Abstract
Recently, Dawson et al identified a previously unrecognized nuclear role of JAK2 in the phosphorylation of histone H3 in hematopoietic cell lines. We searched nuclear JAK2 in total bone marrow (BM) cells and in 4 sorted BM cell populations (CD34+, CD15+, CD41+, and CD71+) of 10 myeloproliferative neoplasia (MPN) patients with JAK2V617F mutation and 5 patients with wild-type JAK2 MPN. Confocal immunofluorescent images and Western blot analyses of nuclear and cytoplasmic fractions found nuclear JAK2 in CD34+ cells of 10 of 10 JAK2-mutated patients but not in patients with wild-type JAK2. JAK2 was predominantly in the cytoplasmic fraction of differentiated granulocytic, megakaryocytic, or erythroid cells obtained from all patients. JAK2V617F up-regulates LMO2 in K562 and in JAK2V617F-positive CD34+ cells. The selective JAK2 inhibitor AG490 normalizes the LMO2 levels in V617F-positive K562 and restores the cyto-plasmic localization of JAK2.
Introduction
The discovery of an acquired somatic mutation in the JAK2 gene resulting in a Val-to-Phe substitution at position 617 (JAK2V617F) has substantially modified our molecular knowledge of myeloproliferative neoplasia (MPN).1-7 Recently, Dawson et al8 identified in hematopoietic cell lines a novel nuclear role of JAK2 in the phosphorylation of Tyr 41 of histone H3, leading to chromatin displacement of heterochromatin protein 1α (HP1α) with a consequent reduction of the potential tumor suppressive functions of (HP1α). It is proposed that these changes result in erratic mitotic recombination and transcription deregulation of several JAK2-regulated genes including LMO2. In this work we find that in contrast to cells with normal JAK2 in which the protein is detected predominantly in the cytoplasm, JAK2 is mostly nuclear in V617F-positive CD34+ cells. However, this nuclear localization is no longer observed in V617F-positive differentiated cells. After expressing JAK2V617F in K562 cells, we observe a similar preferential accumulation of JAK2 in the nucleus in contrast to untransfected and wild-type (wt) JAK2-expressing cells in which the protein is found in the cytoplasm. The mutated-JAK2 nuclear translocation is mainly reverted by the addiction of the JAK2 inhibitor AG490.
Methods
Cell cultures
K562 were grown as previously described.8 Transfection was performed by AMAXA electroporation, and stable cell lines were obtained by puromycin selection (3 μg/mL). Leptomycin B was added to a final concentration of 1μM, and the cells were harvested after 24 hours of incubation. K562 were differentiated with 10nM phorbol 12-myristate 13-acetate (PMA).
Plasmids
pMSCV-puro-JAK2 and pMSCV-puro-JAK2V617F were kindly provided by Dr J. Cools (Center for Human Genetics, KU Leuven, Belgium).
Confocal immunofluorescence microscopy
The BM fractions of CD34+, CD15+, CD41+, CD71+ cells and the K562 cells were prepared as previously described.8 Confocal laser images were captured with a laser scanning LSM 510 META microscope (Zeiss) equipped with a 403 oil-immersion lens.
Patients
BM aspirates and peripheral blood (PB) samples were obtained from 15 patients newly diagnosed with MPN according to the WHO criteria. Informed consent was obtained in accordance with the Declaration of Helsinki, and samples were obtained with approval from the Italian ethics committee.
Cell sorting
BM or PB mononucleated cells were incubated with the antibodies CD15-allophycocyanin, CD41–fluorescein isothiocyanate, CD71–peridinin chlorophyll protein complex, and CD34-phycoerythrin (BD Biosciences), analyzed, and sorted with a fluorescence-activated cell sorter FACSAria (BD Biosciences).
ASO-PCR and RT-PCR
Cell fractionation and immunoblotting
Cytoplasmic and nuclear fractions were prepared using nuclei isolation KIT (Sigma-Aldrich). Equal amounts of cytoplasmic or nuclear fractions or total cell extracts were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with relevant antibodies.
JAK2 inhibitor and antibodies
AG490 was provided by Invivogen. Anti-JAK2 antibody (D2E12 no. 3230; Cell Signaling Technology), anti–b-tubulin (T5201; Sigma-Aldrich), anti–laminin A (Sigma-Aldrich), and Alex Fluor 488–conjugated IgG (Invitrogen) were used at the stated dilutions.
Apoptotic rate
K562 cells were stained with propidium iodide and annexin V before and after 3 hours' incubation with AG490. Cell-cycle parameters were assessed by FACS analysis using FACSCanto II flow cytometer (Becton Dickinson).
Statistical analysis
Student t test was used to evaluate individual differences between means. P ≤ .05 was considered significant.
Results and discussion
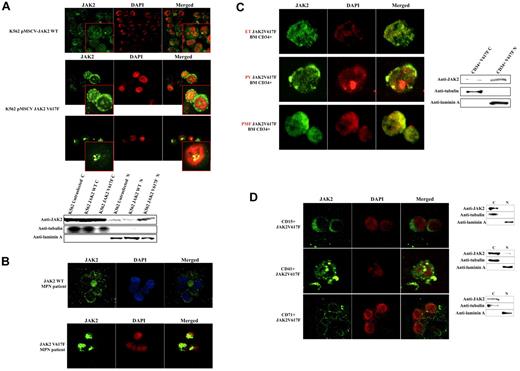
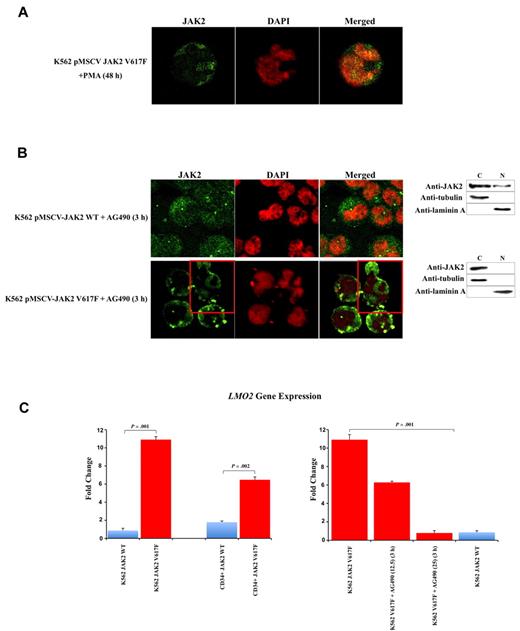
To determine whether the V617F mutation affects the sub-cellular localization of JAK2, we used confocal immunofluorescence (CIF) microscopy to analyze K562 cells stably transfected with pMSCV-puroJAK2V617F or pMSCV-puroJAK2. The results (Figure 1A) confirm the nuclear and cytoplasmic localization of JAK2 in K562 as reported by Dawson et al.8 However, we consistently observed a much stronger nuclear signal in the cells expressing JAK2V617F than in those carrying wt JAK2, suggesting that the mutation leads to a nuclear accumulation of JAK2V617F. By CIF, this accumulation can be seen as a diffused nuclear pattern (Figure 1A top 3 rows, large panels) or as nuclear spots (Figure 1A top 3 rows, insets). This altered subcellular distribution was not affected by the addition of the nuclear export inhibitor leptomycin B (data not shown) and was confirmed by Western blot (WB) analysis of K562 cells (Figure 1A bottom gel lanes). To determine whether there is a preferential nuclear translocation of JAK2V617F in vivo, we analyzed by CIF microscopy the BM cells of 10 JAK2V617F-positive MPN patients (essential thrombocythemia [ET], n = 4, polycythemia vera [PV], n = 3, primary myelofibrosis [PMF], n = 3, allele burden median: 56%, 70%, 72% respectively) and 5 MPN patients with wt JAK2 (PMF n = 2, ET n = 3). We did not observe a significant signal in the nucleus of cells with wt JAK2 (Figure 1B). In contrast, we found a strong nuclear signal in 3% to 5% of total BM mononucleated cells in 10 of 10 JAK2V617F-positive patients, suggesting that, unlike the wt JAK2, JAK2V617F has a predominantly nuclear homing. To identify the phenotype of these cells, we used fluorescence-activated cell sorting (FACS) to isolate CD34+, CD15+, CD41+, and CD71+ fractions from the BM of 3 JAK2V617F-positive MPN patients (1 ET, 1 PV, 1 early PMF). We found nuclear JAK2 only in the fraction containing the CD34+ positive cells (Figure 1C left panels). It should be noted that in these patients the CD34+ cells correspond to approximately 3% to 5% of total BM mononucleated cells. The nuclear localization of JAK2V617F was confirmed by WB analysis (Figure 1C right gel lanes). However, no predominant nuclear signal was detected in differentiated granulocytic, megakaryocytic, and erythroid cells obtained from the patients (n = 15; Figure 1D). Similar results were obtained with PB cells (data not shown). The relocation of the mutated JAK2 to the cytoplasm was confirmed in K562 cells after their differentiation with PMA (Figure 2A). However, the relocation was not complete as observed for the primary BM cells, and nuclear JAK2 was still observed. We believe that this is due to the nature of K562 cells, which are BCR-ABL–positive CML cells, and to the difficulty of obtaining their terminal differentiation in vitro. To determine whether an alteration of JAK2 activity could interfere with the nuclear localization of JAK2, we incubated K562 cells expressing JAK2V617F or JAK2 with the selective JAK2 inhibitor AG490.9,10 After 3 hours of incubation at the IC50 dose of 25μM, CIF images and WB showed a relocalization of JAK2V617F to the cytoplasm in the majority of K562 cells (Figure 2B). Analyses with annexin V and propidium iodide did not reveal any significant change in the apoptotic rate of V617F-positive and wt K562 cells. By QRT-PCR we show that the V617F mutation strongly up-regulates the expression of LMO2 in K562 and in CD34+ cells (Figure 2C left panels).
V617F mutation favors nuclear translocation of JAK2 in K562 and early CD34+ progenitors isolated from BM of MPN patients. This translocation is not observed in differentiated cells. (A) CIF microscopy images of K562 cells stably transfected with pMSCV-JAK2 (top-row images) or pMSCV-JAK2V617F (second- and third-row images) Western blotting of cytoplasmic (C) and nuclear (N) fractions (bottom) confirm that JAK2V617F is more abundant than JAK2 in K562 nuclei. (B) CIF images of BM cells from an MPN patient with wt JAK2 (top row) and an MPN patient with JAK2V617F (bottom row) confirm a nuclear increase of the mutated protein. (C) CIF demonstrates a predominantly nuclear accumulation of JAK2V617F in CD34+ cells isolated from BM of 1 essential thrombocythemia (ET), 1 polycythemia vera (PV), and 1 early primary myelofibrosis (PMF) (left panels). Western blotting of cytoplasmic (C) and nuclear (N) extracts (right) confirm the data. (D) CIF images of CD15+, CD41+, CD71+ cells isolated from a JAK2V617F-positive MPN patient (PV, JAK2 allele burden 71%) and Western blotting of cytoplasmic (C) and nuclear (N) extracts (right). DAPI, 4,6-diamidino-2-phenylindole; Anti-JAK2 (Cell Signaling Technology) monoclonal antibody; anti-tubulin and anti-laminin A (Sigma-Aldrich) monoclonal antibodies.
V617F mutation favors nuclear translocation of JAK2 in K562 and early CD34+ progenitors isolated from BM of MPN patients. This translocation is not observed in differentiated cells. (A) CIF microscopy images of K562 cells stably transfected with pMSCV-JAK2 (top-row images) or pMSCV-JAK2V617F (second- and third-row images) Western blotting of cytoplasmic (C) and nuclear (N) fractions (bottom) confirm that JAK2V617F is more abundant than JAK2 in K562 nuclei. (B) CIF images of BM cells from an MPN patient with wt JAK2 (top row) and an MPN patient with JAK2V617F (bottom row) confirm a nuclear increase of the mutated protein. (C) CIF demonstrates a predominantly nuclear accumulation of JAK2V617F in CD34+ cells isolated from BM of 1 essential thrombocythemia (ET), 1 polycythemia vera (PV), and 1 early primary myelofibrosis (PMF) (left panels). Western blotting of cytoplasmic (C) and nuclear (N) extracts (right) confirm the data. (D) CIF images of CD15+, CD41+, CD71+ cells isolated from a JAK2V617F-positive MPN patient (PV, JAK2 allele burden 71%) and Western blotting of cytoplasmic (C) and nuclear (N) extracts (right). DAPI, 4,6-diamidino-2-phenylindole; Anti-JAK2 (Cell Signaling Technology) monoclonal antibody; anti-tubulin and anti-laminin A (Sigma-Aldrich) monoclonal antibodies.
V617F mutation causes up-regulation of LMO2. The JAK2 inhibitor AG490 replaces JAK2 into cytoplasm and restores LMO2 levels. (A) CIF images show the redistribution of JAK2 and the replacement in the cytoplasm in V617F expressing K562 after PMA differentiation. (B) CIF images (images) and Western blotting of cytoplasmic (C) and nuclear (N) extracts (right) show the redistribution of JAK2 and the replacement in the cytoplasm in the majority of V617F expressing K562 (bottom right) but not in wt cells (top right) after AG490 incubation. (C) Quantitative RT-PCR reveals that V617F mutation strongly up-regulates LMO2 expression in K562 and in CD34+ cells (left graphs). The addiction of AG490 progressively and completely restore LMO2 levels in V617F expressing K562 (right graphs) after 3 hours' incubation. Error bars represent standard deviation. DAPI, 4,6-diamidino-2-phenylindole; Anti-JAK2 (Cell Signaling Technology) monoclonal antibody; anti-tubulin and anti-laminin A (Sigma-Aldrich) monoclonal antibodies.
V617F mutation causes up-regulation of LMO2. The JAK2 inhibitor AG490 replaces JAK2 into cytoplasm and restores LMO2 levels. (A) CIF images show the redistribution of JAK2 and the replacement in the cytoplasm in V617F expressing K562 after PMA differentiation. (B) CIF images (images) and Western blotting of cytoplasmic (C) and nuclear (N) extracts (right) show the redistribution of JAK2 and the replacement in the cytoplasm in the majority of V617F expressing K562 (bottom right) but not in wt cells (top right) after AG490 incubation. (C) Quantitative RT-PCR reveals that V617F mutation strongly up-regulates LMO2 expression in K562 and in CD34+ cells (left graphs). The addiction of AG490 progressively and completely restore LMO2 levels in V617F expressing K562 (right graphs) after 3 hours' incubation. Error bars represent standard deviation. DAPI, 4,6-diamidino-2-phenylindole; Anti-JAK2 (Cell Signaling Technology) monoclonal antibody; anti-tubulin and anti-laminin A (Sigma-Aldrich) monoclonal antibodies.
The link between LMO2 expression and JAK2 inhibition has been reported previously.11-13 In our assay, the addition of AG490 progressively and completely restore LMO2 levels in V617F expressing K562 (Figure 2C right panels).
Our data corroborate recently published results of a nuclear localization of JAK2 in hematopoietic cells, and they also extend these findings by showing that in all subtypes of MPN patients, JAK2V617F accumulates in the nucleus of progenitor CD34+ cells whereas it remains mostly in the cytoplasm of their differentiated progeny. The chromatin alterations due to the preferential accumulation of JAK2V617F in the nucleus correlates with a significant increase in LMO2 expression in cell lines and in sorted CD34+ cells. The selective JAK2 inhibitor AG490 is able to revert nuclear JAK2 and normalize LMO2 levels in vitro, suggesting how the block in JAK2 nuclear translocation could be a new treatment strategy for JAK2 mutated patients. The signals that are required for the translocation of normal and mutated JAK2 to the nucleus remain unknown. It is possible that the activation of the kinase by phosphorylation could be the first one of several modifications that control nuclear translocation, similarly to what happens to the STAT proteins. If this is true, then it is also possible that as the cell undergoes differentiaition these modifications are shut off, leaving mutated JAK2 predominantly in the cytoplasm.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Anthony R. Green (Cambridge Institute for Medical Research and Department of Haematology, University of Cambridge, Cambridge, United Kingdom) for his kind contribution and suggestions.
This work is dedicated to the memory of Prof Bruno Rotoli.
Authorship
Contribution: C.R.R. designed the study, supervised the work, and wrote the manuscript; P.R. and A.A. performed CIF microscopy and QRT-PCR and analyzed the data; M.G. performed the cell sorting; N.E. performed Western blot assays; F.F. captured CIF images; V.M. provided patient samples; V.S. analyzed the data; G.N. wrote the manuscript; and F.P. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Ciro Roberto Rinaldi, Haematology Department, Pilgrim Hospital, United Lincolnshire Hospital NHS Trust, Sibsey Rd - PE21 9QS, Boston, Lincolnshire, United Kingdom; e-mail: Ciro.Rinaldi@ulh.nhs.uk.