Abstract
MicroRNAs are commonly aberrantly expressed in many cancers. Very little is known of their role in T-cell lymphoma, however. We therefore elucidated the complete miRNome of purified T cells from 21 patients diagnosed with Sézary Syndrome (SzS), a rare aggressive primary cutaneous T-cell (CD4+) lymphoma. Unsupervised cluster analysis of microarray data revealed that the microRNA expression profile was distinct from CD4+ T-cell controls and B-cell lymphomas. The majority (104 of 114) of SzS-associated microRNAs (P < .05) were down-regulated and their expression pattern was largely consistent with previously reported genomic copy number abnormalities and were found to be highly enriched (P < .001) for aberrantly expressed target genes. Levels of miR-223 distinguished SzS samples (n = 32) from healthy controls (n = 19) and patients with mycosis fungoides (n = 11) in more than 90% of samples. Furthermore, we demonstrate that the down-regulation of intronically encoded miR-342 plays a role in the pathogenesis of SzS by inhibiting apoptosis, and describe a novel mechanism of regulation for this microRNA via binding of miR-199a* to its host gene. We also provide the first in vivo evidence for down-regulation of the miR-17-92 cluster in malignancy and demonstrate that ectopic miR-17-5p expression increases apoptosis and decreases cell proliferation in SzS cells.
Introduction
Sézary Syndrome (SzS) is a rare aggressive form of primary cutaneous T-cell lymphoma (CTCL) characterized by erythroderma, generalized lymphadenopathy, and the presence of neoplastic cerebriform nucleated CD4+ T cells (Sézary cells) in peripheral blood.1 Patients with SzS typically have a high leukemic burden and a poor prognostic outcome, with an estimated 5-year survival of only 24%.1 The molecular pathogenesis of this devastating disease, however, remains poorly understood.
There is emerging evidence that microRNAs are involved in the pathogenesis of many cancers, including B-cell lymphomas.2 There are, however, very little published data to date on the involvement of microRNAs in human T-cell lymphomas. Therefore, we undertook a comprehensive study to elucidate the miRNome of tumor cells from 21 patients with SzS and normal CD4+ T cells using microarrays containing probes against 655 human microRNAs (miRBase 10.1).
Methods
Patient samples
Peripheral blood was obtained from 21 patients attending either the Skin Tumor Unit, St John's Institute of Dermatology, St Thomas' Hospital, London (patients SzS1-SzS17), or the Department of Dermatology, Leiden University Medical Center, (patients SzS18-SzS21). All patients had a T-cell clone detected in the peripheral blood as determined by T-cell receptor (TCR) gene rearrangement studies and fulfilled the World Health Organization–European Organization for the Research and Treatment of Cancer (WHO-EORTC) diagnostic criteria for SzS.3 Individual patient characteristics are shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). With the exception of blood cells from patients SzS18, SzS19, and SzS21, which were Ficoll-purified peripheral blood mononuclear cells, CD4+ cells were purified from SzS patient peripheral blood using the RosetteSep CD4+ T-cell enrichment kit (StemCell Technologies). Immunomagnetic separation (Miltenyi Biotec) was used to purify CD4+ T cells/CD3+ T cells or CD19+ B cells from the peripheral blood of healthy control donors as indicated in Figures 1 through 3.
A further cohort of SzS patient samples (n = 15), samples of patients with mycosis fungoides (MF) with no peripheral blood involvement (n = 11), and healthy donor controls (n = 12) were used as a validation set for testing the diagnostic ability of microRNA levels by quantitative reverse transcription–polymerase chain reaction (qRT-PCR). CD4+ T cells from these additional patients were also purified by the RosetteSep CD4+ T cell enrichment kit; patient characteristics are given in supplemental Table 2.
All patients gave informed consent to be included in this study in accordance with the Declaration of Helsinki 1975 as revised in 2005. The samples from patients SzS1 to SzS17, SzS22 to SzS36, and MF1 to MF11 were obtained from an ethically approved research tissue bank (St Mary National Research Ethics Committee: 07/H10712/106).
RNA purification and microarray analysis
MicroRNA (and total RNA) was isolated from samples SzS1 to SzS17 using Trizol (Invitrogen) and RNeasy columns as described by the manufacturer (QIAGEN), whereas only microRNA was purified from samples SzS18 to SzS21 using the mirVana kit (Ambion). MicroRNA (approximately 500 ng) were labeled and hybridized to μRNA microarrays as previously described4 using tonsillar material (pooled from 12 healthy donors) as a common reference in a dye-balanced design. The arrays contained 655 human probes (miRBase v.10.1). Probe details can be found at MicroRNA world.5
Image analysis was carried out with BlueFuse v3 software (BlueGnome). Raw image data were global median–normalized within arrays and normalized between arrays using the LIMMA package.6 The normalized log ratios (average of 4 replicates per probe) were used for subsequent analysis in Genespring 7.2 (Agilent Technologies). MicroRNAs were filtered before analysis of variance (ANOVA) to remove those that had a median intensity less than 1.5 × background (200 fluorescence units). ANOVA P values were adjusted using the Benjamini-Hochberg correction method. Differentially expressed genes were tested for their ability to predict sample class using the leave-one-out cross-validation support vector machine (SVM) function in Genespring. All microarray data are available in the GEO public database under accession number GSE21697.7
qRT-PCR
Due to insufficient material, only patient samples which had been CD4+ purified (ie, patients SzS1-SzS17) were used for qRT-PCR and subsequent analysis. CD4+ (n = 7) and CD3+ (n = 6) T cells purified from a total of 13 healthy donors were used as controls. MicroRNA and gene-expression qRT-PCR was carried out using Taqman probes as described by the manufacturer (Applied Biosystems) using 20 ng of microRNA or total RNA per reaction in a Roche LightCycler 480 machine. Triplicate samples were used throughout. Levels of β-2-microglobulin (B2M) and U6 were used as control genes for gene expression and microRNA expression assays, respectively. The mean Ct value of each triplicate was used for analysis by the ΔCt method (ΔCt = mean Ct of control − mean Ct of gene of interest). Expression levels were compared using Mann-Whitney independent t test (GraphPad Prism v.4.0).
To assess the ability of microRNA expression levels to discriminate between SzS and control samples, we used the k-fold cross-validation algorithm based on logistic regression to calculate receiver operator curve (ROC) statistics for these data. Analysis was carried out by 2-class logistic regression modeling with a ridge estimator using the Logistic function in WEKA Version 3.6.1 in the 10-fold cross validation test mode.8
Methylation-specific PCR
Genomic DNA was bi-sulphite treated using the EpiTect kit from QIAGEN. EVL methylation status was measured using a CpG Wiz kit from Millipore and EVL-specific primers as previously described.9 Supplied universally methylated and unmethylated control DNA were used as positive and negative controls. Colorectal cancer cell lines RKO and VACO400, which were previously demonstrated to be methylated and unmethylated, respectively, at the EVL gene, were used as additional controls.9
Luciferase assay
The complete 3′UTR sequence of the EVL gene was amplified from IMAGE clone no. 5222624 by PCR to include XhoI and NotI restriction sites. This was cloned immediately downstream of Renilla luciferase (RL) in the psiCHECK2 vector (Promega) that also encodes for firefly luciferase (FL), which acts as an internal transfection control for experiments. The resulting plasmid (psiCHECK-EVL-3′UTR) was sequence verified. HeLa cells were cotransfected with 2.5 μg of psiCHECK-EVL-3′UTR and 20 nM synthetic RNA oligo (MWG Biotech) encoding either miR-199a* (acaguagucugcacauugguua) or a scrambled version of this sequence (Scramble-miR-199a* [acaguagucugcacauugguua]) with no known homology against any known human microRNA or gene sequence. FL and RL activity was measured 48 hours after transfection in triplicate using a Glomax luminomter (Promega) according to the manufacturer's recommendations. The RL/FL ratio of cells transfected with miR-199a* or Scramble-miR-199a* were compared with cells transfected only with psiCHECK-EVL-3′UTR. Experiments were carried out in triplicate.
Cell transfection, cell proliferation, and apoptosis assays
Jurkat cells were transfected with miR-199a*, Scramble-miR-199a*, or mock transfected (no sequence). SeAx cells were transfected with miRIDIAN miR-342, miR-17-5p, or anti–miR-199a* from Dharmacon, Scramble-miR-199a*, or were mock transfected. All cells were transfected by electroporation using the Amaxa nucleofector machine as described by the manufacturer (Lonza), and transfection efficiency was measured using a green fluorescent protein (GFP)–containing plasmid and by qRT-PCR.
Apoptosis was quantified in triplicate 72 hours after transfection in SeAx cells using the Cell Death Detection ELISA Kit (Roche Applied Science), while cell proliferation was measured using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer's instructions. Experiments were carried out in triplicate.
Results
Most aberrantly expressed microRNAs in SzS are down-regulated
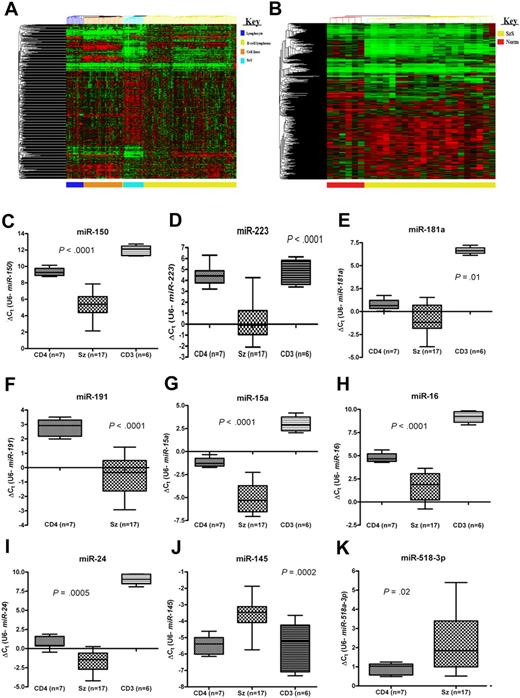
We elucidated the complete (miRBase v.10.1) microRNA profile of CD4+ T cells from 21 patients with SzS and 6 healthy controls. To see how the microRNA profiles of SzS samples related to other hematologic malignancies, we carried out meta-analysis on data generated previously.4,10 Unsupervised cluster analysis of these data revealed that all the clinical lymphoma samples clustered distinctly from cell lines and normal lymphocyte subsets, and that T-cell lymphoma (SzS) samples were distinct from B-cell lymphoma samples (Figure 1A). Using an extended human probe set (miRBase v.10.1; n = 655), unsupervised cluster analysis again placed the SzS samples distinctly from control CD4+ T cells (Figure 1B).
MicroRNAs are aberrantly expressed in SzS. (A) Unsupervised cluster analysis of microRNA expression data (miRBase v. 9.0) for purified lymphocyte subsets (n = 18), B-cell lymphoma samples (n = 98), hematologic cell lines (n = 42), and Sézary Syndrome (SzS) samples (n = 21). (B) Unsupervised cluster analysis of control CD4+ T cells (n = 6) and SzS samples (n = 21) using extended human probe set (miRBase v. 10.1; n = 655). Expression levels of (C) miR-150, (D) miR-223, (E) miR-181a, (F) miR-191, (G) miR-15a, (H) miR-16, (I) miR-24, (J) miR-145, and (K) miR-518a-3p in SzS samples (n = 17) and CD4+ T-cell (n = 7) and CD3+ T-cell (n = 6) controls measured by qRT-PCR. P values relate to SzS versus control CD4+ T cells (Mann-Whitney independent t test). Data shown as box-whisker plots.
MicroRNAs are aberrantly expressed in SzS. (A) Unsupervised cluster analysis of microRNA expression data (miRBase v. 9.0) for purified lymphocyte subsets (n = 18), B-cell lymphoma samples (n = 98), hematologic cell lines (n = 42), and Sézary Syndrome (SzS) samples (n = 21). (B) Unsupervised cluster analysis of control CD4+ T cells (n = 6) and SzS samples (n = 21) using extended human probe set (miRBase v. 10.1; n = 655). Expression levels of (C) miR-150, (D) miR-223, (E) miR-181a, (F) miR-191, (G) miR-15a, (H) miR-16, (I) miR-24, (J) miR-145, and (K) miR-518a-3p in SzS samples (n = 17) and CD4+ T-cell (n = 7) and CD3+ T-cell (n = 6) controls measured by qRT-PCR. P values relate to SzS versus control CD4+ T cells (Mann-Whitney independent t test). Data shown as box-whisker plots.
To identify microRNAs that are aberrantly expressed in SzS, we compared expression levels with controls by ANOVA. This resulted in the identification of 114 microRNAs (adjusted P < .05), only 10 of which were up-regulated in SzS samples (Tables 1–2). To validate the microarray data, 9 of the most up- and down-regulated microRNAs and 6 microRNAs previously associated with malignancy (ie, miR-15a, miR-16, miR-24, miR-17-5p, miR-106a, miR-19a) were measured by qRT-PCR in SzS patient samples (n = 17) and 7 CD4+ and 6 CD3+ T-cell control samples. These data were consistent with the microarray results (Figures 1,Figure 2–3).
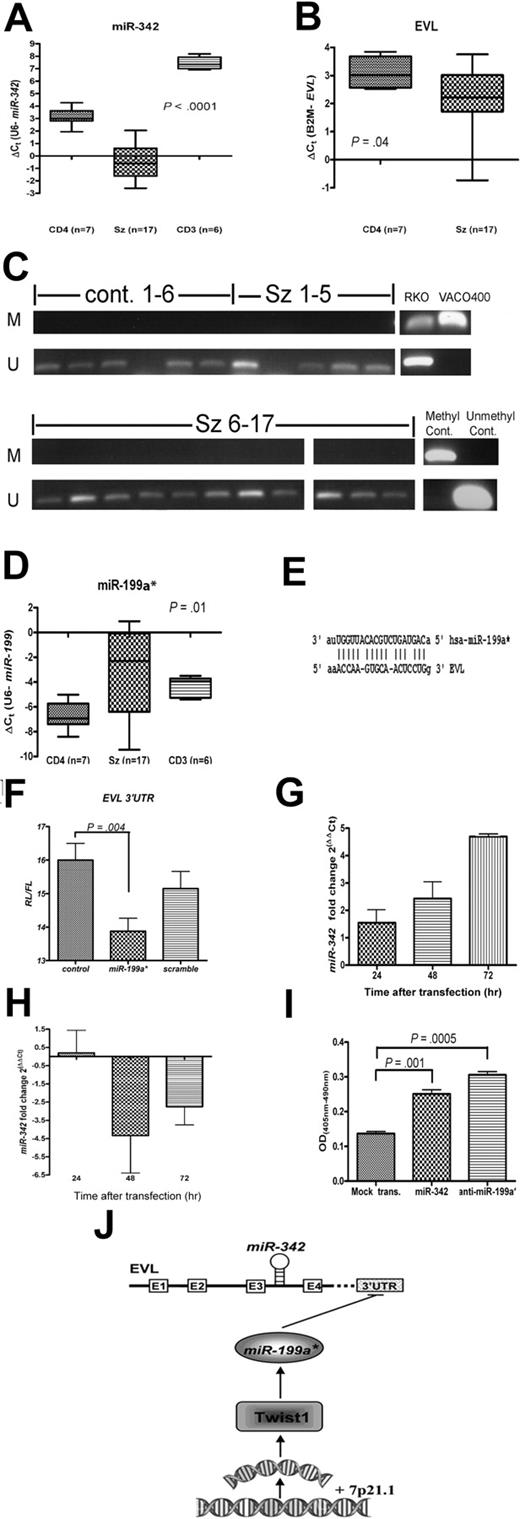
Down-regulation of proapoptotic miR-342 is mediated by miR-199a*, which targets the miR-342-encoding gene EVL. Levels of (A) miR-342 and (B) EVL in SzS samples (n = 17) and CD4+ T-cell (n = 7) and CD3+ T-cell (n = 6) controls measured by qRT-PCR. Data shown as box-whisker plots. (C) CpG-island methylation status of EVL in SzS samples (n = 17) and CD4+ T-cell controls (n = 6) measured by methylation-specific PCR. Universally methylated and unmethylated DNA were used as controls. Colorectal cell lines RKO and VACO400 were methylated and unmethylated, respectively, as previously reported.9 (D) Levels of miR-199a* in SzS samples (n = 17) and CD4+ T-cell (n = 7) and CD3+ T-cell (n = 6) controls measured by qRT-PCR. (E) Predicted binding site for miR-199a* within the 3′UTR sequence of EVL gene. (F) Transfection of miR-199a* in HeLa cell line–suppressed EVL 3′-UTR luciferase reporter activity compared with vector only control or Scramble-miR-199a* sequence. (G) Inhibition of miR-199a* in SeAx cells resulted in increased levels of miR-342 measured by qRT-PCR. Fold change levels shown are relative to Scramble-miR-199a* transfected control (ie, ΔΔCt). (H) Transfection of Jurkat cell line with miR-199a* resulted in decreased levels of miR-342 measured by qRT-PCR. Fold change levels shown are relative to Scramble-miR-199a* transfected control (ie, ΔΔCt). (I) Expression of miR-342 or inhibition of miR-199a* in SeAx cell line resulted in an increase in apoptosis levels compared with mock-transfected control. Values shown are mean values from 3 experiments. (J) Schematic diagram of proposed pathway of miR-342 regulation in SzS.
Down-regulation of proapoptotic miR-342 is mediated by miR-199a*, which targets the miR-342-encoding gene EVL. Levels of (A) miR-342 and (B) EVL in SzS samples (n = 17) and CD4+ T-cell (n = 7) and CD3+ T-cell (n = 6) controls measured by qRT-PCR. Data shown as box-whisker plots. (C) CpG-island methylation status of EVL in SzS samples (n = 17) and CD4+ T-cell controls (n = 6) measured by methylation-specific PCR. Universally methylated and unmethylated DNA were used as controls. Colorectal cell lines RKO and VACO400 were methylated and unmethylated, respectively, as previously reported.9 (D) Levels of miR-199a* in SzS samples (n = 17) and CD4+ T-cell (n = 7) and CD3+ T-cell (n = 6) controls measured by qRT-PCR. (E) Predicted binding site for miR-199a* within the 3′UTR sequence of EVL gene. (F) Transfection of miR-199a* in HeLa cell line–suppressed EVL 3′-UTR luciferase reporter activity compared with vector only control or Scramble-miR-199a* sequence. (G) Inhibition of miR-199a* in SeAx cells resulted in increased levels of miR-342 measured by qRT-PCR. Fold change levels shown are relative to Scramble-miR-199a* transfected control (ie, ΔΔCt). (H) Transfection of Jurkat cell line with miR-199a* resulted in decreased levels of miR-342 measured by qRT-PCR. Fold change levels shown are relative to Scramble-miR-199a* transfected control (ie, ΔΔCt). (I) Expression of miR-342 or inhibition of miR-199a* in SeAx cell line resulted in an increase in apoptosis levels compared with mock-transfected control. Values shown are mean values from 3 experiments. (J) Schematic diagram of proposed pathway of miR-342 regulation in SzS.
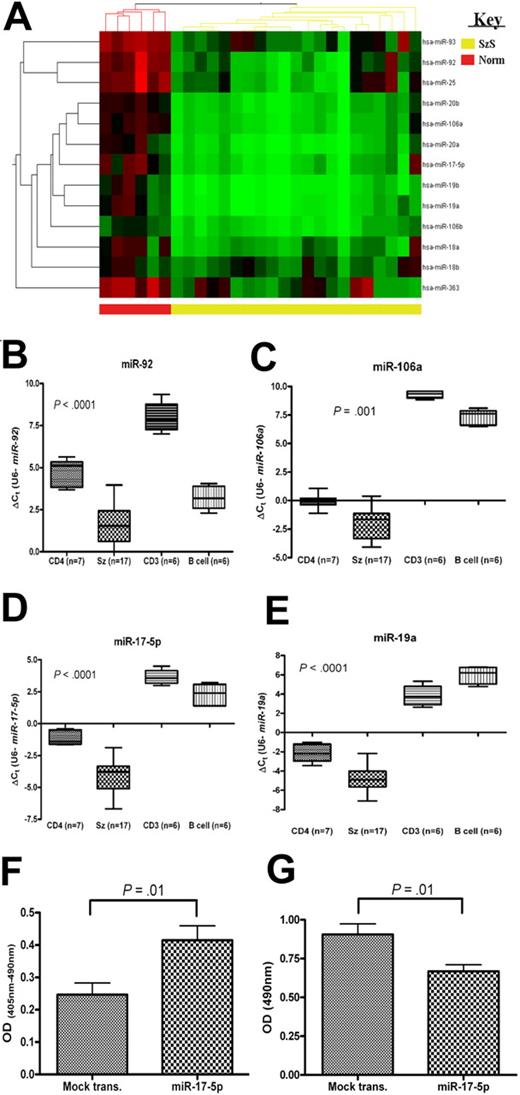
Members of the miR-17-92 (and homologous) clusters are down-regulated and increase apoptosis and decrease cell proliferation in SzS. (A) Heat map depicting levels of members of miR-17-92 and homologous clusters as measured by microarray and levels of (B) miR-92, (C) miR-106a, (D) miR-17-5p, and (E) miR-19a in SzS samples (n = 17), and controls (CD4+ T cells, CD3+ T cells, and B cells), CD4+ T cells (n = 7), CD3+ T cells (n = 6), and B-cell (n = 6) measured by qRT-PCR. P values relate to SzS versus CD4+ (Mann-Whitney independent t test). Data shown as box-whisker plots. Expression of miR-17-5p in a SeAx cell line resulted in an increase in levels of (F) apoptosis and decrease in levels of (G) cell proliferation compared with mock-transfected control. Values shown are mean values from 3 experiments.
Members of the miR-17-92 (and homologous) clusters are down-regulated and increase apoptosis and decrease cell proliferation in SzS. (A) Heat map depicting levels of members of miR-17-92 and homologous clusters as measured by microarray and levels of (B) miR-92, (C) miR-106a, (D) miR-17-5p, and (E) miR-19a in SzS samples (n = 17), and controls (CD4+ T cells, CD3+ T cells, and B cells), CD4+ T cells (n = 7), CD3+ T cells (n = 6), and B-cell (n = 6) measured by qRT-PCR. P values relate to SzS versus CD4+ (Mann-Whitney independent t test). Data shown as box-whisker plots. Expression of miR-17-5p in a SeAx cell line resulted in an increase in levels of (F) apoptosis and decrease in levels of (G) cell proliferation compared with mock-transfected control. Values shown are mean values from 3 experiments.
Many microRNAs are encoded in clusters, and members of these clusters often exhibit the same pattern of expression.11 A higher proportion of SzS-associated microRNAs (70 of 114 [61%]) were encoded in clusters than generally observed (215 of 474 [45%]; source: MiRGen Clusters database12 ). In 23 (74%) of the 31 clusters encoded by SzS-associated microRNAs, all members of the cluster were significantly differentially expressed (Tables 1–2, supplemental Table 3).
MicroRNA expression levels have diagnostic potential for SzS
Using the 10 most discriminatory (based on P value) up- and down-regulated microRNAs (Table 1) for cluster analysis distinguished SzS and control samples (supplemental Figure 1) and correctly predicted diagnosis in 26 of 27 samples (96%) by leave-one-out cross-validation SVM analysis.
To see if microRNA levels measured by qRT-PCR yielded similar results, we analyzed training set data (ie, 17 SzS and 7 CD4+ samples; Figures 1,Figure 2–3) using k-fold cross-validation based on logistic regression analysis. A similar statistical approach was previously used to identify genes with diagnostic potential in SzS.13 The individual expression levels of 4 microRNAs (miR-150, miR-191, miR-15a, and miR-16) correctly predicted diagnosis with 100% accuracy, whereas miR-223 and miR-17-5p were 96% accurate (supplemental Table 4).
To extend these analyses, we measured levels of the 6 microRNAs in an independent validation set consisting of an additional 15 SzS samples, a further 12 healthy controls, and 11 patients with nonerythrodermic MF (patient characteristics are given in supplemental Table 2). Taking the validation set data in isolation, levels of miR-223 correctly discriminated between SzS and control (ie, MF and healthy controls) samples in 34 of 38 cases (90%) with a specificity of 87% and sensitivity of 92% corresponding to an area under the curve (AUC) value of 0.977. Values of the other 5 microRNAs, however, did not discriminate well (supplemental Table 5). Taking all data into account (ie, training and validation set equals 32 SzS samples plus 30 controls), miR-223 levels were 90% accurate with a specificity of 91% and sensitivity of 90% corresponding to an AUC value of 0.938 (supplemental Table 6). Combining data from multiple microRNAs did not improve the performance with this data.
miR-342 expression in SzS is negatively regulated by miR-199a* expression
Because the intronically encoded miR-342 is epigenetically silenced in colorectal cancer due to CpG-island methylation in the promoter region of host EVL gene,9 we wanted to see whether the same mechanism could explain the down-regulation observed in SzS. Both miR-342 and EVL were down-regulated (P < .001 and .04, respectively) in SzS (Figure 2A-B). Using the same EVL-specific primers and colorectal cancer cell line controls as previously described,9 we did not find any evidence for EVL CpG-island promoter hypermethylation in SzS patient samples or control CD4+ T cells (Figure 2C). In addition, we sequenced areas of approximately 1 kbp in patients with SzS representing the promoter regions of both EVL-201/2 and EVL-203 transcripts that included the CpG-island and other identified regulatory sequences,14 but found no mutations (data not shown). In case the intron encoding miR-342 had been deleted or mutated, we also sequenced this region in the SzS samples, but found no mutations or deletions (data not shown).
We next investigated whether any of the SzS-associated microRNAs potentially targeted the EVL gene. Using the miRGen suite of predictive algorithms 12 , we identified miR-199a* (miR-199a-3p), which is up-regulated in SzS (Figure 2D; P = .01), as potentially targeting EVL (Figure 2E). To test this hypothesis, we cloned the 3′UTR sequence of EVL into a luciferase-reporter vector. As can be seen from Figure 2F, transfection with miR-199a* reduced luciferase activity significantly (P = .004), compared with either vector alone or transfection with a scrambled sequence.
To see whether miR-199a* could also directly regulate miR-342 expression in SzS we transfected the SeAx cell line with a miR-199a* inhibitor. This resulted in a greater than 26-fold decrease in the levels of miR-199a* by 72 hours after transfection (data not shown) and an approximately 5-fold increase in endogenous levels of miR-342 (Figure 2G). In contrast, transfection of the SeAx cell line with miR-342 did not significantly affect miR-199a* levels (data not shown). The converse experiment was also carried out by transfecting the Jurkat T-cell line that has high endogenous levels of miR-3424 with miR-199a* which resulted in an approximately 4-fold decrease in miR-342 levels by 48 hours after transfection (Figure 2H).
Reconstitution of down-regulated microRNAs miR-342 or miR-17-5p in Sézary cells inhibits apoptosis
Transfection with either miR-342 or miR-199a* inhibitor resulted in a significant increase in levels of apoptosis (Figure 2I; P ≤ .01), but had no effect on cell proliferation levels (data not shown). In contrast, transfection of SeAx cells with miR-17-5p caused both an increase in apoptosis levels (Figure 3F; P = .01) and also a decrease in levels of cell proliferation (Figure 3G; P = .01).
Discussion
A total of 114 microRNAs were identified as SzS-associated (adjusted P < .05), of which only 10 were up-regulated in SzS samples. A general decrease in microRNA expression in tumors compared with counterpart normal tissue is consistent with previous findings,15,16 although we10 and others17 found the opposite to be the case for B-cell lymphomas.
Using the same array as in this study, we previously identified 60 microRNAs (40 up-regulated and 20 down-regulated) associated (P < .05) with B-cell lymphoma (either DLBCL or FL).10 Surprisingly, a comparison of these microRNAs with SzS-associated microRNAs reveal a great deal of similarity that suggests the presence of a common lymphoma microRNA signature (Tables 1–2). A total of 7 of the 10 microRNAs that were up-regulated in SzS were also significantly up-regulated in B-cell lymphoma, while 18 of the 20 microRNAs that were down-regulated in B-cell lymphoma were also significantly down-regulated in SzS (Tables 1–2).
The cause of aberrant microRNA expression in SzS, in common with most other cancers, is not readily apparent. Recently we detailed a number of recurrent chromosomal aberrations associated with SzS, albeit in a different cohort of patients.18 These abnormalities were generally consistent with microRNA expression in this cohort of patients with SzS (Tables 1–2). For example miR-152, which is up-regulated, is encoded by region of gain (17q21.32) in 70% of SzS cases, whereas the 13q14 locus, frequently deleted in SzS, encodes for miR-15a and miR-16, both of which were down-regulated (P < .001; Figure 1G-H). Similarly, miR-107, miR-146b, and miR-22, which were down-regulated, are encoded by common regions of loss or deletion in SzS (10q23.31, 10q24.32, and 17p13.3, respectively). However, similar to other studies,19 this correlation did not always hold true, as some regions of recurrent gain (ie, 4p16.1, 8q24.1, and 17q25) encode for microRNAs that were down-regulated (miR-95, miR-30, and miR-338, respectively). This suggests that these microRNAs, at least, are regulated by another mechanism. It was recently demonstrated that MYC, which is aberrantly active in SzS,18 down-regulates a number of microRNAs, including miR-30.20 Indeed, all of the microRNAs shown to be repressed by MYC (ie, miR-22, miR-26a, miR-29c, miR-30, miR-146a, and miR-150) were also down-regulated in SzS samples (Table 2).
To investigate whether SzS-associated microRNAs correlated with gene expression patterns, we interrogated the miRGen database 12 for microRNAs predicted to target the 69 genes that we had previously identified as being up-regulated in a separate cohort of patients with SzS.21 Nearly all (97 of 104 [93%]) of down-regulated SzS-associated microRNAs were predicted to target one or more of these genes (median number of targets = 5; Table 2). Furthermore, a comparison between this set of microRNAs with 10 000 randomly generated sets of 104 microRNAs showed a highly significant enrichment for SzS-associated target genes (P < .001). This suggests that microRNAs play an important role in the regulation of gene expression in SzS, although it remains possible that the distinct cohorts of patients with SzS used for this analysis are not directly comparable and further analysis should be carried out.
Early detection and treatment are directly correlated with favorable outcome for SzS, but diagnosis is frequently difficult because cutaneous histology is often not diagnostic in erythrodermic skin disease and atypical T cells can be detected in the peripheral blood of erythrodermic patients with inflammatory conditions.22 To test the diagnostic ability of the SzS-associated microRNAs, we used the top 10 most discriminatory up- and down-regulated microRNAs in a SVM class-prediction model. This resulted in the correct assignment of samples as either SzS or control for 26 of 27 cases (96%), and expression levels of just 3 of these microRNAs (miR-17-5p, miR-150, and miR-15a or miR-16) were sufficient to correctly predict all samples. Although microarray data are informative, it is not readily available in a clinical setting. Therefore, we analyzed microRNA levels measured by qRT-PCR, a technique available to most clinical diagnostic laboratories, using the k-fold cross-validation predictive model. Consistent with the microarray data, individual expression levels of miR-150, miR-15a, miR-16, or miR-191 correctly predicted all samples in this cohort. To validate these findings and include a more clinically relevant control group (ie, nonerythrodermic MF), we expanded these analyses to an independent validation set. This time, only miR-223 levels were found to have useful predictive capacity (90% accuracy, 87% specificity, and 96% sensitivity) that was consistent (90% accuracy, 91% specificity, and 90% sensitivity) when both training and validation sets were combined (ie, 32 SzS and 30 control samples). Very similar results were obtained using a SVM predictive algorithm instead (SMO function in Weka; data not shown). The discrepancy between the 2 cohorts is largely a result of the inclusion of MF samples, which gave similar microRNA expression values to the SzS samples (supplemental Figure 2), possibly due to disruption of the peripheral T-cell repertoire that is often associated with nonerythodermic MF.23
Recently, expression levels of both DNM3 and CDO1 were found to be 100% specific (91 and 82% sensitive, respectively), although this study lacked controls with confounding diagnoses and small numbers were used (11 SzS, 12 healthy controls, and 10 patients with inflammatory disorders).13 Previously, a 5-gene signature was identified that could discriminate between SzS and control samples with 90% accuracy.24 However, this model was not reproducible in the hands of Booken et al,13 and the higher costs and time associated with measuring 5 genes suggest that a single microRNA assay may prove a more useful classifier for SzS diagnosis.
The most discriminatory microRNA identified by ANOVA analysis of the microarray data were miR-342 (adjusted P < .001). This microRNA is intronically encoded within the EVL gene. Both miR-342 and EVL were found to be down-regulated in SzS. It had been shown that the miR-342/EVL locus is epigenetically silenced via CpG-island methylation in colorectal cancer.9 In contrast, we found no evidence of similar methylation in SzS patient samples. A possible explanation for this discrepancy is that unlike colorectal cancer, which express the EVL-203 transcript containing the CpG island in proximity (∼ 30 bp) to its start codon,9 patients with SzS express a much shorter transcript (EVL-201/2; data not shown), which initiates approximately 100 kbp downstream from this regulatory element.
One intriguing possibility to explain down-regulation of miR-342 in SzS was that the host gene, EVL, is itself a target for microRNA regulation. Of particular interest among the microRNAs predicted to target EVL was miR-199a*, which is up-regulated in SzS. We tested this hypothesis by luciferase assay and demonstrated an inverse relationship between levels of miR-199a* and miR-342 in SzS as inhibition of the former resulted in an increase in endogenous levels of miR-342, while expression of miR-199a* in Jurkat cells that naturally express high levels of miR-342 reduced levels of miR-342.
Approximately 40% of microRNAs are encoded in intronic regions of encoding genes.25 Although generally believed to be passively coexpressed with their host mRNA, recent data suggest the regulation of intronic microRNAs is more complex than previously thought.26 Our data suggest a novel and elegantly simple model for the regulation of intronically encoded microRNAs via modulation of expression levels of microRNAs that target the host mRNA. It should be noted, however, that as miR-342 is spliced out of the EVL transcript before being exported to the cytoplasm, miR-199a* must function in the nucleus. There is now compelling evidence that many mature microRNAs are reimported into the nucleus and once there can regulate transcription.27-30 It has recently been demonstrated that miR-199a* is present at higher levels in the nucleus (and nucleolus) than in the cytoplasm of cells.31 This is the first evidence for such a regulatory mechanism operating in biologic systems and provides a new level of complexity to the poorly understood area of microRNA regulation.
Interestingly, miR-199a* is also intronically encoded within the DNM3 gene, which is itself specifically expressed in SzS.13 Furthermore, miR-199a* is directly up-regulated by TWIST1 binding,32,33 which is up-regulated in SzS21 due to frequent gain of the encoding region.18 These data suggest a model for deregulation of miR-342 depicted in Figure 2J.
To investigate the biologic relevance of reduced miR-342 levels in SzS, we reconstituted this microRNA in the SeAx cell line. This resulted in a significant increase in apoptosis levels (P = .001), but did not significantly affect cell proliferation levels as had been reported in colorectal cancer cell lines.9 Significantly, the same effect was observed when miR-199a* expression was suppressed in this model. The presence of distinct yet analogous mechanisms to suppress miR-342 coupled with a common antiapoptotic phenotype suggests that miR-342 could play a tumor-suppressor role in these 2 cancers.
To investigate how miR-342 down-regulation might inhibit apoptosis in SzS, we used the miRGen database to identify putative apoptosis-associated target genes (supplemental Table 7). Of particular interest was TNFSF11 (also known as RANKL or TRANCE; supplemental Figure 3A), an antiapoptotic molecule that was previously identified as being up-regulated in SzS,13,21 that was also up-regulated in our cohort of patients with SzS (P < .001; supplemental Figure 3B). We found that levels of TNFSF11 decreased in SzS cells transfected with either miR-342 or anti–miR-199a* compared with nontransfected controls (supplemental Figure 3C). TNFSF11 is highly expressed in peripheral activated and memory but not resting peripheral T cells.34 This pattern is consistent with our previous observation that miR-342 is more highly expressed in resting than memory T cells.4 It has been suggested that TNFSF11 expression can promote T-cell survival by interacting with dendritic cells, leading to induction of IL-15.34 Interestingly, IL-15 has been shown to protect SzS cells from apoptotic agents.35 Although these data are preliminary, it suggests that miR-342 down-regulation in SzS mediates its antiapoptotic affect via up-regulation of TNFSF11.
The miR-17-92 cluster has been proposed to act as both tumor-suppressor or oncogene depending upon the cellular context, although until now there has been a lack of in vivo evidence to support the former role.36-38 Members of this cluster are widely up-regulated in B-cell lymphomas and solid tumors.4,10,39 However, we found that all 13 microRNAs encoded by this cluster and homologous clusters (miR-106a-363 and miR-106b-25) were down-regulated in SzS samples by microarray analysis (average fold change = 6.1; range, 3.64-11.82). These data were validated by qRT-PCR for miR-106a, miR-19a, miR-17-5p, and miR-92 (Figure 3; P ≤ .001). Although the 13q31 locus is deleted in some solid tumors,19 and it has been reported that miR-17-5p is down-regulated in some breast cancer cell lines,38 as far as we are aware this is the first report to describe in vivo down-regulation of these microRNAs in malignancy. This finding is particularly intriguing because the miR-17-92 cluster, in B-cell lymphomas at least, is up-regulated by MYC-binding,40 which in turn is aberrantly active in SzS.18 To explore the possibility that MYC-binding sites40 were mutated in patients with SzS, we sequenced this and surrounding regions but found no mutations (data not shown). Therefore the causal mechanism for down-regulation of these microRNAs in SzS remains to be determined and is an area we are actively pursuing. Similar to previous reports in breast cancer cell lines,38 ectopic expression of miR-17-5p in SzS cells resulted in a decrease in cell proliferation levels coupled with an increase in levels of apoptosis. Whether or not this finding can be extended to other T-cell lymphomas, or is an anomaly of SzS, remains to be discovered; however, it is clearly an area of research that warrants further investigation
Cumulatively, these data strongly suggest that microRNAs are important in the pathogenesis of SzS and provides exciting new possibilities for the diagnosis and treatment of this disease.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by grants from The Leukemia Research Fund (C.H.L., E.B., J.C., J.B., and J.S.W.), the Julian Starmer-Smith Memorial Fund (C.H.L.), and the Fondation René Touraine (M.S.v.K.). Support from Guys and St Thomas' National Health Service (NHS) Foundation Trust (T.J.M.) is gratefully acknowledged. The authors acknowledge financial support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Center award to Oxford Radcliffe NHS Trust in partnership with the University of Oxford, and Guy's and St Thomas' NHS Foundation Trust in partnership with Kings College London and Kings College Hospital NHS Foundation Trust.
National Institutes of Health
Authorship
Contribution: E.B. designed and performed research, analyzed data, and wrote the paper; T.M., M.S.v.K., I.T., M.H.V., S.J.W., and C.P.T. designed and performed research and contributed patient samples; S.T. analyzed data; N.J.S. contributed vital new reagents and analytical tools; H.M.D., J.C., and D.T. performed research; J.B., J.S.W., F.P., and C.S.R.H. designed research and analyzed data; and C.H.L. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Charles H. Lawrie, Nuffield Department of Clinical Laboratory Sciences, University of Oxford, Level 4, John Radcliffe Hospital, Oxford, OX3 9DU, United Kingdom; e-mail: charles.lawrie@ndcls.ox.ac.uk.