubiquitination and apoptosis in imatinib-sensitive and -resistant CML cells. (A) BV-173 (left) and BV-173R (right) cells were treated with the indicated compound for 72 hours before cell viability and proliferation was estimated by 3-(4,5-dimethylthiazol-2-yl)-2,5,-diphenyl tetrazolium bromide assays. The results represent the average ± SD of 4 replicates. Similar results were obtained in 2 additional independent studies. (B) BV-173R cells were treated with the indicated compound for the interval noted before cell lysates were immunoblotted for caspase 3, caspase 9, and PARP. The major cleaved forms of these proteins are also shown. (C) K562 cells were treated as noted (cells treated with the combination of agents were treated with 17-AAG followed by WP1130) before detergent-soluble cell lysates were prepared for direct Bcr-Abl blotting (top). Lysates were also subjected to Bcr-Abl or Hsp90 immunoprecipitation, followed by immunoblotting as noted. (D) K562 cells were treated as noted before analyzing the Hsp70, Hsp90, and ubiquitin content in the detergent-soluble and -insoluble fractions (fractions were resolved as described in lysate preparation section) by immunoblotting. Equal volumes of detergent-soluble and -insoluble extract were loaded in each lane. Hsp90 blotting served as a protein-loading control for the detergent-soluble fraction.
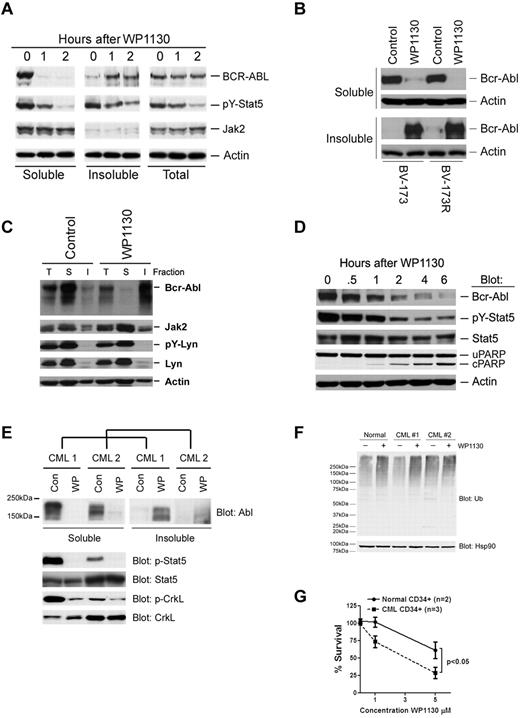
Because ubiquitination can mediate protein degradation30 and intracellular trafficking,31 we examined Bcr-Abl content in WP1130-treated cells. As shown in Figure 3A, WP1130 treatment resulted in a rapid and near complete trafficking of Bcr-Abl from the detergent-soluble to the detergent-insoluble cell fraction. Compartmentalization of Bcr-Abl into the insoluble fraction was associated with loss of phosphorylation of Stat5 without affecting Jak2 (Figure 3A). However, no significant change in the Bcr-Abl protein content in whole-cell extracts was noted. Increased Bcr-Abl protein content in the detergent-insoluble cell fraction was also observed in imatinib-resistant CML cells, such as BV-173R expressing T315I-Bcr-Abl (Figure 3B) and K562R overexpressing Lyn kinase7 (Figure 3C). Neither Jak2 nor Lyn detergent solubility was affected by WP1130 in these cells. In BaF3 transfectants expressing Bcr-Abl, loss of Bcr-Abl from the detergent-soluble fraction was associated with reduced substrate phosphorylation (pY-Stat5) and the onset of apoptosis (PARP cleavage; Figure 3D). Bcr-Abl in primary CML cells from imatinib-resistant patients was also observed to translocate to the detergent-insoluble fraction after WP1130 incubation, and was associated with a loss of Bcr-Abl substrate (pY-Stat5, pY-CrkL) phosphorylation (Figure 3E). The detergent-insoluble fraction is highly enriched in cytoskeletal proteins and components of cellular structures called aggresomes (data not shown).32 These results suggest that WP1130 blocks Bcr-Abl substrate phosphorylation through compartmentalization of Bcr-Abl. To determine whether WP1130 induces differential effects on protein ubiquitination and survival of CD34+ cells from both healthy donors and CML patients, primary CD34+ cells were isolated and treated with WP1130 for 4 hours before total cell lysates were subjected to immunoblotting for ubiquitin. High-molecular-weight ubiquitinated proteins were increased by WP1130 in both normal and leukemic CD34+ cells (Figure 3F). However, CD34+ cells from CML patients were significantly more sensitive to WP1130-mediated apoptosis than cells from healthy donors (Figure 3G). This may be related to the affects of WP1130 on Bcr-Abl compartmentalization and signal transduction. However, because only a 2-fold difference in half-maximal inhibitory concentration (IC50) values was noted between CML (∼ 2.8μM) and normal CD34+ cells (> 5μM), it is unclear whether this compound is a candidate for clinical studies. WP1130 derivatives with greater water solubility and a more restricted DUB-inhibitory profile are being designed and tested. We have also noted that the Bcr-Abl compartmentalization and apoptotic activity of WP1130 are engaged within 1 hour of exposure and cannot be reversed by removal of the compound (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These results suggest that the effects of WP1130 are irreversible and that cells will require only minimal contact time to elicit a response.
WP1130 induces cellular trafficking of Bcr-Abl and inhibits its substrate phosphorylation in CML cells. (A) BV-173 cells were treated with 5μM WP1130 for the intervals indicated before assessing Bcr-Abl and other protein levels in total cell lysates and in the detergent-soluble and -insoluble fractions. Actin was blotted as a protein-loading control. (B) BV-173 and BV-173R cells were treated with 5μM WP1130 for 2 hours before detergent-soluble and -insoluble cell lysates were blotted for Bcr-Abl (K12 antibody) and actin as a protein-loading control. (C) K562R cells were treated with 5μM WP1130 for 2 hours before proteins derived from the total cell lysate (T) and detergent-soluble (S) and insoluble (I) lysates were probed for Bcr-Abl and other proteins or phosphoproteins as indicated. Total cell lysate represents the detergent-soluble and -insoluble lysates combined. Actin served as a protein-loading control. (D) BaF3 cells transformed with eGFP-Bcr-Abl with the T315I mutation were treated with 5μM WP1130 for the intervals noted before detergent-soluble cell lysates were immunoblotted for the indicated protein or phosphoprotein. Actin was blotted as a protein-loading control. Loss of Bcr-Abl from the soluble (cytoplasmic) cell fraction was associated with reduced signaling (pY-Stat5) and the onset of apoptosis (cleaved PARP). (E) Mononuclear cells from 2 CML patients who were progressing on imatinib therapy were treated with 5μM WP1130 for 4 hours before cell lysates representing the combined detergent-soluble and -insoluble fractions were probed for Bcr-Abl by immunoblotting (top). The detergent-soluble cell lysate was also immunoblotted for Bcr-Abl substrate phosphoproteins (pY-Stat5, pY-CrkL), as well as Stat5 and CrkL total protein levels. (F) CD34+ cells from a healthy donor and 2 imatinib-refractory CML patients were treated with 5μM WP1130 for 4 hours before total protein lysates (8 μg) were subjected to ubiquitin immunoblotting. Hsp90 was immunoblotted as a protein-loading control. (G) CD34+ cells isolated from 2 healthy donors and 3 imatinib-refractory CML patients were treated with the indicated concentrations of WP1130 for 24 hours before analysis of cell survival by annexin/propidium iodide staining and flow cytometry. The results represent the average ± SD of 3 replicates. Statistical significance was determined with a paired Student t test.
WP1130 induces cellular trafficking of Bcr-Abl and inhibits its substrate phosphorylation in CML cells. (A) BV-173 cells were treated with 5μM WP1130 for the intervals indicated before assessing Bcr-Abl and other protein levels in total cell lysates and in the detergent-soluble and -insoluble fractions. Actin was blotted as a protein-loading control. (B) BV-173 and BV-173R cells were treated with 5μM WP1130 for 2 hours before detergent-soluble and -insoluble cell lysates were blotted for Bcr-Abl (K12 antibody) and actin as a protein-loading control. (C) K562R cells were treated with 5μM WP1130 for 2 hours before proteins derived from the total cell lysate (T) and detergent-soluble (S) and insoluble (I) lysates were probed for Bcr-Abl and other proteins or phosphoproteins as indicated. Total cell lysate represents the detergent-soluble and -insoluble lysates combined. Actin served as a protein-loading control. (D) BaF3 cells transformed with eGFP-Bcr-Abl with the T315I mutation were treated with 5μM WP1130 for the intervals noted before detergent-soluble cell lysates were immunoblotted for the indicated protein or phosphoprotein. Actin was blotted as a protein-loading control. Loss of Bcr-Abl from the soluble (cytoplasmic) cell fraction was associated with reduced signaling (pY-Stat5) and the onset of apoptosis (cleaved PARP). (E) Mononuclear cells from 2 CML patients who were progressing on imatinib therapy were treated with 5μM WP1130 for 4 hours before cell lysates representing the combined detergent-soluble and -insoluble fractions were probed for Bcr-Abl by immunoblotting (top). The detergent-soluble cell lysate was also immunoblotted for Bcr-Abl substrate phosphoproteins (pY-Stat5, pY-CrkL), as well as Stat5 and CrkL total protein levels. (F) CD34+ cells from a healthy donor and 2 imatinib-refractory CML patients were treated with 5μM WP1130 for 4 hours before total protein lysates (8 μg) were subjected to ubiquitin immunoblotting. Hsp90 was immunoblotted as a protein-loading control. (G) CD34+ cells isolated from 2 healthy donors and 3 imatinib-refractory CML patients were treated with the indicated concentrations of WP1130 for 24 hours before analysis of cell survival by annexin/propidium iodide staining and flow cytometry. The results represent the average ± SD of 3 replicates. Statistical significance was determined with a paired Student t test.
Bcr-Abl is ubiquitinated in WP1130-treated CML cells
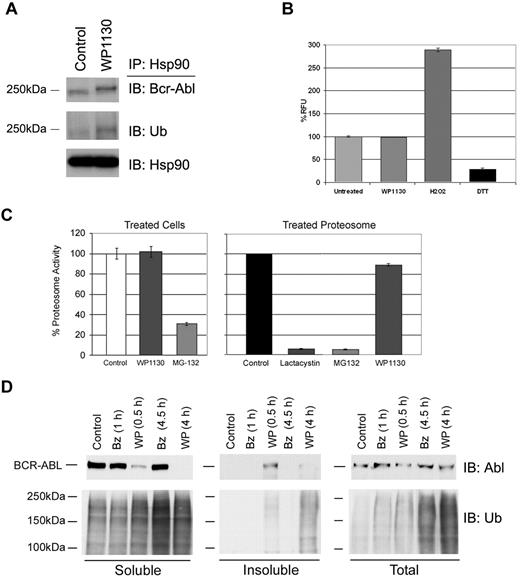
Ubiquitinated protein content can be increased as a consequence of proteasome inhibition, loss of protein chaperone activity, or increased cellular oxidation leading to protein misfolding.33 However, we did not detect the loss of Hsp90 association with Bcr-Abl (Figure 2C and Figure 4A) and did not find elevated ROS in WP1130-treated cells (Figure 4B). Further, WP1130 did not suppress either 20S proteasome activity in intact CML cells or purified 20S proteasome subunit preparations, eliminating the possibility that WP1130 acts as a proteasome inhibitor (Figure 4C). WP1130 was compared with the potent 20S proteasome inhibitor bortezomib to determine whether they both had an impact on protein ubiquitination and Bcr-Abl compartmentalization. Both compounds increased ubiquitinated protein content, but only WP1130 induced accumulation of detergent-insoluble ubiquitinated proteins (Figure 4D). WP1130 treatment led to the reduction of Bcr-Abl from the detergent-soluble fraction and the appearance of Bcr-Abl in the detergent-insoluble fraction, but no change in total Bcr-Abl content was noted. Bortezomib elevated ubiquitinated protein levels but had no affect on Bcr-Abl detergent solubility or protein levels.
WP1130 activity is not mediated through the loss of Hsp90/Bcr-Abl chaperone association, 20S proteasome inhibition, or increased ROS. (A) K562 cells were incubated with 5μM WP1130 for 30 minutes before detergent-soluble cell extracts were subjected to Hsp90 immunoprecipitation followed by immunoblotting for Bcr-Abl, ubiquitin, or Hsp90. (B) K562 cells were incubated with WP1130 (5μM), H2O2 (500μM), or dithiothreitol (1mM) for 2 hours at 37°C. Cells were then washed and replated in medium containing DCFDA for 20 minutes at 37°C. DCFDA fluorescence was read and used as a measure of ROS production in treated samples. The results represent the average ± SD of triplicate assays. Similar results were obtained in 2 additional CML cell lines, WDT-2 and BV-173. (C left) K562 cells were left untreated or treated with 5μM of the indicated compound for 2 hours at 37°C. Protein lysates were incubated with proteasome substrate and activity was determined by fluorogenic substrate cleavage. (C right) Purified 20S proteasome was incubated with 5μM MG-132, lactacystin, or WP1130 before assaying proteasome activity as described above. The results represent the average ± SD of triplicate assays. Similar results were obtained with 2 additional CML cell lines. (D) K562 cells were treated with 50 nM bortezomib (Bz) or 5μM WP1130 (WP) before equal volumes of lysate from the detergent-soluble, detergent-insoluble, and total cell lysate were immunoblotted for Bcr-Abl (top) or ubiquitin (bottom). Both Bz and WP increased ubiquitin content, but only WP affected Bcr-Abl and detergent-insoluble ubiquitinated protein levels.
WP1130 activity is not mediated through the loss of Hsp90/Bcr-Abl chaperone association, 20S proteasome inhibition, or increased ROS. (A) K562 cells were incubated with 5μM WP1130 for 30 minutes before detergent-soluble cell extracts were subjected to Hsp90 immunoprecipitation followed by immunoblotting for Bcr-Abl, ubiquitin, or Hsp90. (B) K562 cells were incubated with WP1130 (5μM), H2O2 (500μM), or dithiothreitol (1mM) for 2 hours at 37°C. Cells were then washed and replated in medium containing DCFDA for 20 minutes at 37°C. DCFDA fluorescence was read and used as a measure of ROS production in treated samples. The results represent the average ± SD of triplicate assays. Similar results were obtained in 2 additional CML cell lines, WDT-2 and BV-173. (C left) K562 cells were left untreated or treated with 5μM of the indicated compound for 2 hours at 37°C. Protein lysates were incubated with proteasome substrate and activity was determined by fluorogenic substrate cleavage. (C right) Purified 20S proteasome was incubated with 5μM MG-132, lactacystin, or WP1130 before assaying proteasome activity as described above. The results represent the average ± SD of triplicate assays. Similar results were obtained with 2 additional CML cell lines. (D) K562 cells were treated with 50 nM bortezomib (Bz) or 5μM WP1130 (WP) before equal volumes of lysate from the detergent-soluble, detergent-insoluble, and total cell lysate were immunoblotted for Bcr-Abl (top) or ubiquitin (bottom). Both Bz and WP increased ubiquitin content, but only WP affected Bcr-Abl and detergent-insoluble ubiquitinated protein levels.
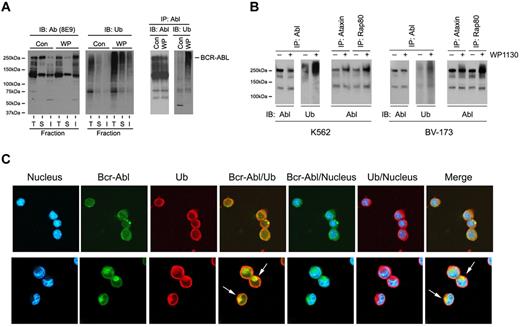
To determine whether Bcr-Abl was ubiquitinated in WP1130-treated cells, we treated CML cells with WP1130 for short intervals (30 minutes) to allow recovery of Bcr-Abl from the detergent-soluble fraction (for direct immunoprecipitation). Bcr-Abl immunoblotting after immunoprecipitation from WP1130-treated cells demonstrated a moderate reduction in Bcr-Abl recovery from the detergent-soluble extract (Figure 5A left), but a marked increase in its ubiquitination. Blotting confirmed a significant increase of Bcr-Abl and ubiquitinated proteins in the insoluble fraction from WP1130-treated cells (Figure 5A right). To determine whether Bcr-Abl modification by WP1130 was due to the transfer of specific ubiquitin polymers to the kinase, soluble cell lysates were subjected to immunoprecipitation with agarose beads conjugated to proteins with high affinity for K48-linked (ataxin) or K63-linked (Rap80) ubiquitin polymers,34 and protein eluates were immunoblotted for Bcr-Abl. Recovery of Bcr-Abl/K63-linked ubiquitin polymers was more prominent in WP1130-treated cells (Figure 5B), suggesting that increased K63-linked ubiquitin polymers on Bcr-Abl underlies the signal for its translocation into detergent-insoluble complexes.
WP1130 stimulates Bcr-Abl ubiquitination and trafficking in CML cells. (A) WDT-2 cells were treated with vehicle alone (control) or 5μM WP1130 (WP) for 30 minutes at 37°C before an equal volume of cell lysates were resolved into total (T), detergent-soluble (S), or detergent-insoluble (I) fractions and probed for Bcr-Abl (left) or ubiquitin (center). Equal protein (400 μg) detergent-soluble cell lysate from WP1130-treated or control cells was treated with 1% SDS at 60°C, diluted to 0.1% SDS, and subjected to immunoprecipitation with anti-Abl (K12). The immunoprecipitate was washed, resolved on gels, and immunoblotted for Abl (left) or ubiquitin (right). (B) K562 or BV-173 cells were treated as described in panel A, and equal amounts of detergent-soluble cell lysates were subjected to Abl immunoprecipitation and blotting for Abl and ubiquitin (as described in panel A right). An aliquot of the same lysate (200 μg) from control and treated cells was also subjected to affinity enrichment for K48-linked (ataxin) or K63-linked (Rap80) ubiquitin polymers. Bound protein was eluted and subjected to Abl immunoblotting. (C) eGFP-Bcr-Abl–transformed BaF3 cells were treated with vehicle alone (top panel) or with 5μM WP1130 for 4 hours (bottom panel) before cells were fixed, cytospun onto slides, and permeabilized. After blocking reactive sites, slides were incubated with anti-ubiquitin antibody (1:100), washed, and antigen detected with Alexa Fluor antibodies. The slides were washed again and stained for nucleus detection with Hoechst 33342. Images were acquired using an Olympus confocal microscope FV-500 and merged with Photoshop Version 6.0 software. The arrows depict Bcr-Abl/ubiquitin colocalization, and their juxtanuclear localization resembles aggresomes.
WP1130 stimulates Bcr-Abl ubiquitination and trafficking in CML cells. (A) WDT-2 cells were treated with vehicle alone (control) or 5μM WP1130 (WP) for 30 minutes at 37°C before an equal volume of cell lysates were resolved into total (T), detergent-soluble (S), or detergent-insoluble (I) fractions and probed for Bcr-Abl (left) or ubiquitin (center). Equal protein (400 μg) detergent-soluble cell lysate from WP1130-treated or control cells was treated with 1% SDS at 60°C, diluted to 0.1% SDS, and subjected to immunoprecipitation with anti-Abl (K12). The immunoprecipitate was washed, resolved on gels, and immunoblotted for Abl (left) or ubiquitin (right). (B) K562 or BV-173 cells were treated as described in panel A, and equal amounts of detergent-soluble cell lysates were subjected to Abl immunoprecipitation and blotting for Abl and ubiquitin (as described in panel A right). An aliquot of the same lysate (200 μg) from control and treated cells was also subjected to affinity enrichment for K48-linked (ataxin) or K63-linked (Rap80) ubiquitin polymers. Bound protein was eluted and subjected to Abl immunoblotting. (C) eGFP-Bcr-Abl–transformed BaF3 cells were treated with vehicle alone (top panel) or with 5μM WP1130 for 4 hours (bottom panel) before cells were fixed, cytospun onto slides, and permeabilized. After blocking reactive sites, slides were incubated with anti-ubiquitin antibody (1:100), washed, and antigen detected with Alexa Fluor antibodies. The slides were washed again and stained for nucleus detection with Hoechst 33342. Images were acquired using an Olympus confocal microscope FV-500 and merged with Photoshop Version 6.0 software. The arrows depict Bcr-Abl/ubiquitin colocalization, and their juxtanuclear localization resembles aggresomes.
To further assess Bcr-Abl ubiquitination and cellular distribution, eGFP-Bcr-Abl BaF3 transformants were examined by confocal microscopy. Treatment with WP1130 resulted in the clustering of Bcr-Abl and ubiquitin in juxtanuclear complexes (white arrows) resembling aggresomes.32 Biochemical studies support the inclusion of other proteins defining the aggresome in complex with Bcr-Abl from WP1130-treated cells32,35 (data not shown). These results suggest that WP1130 stimulates ubiquitination of Bcr-Abl (primarily with K63-linked polymers), leading to its transfer to the aggresome.
WP1130 inhibits DUB activity
A cross-conjugated α,β-unsaturated dienone with 2 sterically accessible electrophilic β-carbons is a molecular determinant of isopeptidase or DUB-inhibitor activity.36 The presence of these determinants in WP1130 prompted an investigation of its potential DUB-inhibitor activity. Lysates derived from untreated or WP1130-treated cells were incubated with purified ubiquitin polymers (Ub1-Ub5), and the relative recovery of polymers was assessed by immunoblotting. Lysates derived from WP1130-treated CML cells showed a partial protection of both K48- and K63-linked ubiquitin polymers from disassembly, supporting the possibility of DUB inhibition by WP1130 (Figure 6A). To more closely examine DUB inhibition, lysates from untreated, WP1130-treated (1-8 hours), or NEM-treated37 CML cells were incubated with fluorescent DUB substrate (Ub-AMC) and activity was monitored over time. Lysates derived from cells treated with the nonspecific DUB inhibitor NEM completely blocked substrate hydrolysis (supplemental Figure 2A), whereas lysates from WP1130-treated CML cells showed limited effects on Ub-AMC hydrolyzing activity. These results suggested that WP1130-mediated ubiquitin changes may be mediated through inhibition of a limited subset of DUBs.
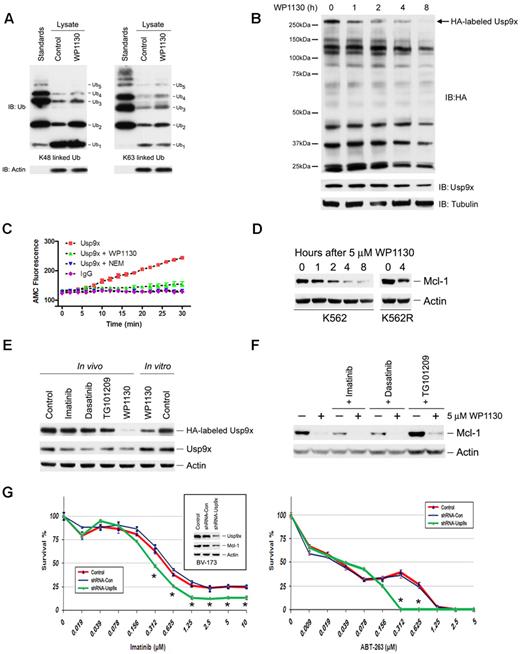
WP1130 affects cellular DUB activity. (A) Lysates (5 μg) from untreated (control) or WP1130-treated (5μM, 4 hours) cells were incubated with 1 μg of K48-linked (left) or K63-linked (right) free chains of polyubiquitin (Ub1-5) for 10 minutes at 37°C. The extent of free chain hydrolysis by active DUB in the each lysate was examined by ubiquitin immunoblotting. Ubiquitin polymer standards (Ub1-5) were resolved on the left and actin was immunoblotted as a protein-loading control. (B) K562 cells treated with 5μM WP1130 (0 to 8 hours) were lysed in DUB-labeling buffer as described in “Ub-AMC protease assay.” Clarified supernatant (20 μg) was incubated with 200nM HA-UbVs (Boston Biochem) for 1 hour at 37°C. HA immunoblotting was used to assess changes in DUB activity (top). Tubulin was probed as a protein-loading control (bottom). The level of the upper HA-labeled band (arrow) was reduced by WP1130 and was determined to represent Usp9x (molecular weight, 293 kDa). The blot was reprobed for Usp9x as a measure of its protein-loading level (middle blot). (C) Usp9x was immunoprecipitated from K562 cells, washed, and incubated in DUB assay buffer containing NEM (5mM), WP1130 (5μM), or DMSO in a 100-μL reaction volume for 30 minutes at 37°C in 96-well fluorometry plates. K562 cell lysates immunoprecipitated with rabbit IgG were used as a control. After incubation, 500nM Ub-AMC was added to the reaction and the release of AMC-fluorescence was recorded over time. The representative fluorescence change over time is shown. The Usp9x activity results represent the average ± SD of triplicate assays. Similar results were obtained in 2 additional independent experiments. (D) K562 and K562R cells were treated with WP1130 for the intervals noted before total cell lysates were subjected to Mcl-1 and actin immunoblotting. (E) In vivo K562 cells were left untreated or treated with 5μM imatinib, 0.5μM dasatinib, 0.5μM TG101209, or 5μM WP1130 for 4 hours before cell lysates were analyzed for Usp9x activity by incubation with HA-UbVs and HA immunoblotting (as described in panel B). In vitro K562 cell lysates were treated with DMSO (control) or 5μM WP1130 for 30 minutes at 37°C before assessing Usp9x activity by HA-UbVs labeling followed by HA blotting. The top portion of the HA immunoblot containing Usp9x is shown. The membrane was stripped and immunoblotted for Usp9x and actin. (F) WDT-2 cells were treated with WP1130 alone or pretreated with imatinib (5μM), dasatinib (0.5μM), or TG101209 (0.5μM) for 1 hour before additional incubation in the presence of WP1130. All cells were harvested 4 hours after WP1130 treatment and total cell lysates were subjected to Mcl-1 immunoblotting. Actin was immunoblotted as a protein-loading control (Note: more protein was loaded in samples obtained from TG101209-treated cells). (G) Left inset, BV-173 cells were untreated (Control) or infected with LVX-shRNA (shRNA-Con) or LVX-shRNA-Usp9x (shRNA-Usp9x) as described in “shRNA and Usp9x silencing.” After puromycin selection, Usp9x and Mcl-1 levels were examined by immunoblotting. Actin was immunoblotted as a protein-loading control. Left, Control and shRNA expressing BV-173 cells (as indicated) were treated with the indicated concentration of imatinib for 72 hours before assessing viability by MTT staining. The results represent the average ± SD of 4 replicates. Statistical significance was determined with a paired Student t test. *P < .05 for shRNA-Usp9x compared with Control or shRNA-Con. Right, BV-173 cells (control and shRNA-expressing as noted) were treated with the indicated concentration of ABT-263 for 72 hours before assessing viability as described in the center panel. The results represent the average ± SD of 4 replicates. Statistical significance was determined with a paired Student t test. *P < .05 for shRNA-Usp9x compared with Control or shRNA-Con.
WP1130 affects cellular DUB activity. (A) Lysates (5 μg) from untreated (control) or WP1130-treated (5μM, 4 hours) cells were incubated with 1 μg of K48-linked (left) or K63-linked (right) free chains of polyubiquitin (Ub1-5) for 10 minutes at 37°C. The extent of free chain hydrolysis by active DUB in the each lysate was examined by ubiquitin immunoblotting. Ubiquitin polymer standards (Ub1-5) were resolved on the left and actin was immunoblotted as a protein-loading control. (B) K562 cells treated with 5μM WP1130 (0 to 8 hours) were lysed in DUB-labeling buffer as described in “Ub-AMC protease assay.” Clarified supernatant (20 μg) was incubated with 200nM HA-UbVs (Boston Biochem) for 1 hour at 37°C. HA immunoblotting was used to assess changes in DUB activity (top). Tubulin was probed as a protein-loading control (bottom). The level of the upper HA-labeled band (arrow) was reduced by WP1130 and was determined to represent Usp9x (molecular weight, 293 kDa). The blot was reprobed for Usp9x as a measure of its protein-loading level (middle blot). (C) Usp9x was immunoprecipitated from K562 cells, washed, and incubated in DUB assay buffer containing NEM (5mM), WP1130 (5μM), or DMSO in a 100-μL reaction volume for 30 minutes at 37°C in 96-well fluorometry plates. K562 cell lysates immunoprecipitated with rabbit IgG were used as a control. After incubation, 500nM Ub-AMC was added to the reaction and the release of AMC-fluorescence was recorded over time. The representative fluorescence change over time is shown. The Usp9x activity results represent the average ± SD of triplicate assays. Similar results were obtained in 2 additional independent experiments. (D) K562 and K562R cells were treated with WP1130 for the intervals noted before total cell lysates were subjected to Mcl-1 and actin immunoblotting. (E) In vivo K562 cells were left untreated or treated with 5μM imatinib, 0.5μM dasatinib, 0.5μM TG101209, or 5μM WP1130 for 4 hours before cell lysates were analyzed for Usp9x activity by incubation with HA-UbVs and HA immunoblotting (as described in panel B). In vitro K562 cell lysates were treated with DMSO (control) or 5μM WP1130 for 30 minutes at 37°C before assessing Usp9x activity by HA-UbVs labeling followed by HA blotting. The top portion of the HA immunoblot containing Usp9x is shown. The membrane was stripped and immunoblotted for Usp9x and actin. (F) WDT-2 cells were treated with WP1130 alone or pretreated with imatinib (5μM), dasatinib (0.5μM), or TG101209 (0.5μM) for 1 hour before additional incubation in the presence of WP1130. All cells were harvested 4 hours after WP1130 treatment and total cell lysates were subjected to Mcl-1 immunoblotting. Actin was immunoblotted as a protein-loading control (Note: more protein was loaded in samples obtained from TG101209-treated cells). (G) Left inset, BV-173 cells were untreated (Control) or infected with LVX-shRNA (shRNA-Con) or LVX-shRNA-Usp9x (shRNA-Usp9x) as described in “shRNA and Usp9x silencing.” After puromycin selection, Usp9x and Mcl-1 levels were examined by immunoblotting. Actin was immunoblotted as a protein-loading control. Left, Control and shRNA expressing BV-173 cells (as indicated) were treated with the indicated concentration of imatinib for 72 hours before assessing viability by MTT staining. The results represent the average ± SD of 4 replicates. Statistical significance was determined with a paired Student t test. *P < .05 for shRNA-Usp9x compared with Control or shRNA-Con. Right, BV-173 cells (control and shRNA-expressing as noted) were treated with the indicated concentration of ABT-263 for 72 hours before assessing viability as described in the center panel. The results represent the average ± SD of 4 replicates. Statistical significance was determined with a paired Student t test. *P < .05 for shRNA-Usp9x compared with Control or shRNA-Con.
To examine the impact of WP1130 on specific DUB activity, CML cells were treated with WP1130 and lysates were incubated with the HA-tagged, irreversible DUB substrate HA-UbVs, as described previously.37 The covalent modification of DUBs by this construct allows analysis of DUB activity by HA immunoblotting. Inhibition of DUB activity by WP1130 may be detected by a reduction in HA labeling of WP1130-sensitive DUBs. As shown in Figure 6B, lysates from WP1130-treated cells showed a reduced capacity to label one prominent high–molecular weight DUB identified as Usp9x (Figure 6B arrow). Usp9x is predominantly expressed in the cytoplasmic, detergent-soluble fraction, and WP1130 treatment did not affect Usp9x recovery or protein content (Figure 6C bottom). To determine whether WP1130 directly affects Usp9x activity, K562 cell lysates were incubated with WP1130 for 1 hour before incubation with HA-UbVs DUB substrate and analysis by HA blotting. Usp9x labeling was completely blocked in the presence of WP1130, suggesting direct Usp9x inhibition (supplemental Figure 2B). Other DUBs were also affected by WP1130, suggesting that WP1130 targets additional DUBs with as yet unknown specificity or selectivity. To confirm direct DUB inhibition, Usp9x was immunoprecipitated, washed extensively, and incubated with WP1130 before assessing DUB activity using the fluorescent DUB substrate Ub-AMC (supplemental Figure 2A). The rate of substrate hydrolysis was measured and compared in DMSO and WP1130 incubation assays to estimate the percentage of DUB inhibition by WP1130. Normal IgG immunoprecipitates and treatment of Usp9x immunoprecipitates with NEM (Figure 6C) did not result in substrate hydrolysis compared with control reactions. WP1130 reduced Usp9x activity by > 80% (Figure 6C), demonstrating that WP1130 functions as a subset-specific DUB inhibitor with activity against Usp9x. Other DUB activities were completely insensitive to WP1130 inhibition when assessed by HA-UbVs labeling (supplemental Figure 3A) or in DUB enzyme assays with Ub-AMC substrate (supplemental Figure 3B).
Recent studies have demonstrated that Usp9x controls Mcl-1 deubiquitination and degradation.21 To determine whether Usp9x inhibition by WP1130 affects Mcl-1 levels in CML cells, total cell extracts were immunoblotted for Mcl-1. WP1130 reduced Mcl-1 levels (Figure 6D) in a temporal fashion that paralleled Usp9x inhibition, and was active in both imatinib-sensitive (K562) and imatinib-resistant (K562R) CML cells.7 To determine whether Bcr-Abl or Jak2 kinase inhibition could also reduce Usp9x activity, CML cells were treated with WP1130, a Bcr-Abl–selective inhibitor (imatinib), a Jak2-selective inhibitor (TG101209), or a multi-kinase inhibitor (dasatinib) for 4 hours before cell lysates were subjected to HA-UbVs labeling and direct Usp9x immunoblotting. In vitro treatment of cell lysates with WP1130 (5μM, 30 minutes) was also included to confirm direct Usp9x inhibition (Figure 6E last 2 lanes). WP1130 effectively reduced Usp9x activity, whereas none of the kinase inhibitors examined had significant Usp9x-inhibitory activity. The effect of kinase inhibition on Mcl-1 protein levels was also examined in WDT-2 CML cells and compared with the effects of WP1130 alone or in combination with kinase inhibitors. As shown in Figure 6F, WP1130 treatment resulted in a rapid reduction in Mcl-1 protein levels and was associated with Usp9x inhibition (supplemental Figure 3A). Bcr-Abl kinase inhibitors (imatinib and dasatinib) also reduced Mcl-1 levels, but to a lesser extent than that detected in WP1130-treated cells. Interestingly, the Jak2 inhibitor TG101209 did not reduce Mcl-1 protein levels, suggesting that Bcr-Abl, but not Jak2, plays a role in the control of Mcl-1 expression in CML cells. WP1130 caused a greater reduction in Mcl-1 levels than kinase inhibitors alone, suggesting that the combined effects of WP1130 on Bcr-Abl signaling and Usp9x activity in CML cells induces greater Mcl-1 down-regulatory activity than that achieved through Bcr-Abl kinase inhibition alone.
BV-173 cells expressing shRNA directed against Usp9x were used to determine the effects of this DUB on apoptosis initiated by Bcr-Abl kinase inhibition (imatinib) or by a BH3 mimetic (ABT-263), as described previously.21 Usp9x-shRNA–expressing cells showed reduced levels of Mcl-1 and ∼ 2-fold increased sensitivity to imatinib compared with controls (IC50 ∼ 300nM in shRNA-Usp9x–expressing cells vs ∼ 600nM in shRNA controls; Figure 6G). Cells with reduced levels of Usp9x were also more sensitive to intermediate concentrations of ABT-263.
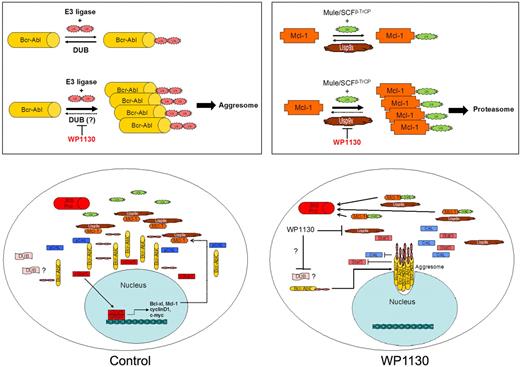
A summary of the effects of WP1130 on CML cells is shown in Figure 7. WP1130 induces Bcr-Abl ubiquitination and directly inhibits Usp9x activity. However, although WP1130 inhibits Usp9x activity, this does not appear to be responsible for the observed increase in Bcr-Abl ubiquitination. These 2 activities led to a reduction in Bcr-Abl signaling and induction of apoptosis, because aggresomal Bcr-Abl was unable to phosphorylate downstream proteins, whereas Usp9x inhibition reduced apoptotic thresholds through its control of Mcl-1 protein levels. This mechanism of control over tumor cell survival and signaling through changes in the activity of ubiquitin cycle–regulatory proteins has not been described previously and may be of significance in tumors that are dependent on continuous kinase activity and elevated Usp9x activity.
Overview of WP1130 activity in CML cells. (Top left) Bcr-Abl is ubiquitinated with K63-linked ubiquitin polymers, but balanced E3 ligase and DUB activities prevent its accumulation and detection. WP1130 prevents deubiquitination of Bcr-Abl (or increases E3 ligase activity), resulting in detectable ubiquitination of Bcr-Abl and its trafficking to the aggresome. (Top right) Mcl-1 undergoes balanced ubiquitination and deubiquitination to regulate its stability and entry into the proteasome. WP1130 inhibits Usp9x activity, resulting in increased Mcl-1 ubiquitination and degradation by the proteasome. (Bottom left) In CML cells, Bcr-Abl phosphorylates multiple substrates, leading to transformation and increased expression of Mcl-1 and other anti-apoptotic proteins. Balanced ubiquitination/deubiquitination reactions result in steady-state protein levels. (Bottom right) WP1130 increases Bcr-Abl ubiquitination, resulting in its translocation to the aggresome, where it is unable to maintain signal transduction and gene expression. WP1130 directly inhibits Usp9x activity, resulting in the ubiquitination and proteasomal destruction of Mcl-1. Loss of Bcr-Abl signaling and Mcl-1 apoptotic protection results in CML cell apoptosis.
Overview of WP1130 activity in CML cells. (Top left) Bcr-Abl is ubiquitinated with K63-linked ubiquitin polymers, but balanced E3 ligase and DUB activities prevent its accumulation and detection. WP1130 prevents deubiquitination of Bcr-Abl (or increases E3 ligase activity), resulting in detectable ubiquitination of Bcr-Abl and its trafficking to the aggresome. (Top right) Mcl-1 undergoes balanced ubiquitination and deubiquitination to regulate its stability and entry into the proteasome. WP1130 inhibits Usp9x activity, resulting in increased Mcl-1 ubiquitination and degradation by the proteasome. (Bottom left) In CML cells, Bcr-Abl phosphorylates multiple substrates, leading to transformation and increased expression of Mcl-1 and other anti-apoptotic proteins. Balanced ubiquitination/deubiquitination reactions result in steady-state protein levels. (Bottom right) WP1130 increases Bcr-Abl ubiquitination, resulting in its translocation to the aggresome, where it is unable to maintain signal transduction and gene expression. WP1130 directly inhibits Usp9x activity, resulting in the ubiquitination and proteasomal destruction of Mcl-1. Loss of Bcr-Abl signaling and Mcl-1 apoptotic protection results in CML cell apoptosis.
Discussion
In this study, we analyzed the mechanism of action of WP1130, a small molecule with previously reported Bcr-Abl down-regulatory activity.20 By screening multiple kinases expressed and activated in CML cells, we could detect only Bcr-Abl down-regulation in CML cells (Figure 1A). Thorough examination showed that WP1130 action is initiated in the cytoplasmic fraction of the cell (Figure 2D), which may partially explain the selectivity for the predominantly cytoplasmic Bcr-Abl protein. Other kinases, such as Jak2, Lyn, and PI3-K, are reported to be detected in the membrane fraction or associated with transmembrane receptors. Analysis of several tumor types supports the observed selectivity of WP1130 for specific cytoplasmic kinases, because transmembrane kinases such as the HER family, c-kit, and Flt-3 are insensitive to WP1130. However, in hematologic tumors that do not express Bcr-Abl, Jak2 undergoes rapid ubiquitination and aggresomal trafficking in response to WP1130. The reason for this change in kinase target in Bcr-Abl–negative cells is unknown, but may be due to differential expression and activity of DUB in Bcr-Abl–transformed cells (N.J.D., L.F.P., manuscript in preparation). Jak2 ubiquitination also explains the reported Jak/Stat-inhibitory activity of WP1130 (V.K., N.J.D., manuscript in preparation) and less-active derivatives such as WP1066 and WP1034.38-41 Based on the activities reported here and on previous observations, WP1130 initiates a series of events that results in the ubiquitination of proteins such as Bcr-Abl with K63-linked polymers. This form of protein modification typically signals for proteins to traffic or transfer to organelles, and, in the case of Bcr-Abl, the aggresome appears to be the main site for transfer.31 Aggresomes are typically formed in response to protein misfolding or overload and are proposed to be cytoprotective by reducing the potential for excess protein to interfere with cellular metabolism.32,42 However, in the case of Bcr-Abl, loss of signaling, even for short intervals, is associated with the onset of CML cell apoptosis.43 In this regard, WP1130-initiated compartmentalization of Bcr-Abl converts a cytoprotective process into a cytodestructive one. Based on the observed inhibition of DUB activity by WP1130, it appears that DUB targets may be associated with the observed WP1130 activity. However, other potential targets must also be considered, and several approaches are being explored to define mediators underlying aggresome formation in WP1130-treated cells. We have also noted that WP1130, like imatinib,44 triggers CML cell autophagy, but this is not sufficient to fully cyto-protect CML cells from DUB inhibition–mediated apoptosis. However, as demonstrated with imatinib,44 inhibition of WP1130-induced autophagy increases CML cell sensitivity to WP1130-mediated apoptosis (data not shown), suggesting that cellular stress applied through direct (imatinib) or indirect (WP1130) loss of Bcr-Abl signaling can engage the autophagic pathway to partially subvert CML cell death. Suppressing autophagy may be an effective strategy to increase CML cell sensitivity to both kinase and DUB inhibitor–mediated apoptosis.
The increase in ubiquitinated protein content in WP1130-treated cells appears to be mediated primarily through inhibition of DUB activity, but other activities may also be involved.45,46 We did note an increase in Hsp70 levels afterWP1130 treatment, suggesting a potential role for Hsp90 inhibition in the cellular response. However, we did not detect the reduction in Hsp90:Bcr-Abl complexes (Figure 2C) that is reported to occur as a consequence of loss of Hsp90 chaperone activity. One possible explanation for this response is that misfolded proteins accumulate as a result of DUB inhibition and increased protein ubiquitination in WP1130-treated cells. The cell induces protein chaperones (such as Hsp70) in an attempt to restrict the formation of toxic protein aggregates.45 The kinetics of Hsp70 induction (Figure 2) and the onset of aggresome formation (Figure 5) support this possibility.
We did not observe a change in the activity of the Bcr-Abl–associated E3 ligase c-Cbl after WP1130 treatment, and its involvement in regulating Bcr-Abl ubiquitination appears remote, because we detected a predominant linkage with K63-linked ubiquitin polymers on Bcr-Abl that are not considered substrates for c-Cbl in ubiquitin ligase reactions.47 Bcr-Abl trafficking, not its proteasomal degradation, appears to underlie the major impact of WP1130 on CML cells and is aligned with previously described activities associated with increased K63-specific ubiquitin polymers on target proteins.31 Because Ubc13 (E2) is the only known enzyme involved in forming K63-linked polyubiquitin chains,48 this restricts the number of potential upstream components that provide K63-linked ubiquitin polymers for subsequent transfer to Bcr-Abl. Additional studies are being performed to determine the role of Ubc13 and other ubiquitin-conjugating or -activating enzymes in WP1130 activity against Bcr-Abl.
We have identified a subset of DUB enzymes that may underlie some of the protein ubiquitination and apoptotic activity of WP1130. Although we did not detect a major change in total DUB activity in CML cells after WP1130 administration (Figure 6B and supplemental Figure 2), we were able to detect inhibition of specific DUBs such as Usp9x. Usp9x activity was sensitive to WP1130 in assays from intact cells, cell lysates, and enzyme preparations at concentrations necessary to induce Bcr-Abl trafficking and apoptosis. Recent reports have suggested that Usp9x increases Mcl-1 stability, extending its half-life by preventing its proteasomal destruction through de-ubiquitination.21 This activity and the up-regulation of Usp9x are associated with poor prognosis in multiple myeloma patients, and may underlie chemoresistance in several additional cancers.21 We were able to demonstrate a reduction in Mcl-1 levels in WP1130-treated cells that paralleled inhibition of Usp9x activity (Figure 6). Mcl-1 down-regulation combined with loss of Bcr-Abl signaling activity may be key contributors to the induction of apoptosis in WP1130-treated CML cells.
To determine the impact of Usp9x inhibition on CML cell survival, we silenced Usp9x in CML cells and demonstrated Mcl-1 down-regulation, which increased sensitivity to imatinib and other apoptotic stimuli (Figure 6G), but no affect on Bcr-Abl ubiquitination or its cellular localization was observed (data not shown). Other reports have suggested an association between Bcr-Abl and Usp9x through secondary intermediates (Shc15 ). However, we did not detect Usp9x in Bcr-Abl pull-downs from control or WP1130-treated cells (data not shown). Whereas Usp9x expression and activity are highly relevant to Mcl-1 control in multiple tumors, they may also be of high significance to the sustained viability of CML stem cells that are only moderately responsive to Bcr-Abl kinase inhibition. Multiple studies have shown that early leukemogenic progenitors overexpress Mcl-1 as a consequence of activation of several upstream cascades, including Bcr-Abl and cytokine-mediated Stat activation.24 In the present study, kinase inhibitors did not block Usp9x activity in CML cells (Figure 6G), but did affect Mcl-1 protein levels, possibly through modulation of Mcl-1 gene expression. Until recently, the role of Usp9x as a modulator of Mcl-1 activity in transformed cells was unknown, but inhibitors of Usp9x may be of therapeutic importance in several settings. WP1130 may be highly suited for CML-based therapy because it has indirect effects on Bcr-Abl signaling through kinase sequestration into aggresomes and direct inhibition of Usp9x activity, which may be essential in stabilizing CML stem cell survival. In light of the importance of Usp9x in the control of Mcl-1 levels, compounds such as WP1130 may be useful in overcoming the apoptotic resistance associated with Usp9x activity and Mcl-1 protection. Further, CD34+ cells from CML patients are more sensitive to WP1130 than CD34+ cells derived from healthy donors (Figure 3G), suggesting that WP1130 or similar compounds may be useful in CML patients with residual or imatinib-refractory disease.
Inhibition of other DUBs may also play a role in WP1130 activity. We have noted a broader spectrum of DUB inhibition by WP1130 in other tumor cell types (data not shown). It is possible that simultaneous inhibition of several DUBs may be necessary to induce changes in Bcr-Abl ubiquitination and trafficking. Other reagents that measure DUB activity may yield a more comprehensive assessment of DUB activity and sensitivity to WP1130 in various tumors.49,50 In addition, an unbiased determination of direct targets of WP1130 may be assessable through other techniques and will be important in understanding its activity.51 In either case, our studies with WP1130 identified a novel pathway to kinase inhibition and apoptosis in CML cells. Additional studies of the key components in this pathway and the role of DUB inhibition in this process may highlight important targets for future cancer therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
These studies were supported by a University of Michigan Cancer Center Research Support Grant (to N.J.D.) and by University of Michigan Cancer Center Start Up Funds (to M.T.).
Authorship
Contribution: H.S. and V.K. performed the majority of the research, analyzed data, and wrote parts of the manuscript; L.F.P. and D.F. performed specific assays and analyzed data; W.G.B. and G.B. performed research; M.T. provided clinical samples and analyzed data; and N.J.D. directed the scope of the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicholas J. Donato, PhD, Dept of Internal Medicine, Division of Hematology/Oncology, 1500 E Medical Center Dr, University of Michigan Comprehensive Cancer Center, Ann Arbor, MI 48109; e-mail: ndonato@med.umich.edu.
References
Author notes
H.S. and V.K. contributed equally to this study.