Abstract
The neutrophil nicotinamide adenine dinucleotide phosphate-oxidase is a multisubunit enzyme (comprising gp91phox, p22phox, p67phox, p40phox, p47phox, and Rac) that plays a vital role in microbial killing. The recent discovery of a chronic granulomatous disease patient who expresses a mutant p40phox subunit, together with the development of mouse models of p40phox function, indicate phosphatidylinositol 3-phosphate binding to the PX domain of p40phox is an important signal for oxidase activation. However, the presence of other conserved residues and domains in p40phox suggest further regulatory roles for this protein. To test this, we introduced wild-type and mutated versions of p40phox into fully differentiated mouse neutrophils by retroviral transduction of p40phox−/− bone marrow progenitors and repopulation of the bone marrow compartment in radiation chimaeras. Phosphorylation of p40phox on threonine 154, but not serine 315, was required for full oxidase activation in response to formylated bacterial peptide fMLP, serum-opsonized S aureus, and immunoglobulin-opsonized sheep red blood cells. A functional SH3 domain was not required for oxidase activation, and deletion of the entire domain resulted in enhanced oxidase responses. Phosphorylation of threonine 154 in response to S aureus was mediated by protein kinase Cδ and was required for full translocation of p47phox to phagosomes. These results define an important new element in the physiological activation of the oxidase.
Introduction
The neutrophil nicotinamide adenine dinucleotide phosphate [NADPH] oxidase plays an important role in the mechanisms that neutrophils use to kill pathogens.1,2 In the resting state of the neutrophil, the components of the NADPH oxidase are segregated between membrane-bound cytochrome b558, comprising gp91phox and p22phox, and 4 cytosolic components, p47phox, p67phox, p40phox, and guanosine diphosphate (GDP)-bound Rac1/2.3-6 Upon neutrophil stimulation by chemoattractants, cytokines, or pathogens, the cytosolic components translocate to the membrane and interact with the cytochrome, with each other, and with phospholipids, resulting in formation of an active NADPH oxidase complex.7-10 This active complex catalyses the transfer of electrons across the membrane, from NADPH to O2, to generate the superoxide anion, which can then be converted to other reactive oxygen species (ROS) that together form one of the key weapons that are used to kill pathogens. The importance of the NADPH oxidase is highlighted by chronic granulomatous disease (CGD), which is characterized by genetic deficiency in one of the oxidase components, leading to recurrent bacterial and fungal infections.11
The key steps underlying oxidase activation include the guanosine-5′-triphosphate loading of Rac, allowing its translocation to the membrane and its interaction with the tetratricopeptide repeat domain of p67phox; the interaction of p67phox with gp91phox; and the phosphorylation of p47phox.8,12,13 Phosphorylation of p47phox results in the relief of an auto-inhibitory conformation and exposure of the tandem SH3 domain to allow its interaction with p22phox. The C-terminus of p47phox also binds to the C-terminal SH3 domain of p67phox, and thus p47phox phosphorylation is thought to play a major role in orchestrating assembly and subsequent activation of the oxidase. At the membrane, the phox homology (PX) domain of p47phox interacts with phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2) and phosphatidic acid,14,15 which may also contribute to efficient complex assembly and activation,16 though the physiological importance of these interactions have yet to be defined.
The p40phox subunit is thought to be an obligate binding partner for p67phox, but its role in oxidase activation has been controversial.17,18 However, the recent development of mouse models that either lack p40phox (p40phox−/−)19 or carry a point mutation in the p40phox PX domain (p40phoxR58A/−),20 together with the recent discovery of a CGD patient expressing a PX domain mutant of p40phox (p40phoxR105Q/−),21 has now firmly established the importance of this subunit in oxidase activation by physiological stimuli. Further, this work has shown that PtdIns3P binding to the PX domain of p40phox plays an important role in oxidase activation to several agonists, in particular during the assembly of the oxidase on phagosomes,20,21 though the molecular mechanism through which this interaction regulates oxidase activity is still uncertain. Moreover, oxidase defects in p40phoxR58A/−neutrophils are not as severe as those in p40phox−/− neutrophils, in particular the extracellular ROS responses to the formylated bacterial peptide (fMLP) are unaffected in the former but severely reduced in the latter.19 Unfortunately, loss of p40phox also reduces expression of its binding partner, the p67phox subunit (by approx 55%), and, therefore, since p67phox is an essential component of the oxidase, it is difficult to assess the extent to which the p40phox−/− phenotype is caused by non-PX domain functions of p40phox.19
p40phox also contains a conserved SH3 domain and 2 conserved phosphorylation sites, threonine (T) 154 and serine (S) 315.22 Good evidence has been provided that phosphorylation of p40phox on both of these sites occurs during stimulation of neutrophils or neutrophil-like cells by phorbol 12-myristate 13-acetate (PMA) or fMLP.22-24 Further, both of these sites can be phosphorylated in vitro by protein kinase C (PKC), an enzyme known to be important in the phosphorylation of p47phox and in the activation of the oxidase.22 In a cell-free system, using recombinant, phosphorylated p47phox as the activator of the NADPH oxidase, p40phox phosphorylated on T154 was shown to inhibit oxidase activity,25 but the in vivo functions of either T154 or S315 phosphorylation remain to be defined.
The physiological function of the SH3 domain of p40phox is also unclear. A structure has been resolved for the SH3 domain of p40phox in complex with the polyproline (PP) motif of p47phox.26 Among the key residues in the SH3 domain of p40phox that interact with the PP motif of p47phox is tryptophan (W) 207, a residue that is evolutionarily highly conserved and that interacts with the backbone of arginine (R) 368 in p47phox through hydrophobic interactions. p40phox-W207 to R mutation, which is predicted to disrupt the interaction with its PP binding target in vivo, fails to reconstitute full NADPH oxidase activity in permeabilized “core” neutrophils in response to PMA, as well as in the COSphox system in response to immunoglobulin-opsonized sheep red blood cells (IgG-SRBCs).27,28 Further, using a cell-free system for oxidase activation, the SH3 domain of p40phox was shown to interact with the PP motif of p22phox and through this interaction was able to substitute for p47phox.29 The W207R mutation was also found to disrupt this interaction. However, the in vivo binding target(s) of the SH3 domain of p40phox, as well as its true physiological function, still remain to be established.
A major obstacle to fully understanding p40phox function and indeed, more generally, the signaling mechanisms that underpin NADPH oxidase activation is the limited half-life of neutrophils ex vivo and the difficulty in introducing defined proteins in a robust and controlled manner. To try to overcome this obstacle, we set up a system for retroviral transduction of bone marrow progenitors and reconstitution of the bone marrow in irradiated recipient mice to introduce wild-type (wt) or mutated versions of p40phox in neutrophils on a p40phox−/− genetic background. We then performed functional studies to elucidate the role of the SH3 domain and phosphorylation sites of p40phox in NADPH oxidase activation in fully differentiated, mouse bone marrow–derived neutrophils.
Methods
Materials
fMLP, PMA, luminol, and horseradish peroxidase (HRP) were from Sigma-Aldrich. Murine tumor necrosis factor (TNF)α was from R&D Systems. All other cytokines were from Peprotech. Dulbecco's phosphate-buffered saline (PBS) with Ca2+ and Mg2+ was from Sigma-Aldrich. Tissue culture reagents were from Invitrogen. All buffer components were from Sigma-Aldrich and were endotoxin-free or low-endotoxin, as available.
Mouse strains
For mouse bone marrow–reconstitution experiments, donor mice were p40phox−/− mice19 or PKCδ−/− mice.30 Recipient mice were Rag20IL2rg0 mice.31 The p40phoxR58A/R58A mice were described previously.32 All animals were housed in the small animal barrier unit at the Babraham Institute under specific pathogen-free conditions or in individually ventilated cages in the small animal unit. This work was approved by Home Office Project License PPL 80/1875 and PPL 80/2335. All procedures were carried out under Home Office Personal Individual License PIL 80/10 089.
Cell culture
Plat-E cells33 were cultured in Dulbecco Modified Eagle Medium containing 4500 mg/L glucose, l-glutamine, and pyruvate and supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Fetal liver (FL) cells were cultured in StemPro-34 medium supplemented with the provided nutrient supplement, 1% Glutamax, and 1% penicillin/streptomycin/amphotericin B (StemPro-34 complete medium). In addition, a cocktail of cytokines was used to support hematopoietic stem and progenitor cell proliferation: 100 ng/mL mSCF, 100 ng/mL thrombopoietin, 10 ng/mL human interleukin-6, 6 ng/mL murine interleukin-3, and 20 ng/mL Flt3-ligand (Martin Turner, Babraham Institute, personal communication).
Retroviral vectors
The bicistronic retroviral transfer vector pMIG R1-internal ribosome entry site-green fluorescent protein (IRES-GFP) has been described previously.34 A full-length murine p40phox IMAGE cDNA clone was obtained from RZPD Deutsches Ressourcenzentrum für Genomforschung GmbH and sequence verified. To obtain pMIG R1-p40phox-IRES-GFP, polymerase chain reaction (PCR) fragments of p40phox containing engineered restriction sites BglII and EcoRI were digested and cloned into pMIG R1-IRES-GFP. To create pMIG R1-p40phox ΔSH3-IRES-GFP, PCR fragments were generated from p40phox 5′ and 3′ of the SH3 domain. PCR fragments were digested with BglII and SnaBI (5′ fragment) or SnaBI and EcoRI (3′ fragment) and cloned into pMIG R1-IRES-GFP. All point mutants of p40phox that were used in this study were created by site-directed mutagenesis (Stratagene), according to the manufacturer's instructions. Each mutant was verified by sequencing of the p40phox coding sequence and recloned into their original vectors.
Production of retroviral supernatants
Retroviral vectors were produced by transient transfection of the packaging cell line Plat-E, using Lipofectamine 2000 (Invitrogen), with minor modifications. Briefly, transfer vector DNA and Lipofectamine were combined in Optimem medium in a ratio of 1:3 (wt/vol) for 20 minutes at room temperature (RT). Optimem/DNA/Lipofectamine were then added to cells suspended in Plat-E medium lacking antibiotics. Per 1.2 × 107 cells, 16.8 μg of DNA and 50.4 μL Lipofectamine was used. Twenty-four hours after transfection, medium containing DNA-Lipofectamine 2000 complexes was removed and replaced by StemPro-34 complete medium. Forty-eight hours after transfection, the virus-containing supernatant was harvested and centrifuged at 800 × g for 5 minutes at 4°C to pellet cell debris. The supernatant was filtered through a 0.45-μm polyamide filter (Sartorius) and directly used for transduction of FL cells.
Reconstitution of the mouse bone marrow compartment
p40phox−/− FL cells were harvested from E14.5 embryos, cultured for 24 hours, and transduced on non-tissue culture–treated plates coated with 12.5 ng/mL Retronectin (Takara Bio Inc.) at a concentration of 5 × 105 cells/mL. Seventeen hours after transduction, cells were washed in Hank's Buffered Salt Solution, resuspended in PBS/10% FBS, and injected into 8-week-old lethally irradiated recipient mice (1-2 × 106 FL cells/mouse). Recipients were irradiated the day before injection (2 × 5 Gy from a 137Cs source, separated by 3 hours). wt or PKCδ−/− bone marrow was isolated under sterile conditions and suspended in PBS/10% FBS. To increase the number of experimental animals, 5 × 106 bone marrow cells were injected per lethally irradiated recipient mouse. To monitor the efficiency of reconstitution of the bone marrow compartment by transduced FL cells, samples of peripheral blood were stained with neutrophil indicator Gr1-phycoerythrin (BD Biosciences) and subjected to flow cytometric analysis of GFP marker gene expression.
Isolation of neutrophils
Mouse neutrophils (bone marrow–derived neutrophils [BMNs]) were isolated from bone marrow as previously described.32 Purity of neutrophils was determined by flow cytometric analysis of their distinct forward scatter/side scatter properties and was generally between 70% and 90%. Cells were resuspended at a concentration of 6.25 × 106/mL in Dulbecco's PBS with Ca2+ and Mg2+, 1 g/L glucose, and 4 mM sodium bicarbonate (D-PBS+) and primed with 1000 U/mL mouse TNFα and 100 ng/mL granulocyte-macrophage colony-stimulating factor for 1 hour at 37°C with occasional gentle mixing.
Preparation of particles
Staphylococcus aureus (S aureus) bacteria from the Wood 46 strain were cultured and opsonized as described.32 A quantity of 10 μL of SRBCs in Alsevers (TCS Biosciences) were washed (4 minutes, 4000 × g) and resuspended in 1 mL of D-PBS+ containing 0.1% fatty acid- and endotoxin-free bovine serum albumin (D-PBS++). SRBCs were opsonized by using a subagglutinating concentration of rabbit anti-SRBC IgG while rotating end-over-end for 20 minutes at RT. SRBCs were washed twice and resuspended in 800 μL of D-PBS++.
Measurement of ROS production
Where indicated, primed BMNs (typically isolated from a single reconstituted mouse) were pre-incubated with vehicle control (0.1% dimethyl sulfoxide), selective PKC inhibitors bisindolylmaleimide I (BIM-1), Gö 6976, or Gö 6983 (1μM; Calbiochem), or PI3K inhibitor wortmannin (100 nM or 300 nM) for 10 minutes before stimulation. Rate kinetics of ROS production were measured using a luminol-based assay in polystyrene 96-well plates (Berthold Technologies Ltd) as described previously.32 Assays were conducted in the absence or presence of exogenously added HRP (18.75 U/mL) to measure intracellular or total ROS, respectively. fMLP was used at 10μM, PMA at 100 nM, and particles were used at a BMN/particle ratio of 1:20. Light emission was recorded by a Berthold Microlumat Plus luminometer (Berthold Technologies). Data output is relative light units per second or total relative light units integrated over the indicated periods of time. To compare the average responses of wt and mutated p40phox across different agonists, a logarithmic transformation of the raw data were performed before analysis to stabilize the errors, followed by a repeated measures analysis of variance (ANOVA), taking into account the variability between the responses to different agonists.
S aureus phagocytosis assay
S aureus phagocytosis assays were performed as described previously.32 Fixed neutrophils were stained for phox components p40phox (Millipore) or p47phox (Millipore; 1 hour, RT). After washing in PBS/0.5% bovine serum albumin, cells were stained with AlexaFluor405- or AlexaFluor568-conjugated goat anti-rabbit antibodies. Coverslips were mounted on glass microslides with Aqua-Polymount antifading solution (Poly-Science Inc) and fluorescence visualized using a Zeiss LSM 510 META point-scanning confocal microscope with an integrated camera, using a Plan/Apochromat 63×/1.4 oil objective. Cytosolic phox levels and phagosomal accumulation were quantified using LSM 510 software. Given the variability in the data obtained, heterogeneous random error was assumed and a logarithmic transformation of the data was carried out before analysis to stabilize any errors. Outliers were then removed, and a 2-way ANOVA was performed to determine whether the differences between the means of the datasets were statistically significant, taking into account the experimental variability.
Western blotting
Where indicated, primed BMNs were pretreated with vehicle control or inhibitors and stimulated with agonists as described in section Measurement of ROS production. A quantity of 5 × 105 BMNs were sonicated into 1× sodium dodecyl sulfate (SDS) loading buffer, subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred, and immunoblotted for phospho-p40phox (T154; Cell Signaling Technology), p40phox, p47phox, or p67phox (Millipore). Signal was detected by enhanced chemiluminescence and quantified using Aida Image Analyser 2.2.
Results
A method for protein expression in fully differentiated neutrophils
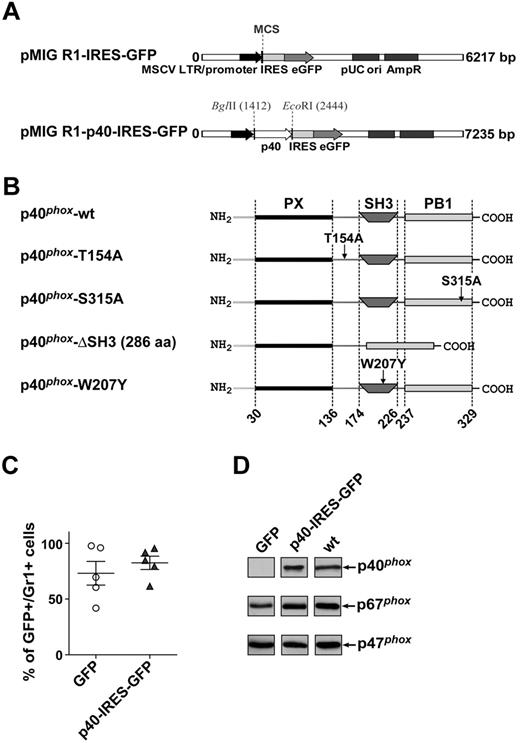
We previously showed that p40phox−/− mice exhibit a CGD-like phenotype, with profound defects in neutrophil ROS responses to both soluble and particulate stimuli.19 Neutrophils from these mice also show a substantial reduction in expression of p67phox. We aimed to use p40phox−/− tissue as a vehicle for the expression and functional analysis of p40phox mutants in fully differentiated neutrophils. To this end, we used retroviral transduction of p40phox−/− FL cells followed by repopulation of the bone marrow compartment of lethally irradiated recipient mice. We established this method by first expressing wt, untagged p40phox in p40phox−/− FL cells using the retroviral vector pMIG R1-IRES-GFP34 (the retroviral constructs and p40phox mutants used in this study are listed in Figure 1A-B). These transduced FL cells were then used to repopulate irradiated mice and to isolate fully differentiated neutrophils from their bone marrow compartment. Flow cytometric analysis of peripheral blood from these chimeric mice revealed that the average percentage of Gr1-positive cells expressing GFP varied between 73.1% and 82.5%, indicating successful reconstitution of the bone marrow compartment by transduced FL cells (Figure 1C). We also analyzed expression of phox components in the BMNs isolated from these mice and found that p40phox introduced by pMIG R1-IRES-GFP expressed at levels similar to endogenous p40phox in wt neutrophils (Figure 1D). Importantly, p67phox expression was also rescued to levels that were equivalent to endogenous p67phox in wt neutrophils (Figure 1D).
Reconstitution of protein expression in mouse neutrophils by untagged, wt p40phox. (A) Retroviral vectors used for mouse bone marrow reconstitution. Maps of empty vector pMIG R1-IRES-GFP (top) and a construct containing untagged, wt p40phox (bottom). (B) Position of mutated residues in the primary protein structure of p40phox. (C) Flow cytometric analysis of GFP expression in circulating neutrophils, as a measure of the efficiency of reconstitution of peripheral blood and bone marrow by transduced donor cells. Mouse FL cells on a p40phox−/− background were transduced with retroviral supernatants and injected into irradiated recipients. Four weeks after injection, allowing sufficient time to repopulate the bone marrow compartment, peripheral blood samples were subjected to flow cytometric analysis of GFP-positive neutrophils, using Gr1 as an indicator for neutrophils. Each data point represents one individual mouse. Data are mean ± SEM. (D) Expression of p40phox (top), p67phox (middle), and p47phox (bottom) in BMNs isolated from p40phox−/− mice heterologously expressing GFP or untagged p40phox (p40-IRES-GFP) or wt mice. Neutrophils were sonicated into SDS sample buffer, subjected to SDS-PAGE (4 × 105 cells/lane), and immunoblotted for phox components as described in “Methods.” Shown is a representative blot of 2, performed in duplicate.
Reconstitution of protein expression in mouse neutrophils by untagged, wt p40phox. (A) Retroviral vectors used for mouse bone marrow reconstitution. Maps of empty vector pMIG R1-IRES-GFP (top) and a construct containing untagged, wt p40phox (bottom). (B) Position of mutated residues in the primary protein structure of p40phox. (C) Flow cytometric analysis of GFP expression in circulating neutrophils, as a measure of the efficiency of reconstitution of peripheral blood and bone marrow by transduced donor cells. Mouse FL cells on a p40phox−/− background were transduced with retroviral supernatants and injected into irradiated recipients. Four weeks after injection, allowing sufficient time to repopulate the bone marrow compartment, peripheral blood samples were subjected to flow cytometric analysis of GFP-positive neutrophils, using Gr1 as an indicator for neutrophils. Each data point represents one individual mouse. Data are mean ± SEM. (D) Expression of p40phox (top), p67phox (middle), and p47phox (bottom) in BMNs isolated from p40phox−/− mice heterologously expressing GFP or untagged p40phox (p40-IRES-GFP) or wt mice. Neutrophils were sonicated into SDS sample buffer, subjected to SDS-PAGE (4 × 105 cells/lane), and immunoblotted for phox components as described in “Methods.” Shown is a representative blot of 2, performed in duplicate.
Retroviral expression of p40phox rescues ROS responses in p40phox−/− neutrophils
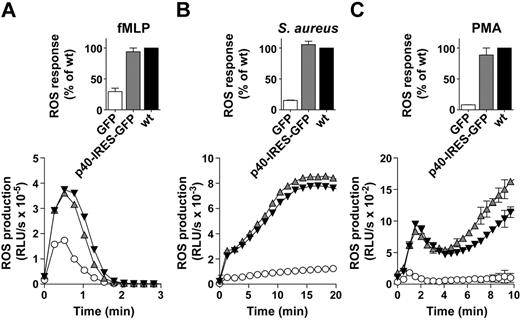
We characterized ROS responses in p40phox−/− neutrophils reconstituted with p40phox-IRES-GFP. We measured the rate of ROS formation in TNFα/granulocyte-macrophage colony stimulating factor primed BMNs stimulated with the chemotactic peptide fMLP or serum-opsonized S aureus. We also measured ROS formation stimulated by the phorbol ester PMA, which is envisaged to bypass receptor activation. Expression of untagged p40phox fully rescued the ROS responses to fMLP, S aureus, and PMA (Figure 2A-C). Expression of a GFP-tagged p40phox only partially rescued these responses, indicating an N-terminal GFP-tag partially inhibits p40phox function (data not shown). We have also noted that neutrophils from p40phox+/− heterozygotes displayed normal ROS responses to a variety of different agonists, despite reductions in p40phox expression, suggesting these responses are insensitive to approximately 50% reductions in the levels of p40phox (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Taken together, these results indicate that the introduction of untagged p40phox mutants, through retroviral transduction of progenitor cells and subsequent repopulation of the bone marrow compartment, may be a useful tool for assessing their impact on neutrophil oxidase activation.
Untagged p40phox fully rescues NADPH oxidase activity in p40phox−/− neutrophils. A quantity of 1 × 106 wt BMNs (▾) or p40phox−/− BMNs expressing GFP (○) or untagged p40phox (p40-IRES-GFP;  ) were pre-incubated with luminol/HRP before addition to fMLP (A) or PMA (C) or pre-incubated with luminol before addition to serum-S aureus (B), as described in the Methods. Shown are total integrated ROS responses for 2 independent experiments (mean ± range), expressed as a percentage of the response in wt BMNs, as well as profiles of kinetics of ROS production, measured in relative light units per second (RLU/s) (mean ± range), from 1 representative experiment performed in duplicate.
) were pre-incubated with luminol/HRP before addition to fMLP (A) or PMA (C) or pre-incubated with luminol before addition to serum-S aureus (B), as described in the Methods. Shown are total integrated ROS responses for 2 independent experiments (mean ± range), expressed as a percentage of the response in wt BMNs, as well as profiles of kinetics of ROS production, measured in relative light units per second (RLU/s) (mean ± range), from 1 representative experiment performed in duplicate.
Untagged p40phox fully rescues NADPH oxidase activity in p40phox−/− neutrophils. A quantity of 1 × 106 wt BMNs (▾) or p40phox−/− BMNs expressing GFP (○) or untagged p40phox (p40-IRES-GFP;  ) were pre-incubated with luminol/HRP before addition to fMLP (A) or PMA (C) or pre-incubated with luminol before addition to serum-S aureus (B), as described in the Methods. Shown are total integrated ROS responses for 2 independent experiments (mean ± range), expressed as a percentage of the response in wt BMNs, as well as profiles of kinetics of ROS production, measured in relative light units per second (RLU/s) (mean ± range), from 1 representative experiment performed in duplicate.
) were pre-incubated with luminol/HRP before addition to fMLP (A) or PMA (C) or pre-incubated with luminol before addition to serum-S aureus (B), as described in the Methods. Shown are total integrated ROS responses for 2 independent experiments (mean ± range), expressed as a percentage of the response in wt BMNs, as well as profiles of kinetics of ROS production, measured in relative light units per second (RLU/s) (mean ± range), from 1 representative experiment performed in duplicate.
Mutation of T154 but not S315 in p40phox results in substantially reduced ROS responses to both soluble and particulate stimuli
We investigated the function of the 2 documented phosphorylation sites of p40phox, threonine 154, and serine 31522 by creating bone marrow chimaeras expressing p40phox-T154A-IRES-GFP or p40phox-S315A-IRES-GFP in their hematopoietic lineage. BMNs were isolated from these mice, and we compared both their p40phox expression levels and ROS responses to equivalently generated BMNs expressing either wt p40phox-IRES-GFP or IRES-GFP alone (Figure 3). Both p40phox-T154A and p40phox-S315A were expressed to very similar levels as p40phox, and, importantly, both mutants also rescued expression of endogenous p67phox (Figure 3A). Substitution of threonine 154 to alanine led to significant decreases in p40phox-dependent ROS responses to fMLP (40%), IgG-SRBCs (67%), and S aureus (63%) but no clear defect in the response to PMA (Figure 3B-E). In contrast, substitution of serine 315 to alanine resulted only in a small decrease in the ROS response to PMA (32%) but had no effect on the ROS responses to each of the physiological agonists tested (Figure 3B-E).
Phosphorylation of p40phox on T154, but not S315, is essential for NADPH oxidase activity in mouse neutrophils. (A) Lysates from p40phox−/− BMNs reconstituted with GFP (empty vector [ev]), wt p40phox (wt), p40phox-T154A (T154A), or p40phox-S315A (S315A) were subjected to SDS-PAGE and immunoblotted for phox components as described in “Methods.” A representative blot of 3 independent experiments is shown. Levels of p40phox and p67phox were normalized against levels of p47phox, which was used as a loading control. Histograms represent relative levels of p40phox (left) or p67phox (right) as a percentage of wt. Data are mean ± SEM (n = 3 independent experiments performed in duplicate). (B-E) p40phox−/− BMNs expressing GFP (empty vector [ev]; ○), wt p40phox (▾), p40phox-T154A ( ), or p40phox-S315A (■) were pre-incubated with luminol/HRP before addition to fMLP (B) or PMA (E) or with luminol before addition to S aureus (C) or IgG-SRBCs (D), as described in “Methods.” ROS responses were measured by chemiluminescence as described in “Methods.” Shown are profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for 3-6 independent experiments (mean ± SEM), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. **P = .006; ***P = .000; as determined by a paired Student t test.
), or p40phox-S315A (■) were pre-incubated with luminol/HRP before addition to fMLP (B) or PMA (E) or with luminol before addition to S aureus (C) or IgG-SRBCs (D), as described in “Methods.” ROS responses were measured by chemiluminescence as described in “Methods.” Shown are profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for 3-6 independent experiments (mean ± SEM), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. **P = .006; ***P = .000; as determined by a paired Student t test.
Phosphorylation of p40phox on T154, but not S315, is essential for NADPH oxidase activity in mouse neutrophils. (A) Lysates from p40phox−/− BMNs reconstituted with GFP (empty vector [ev]), wt p40phox (wt), p40phox-T154A (T154A), or p40phox-S315A (S315A) were subjected to SDS-PAGE and immunoblotted for phox components as described in “Methods.” A representative blot of 3 independent experiments is shown. Levels of p40phox and p67phox were normalized against levels of p47phox, which was used as a loading control. Histograms represent relative levels of p40phox (left) or p67phox (right) as a percentage of wt. Data are mean ± SEM (n = 3 independent experiments performed in duplicate). (B-E) p40phox−/− BMNs expressing GFP (empty vector [ev]; ○), wt p40phox (▾), p40phox-T154A ( ), or p40phox-S315A (■) were pre-incubated with luminol/HRP before addition to fMLP (B) or PMA (E) or with luminol before addition to S aureus (C) or IgG-SRBCs (D), as described in “Methods.” ROS responses were measured by chemiluminescence as described in “Methods.” Shown are profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for 3-6 independent experiments (mean ± SEM), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. **P = .006; ***P = .000; as determined by a paired Student t test.
), or p40phox-S315A (■) were pre-incubated with luminol/HRP before addition to fMLP (B) or PMA (E) or with luminol before addition to S aureus (C) or IgG-SRBCs (D), as described in “Methods.” ROS responses were measured by chemiluminescence as described in “Methods.” Shown are profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for 3-6 independent experiments (mean ± SEM), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. **P = .006; ***P = .000; as determined by a paired Student t test.
We investigated the role of phosphorylation on threonine 154 further by creating mouse chimaeras expressing p40phox-T154E-IRES-GFP. The substitution of threonine by an acidic glutamate residue is often found to mimic the charge created by phosphorylation; however, we found p40phox-T154E behaved almost identically to p40phox-T154A in all of the BMN ROS assays we conducted, suggesting this substitution was unable to mimic phosphorylation in this context (supplemental Figure 2).
The SH3 domain of p40phox does not play an essential role in NADPH oxidase activation
We also investigated the function of the SH3 domain of p40phox in mouse neutrophils by creating bone marrow chimaeras expressing a mutant of p40phox lacking this domain (p40phox-ΔSH3-IRES-GFP) or carrying a mutation predicted to disrupt its binding to PP targets (p40phox-W207Y-IRES-GFP). We had initially attempted to express p40phox-W207R, because previous work had established that the tryptophan to arginine substitution blocks p40phox SH3 activity,27,28 but this mutant was poorly expressed in trial experiments (data not shown). We therefore made an alternative tryptophan 207 to tyrosine mutation (W207Y), introducing an acidic hydroxyl group predicted to disrupt hydrophobic interactions between the SH3 domain of p40phox and a PP target (supplemental Figure 3) and which also preserved expression of the protein (see below).
Both p40phox-W207Y and p40phox-ΔSH3 were expressed in p40phox−/− BMNs to similar levels as wt p40phox (Figure 4A). In addition, both mutants of p40phox rescued expression of endogenous p67phox (Figure 4A). The p40phox-dependent ROS response to PMA was reduced by approximately 30%-40% in BMNs expressing p40phox-W207Y or p40phox-ΔSH3 (Figure 4E). However, ROS responses to fMLP, S aureus, and IgG-SRBCs were unaffected in cells expressing p40phox-W207Y and significantly elevated in cells expressing p40phox-ΔSH3 (Figure 4B-D).
Mutation or deletion of the SH3 domain of p40phox does not impair ROS production in mouse neutrophils. (A) p40phox−/− BMNs reconstituted with GFP (empty vector [ev]), wt p40phox (wt), p40phox-ΔSH3 (ΔSH3), or p40phox-W207Y (W207Y) were sonicated, subjected to SDS-PAGE, and immunoblotted for phox components as described in “Methods.” A representative blot of 2 independent experiments is shown. Levels of p40phox and p67phox were normalized against levels of p47phox, which was used as a loading control. Histograms represent relative levels of p40phox (left) or p67phox (right) as a percentage of wt. Data are mean ± range (n = 2 independent experiments performed in duplicate). (B-E) p40phox−/− BMNs expressing GFP (ev; ○), wt p40phox (▾), p40phox-ΔSH3 ( ), or p40phox-W207Y (■) were used in assays for ROS production as described in Figure 3. Shown are through profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for a minimum of 3 independent experiments (mean ± SEM; ev: n = 8-9; wt: n = 9; ΔSH3: n = 9; W207Y: n = 3), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. +P = .009; as determined by a repeated measures ANOVA.
), or p40phox-W207Y (■) were used in assays for ROS production as described in Figure 3. Shown are through profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for a minimum of 3 independent experiments (mean ± SEM; ev: n = 8-9; wt: n = 9; ΔSH3: n = 9; W207Y: n = 3), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. +P = .009; as determined by a repeated measures ANOVA.
Mutation or deletion of the SH3 domain of p40phox does not impair ROS production in mouse neutrophils. (A) p40phox−/− BMNs reconstituted with GFP (empty vector [ev]), wt p40phox (wt), p40phox-ΔSH3 (ΔSH3), or p40phox-W207Y (W207Y) were sonicated, subjected to SDS-PAGE, and immunoblotted for phox components as described in “Methods.” A representative blot of 2 independent experiments is shown. Levels of p40phox and p67phox were normalized against levels of p47phox, which was used as a loading control. Histograms represent relative levels of p40phox (left) or p67phox (right) as a percentage of wt. Data are mean ± range (n = 2 independent experiments performed in duplicate). (B-E) p40phox−/− BMNs expressing GFP (ev; ○), wt p40phox (▾), p40phox-ΔSH3 ( ), or p40phox-W207Y (■) were used in assays for ROS production as described in Figure 3. Shown are through profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for a minimum of 3 independent experiments (mean ± SEM; ev: n = 8-9; wt: n = 9; ΔSH3: n = 9; W207Y: n = 3), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. +P = .009; as determined by a repeated measures ANOVA.
), or p40phox-W207Y (■) were used in assays for ROS production as described in Figure 3. Shown are through profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for a minimum of 3 independent experiments (mean ± SEM; ev: n = 8-9; wt: n = 9; ΔSH3: n = 9; W207Y: n = 3), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. +P = .009; as determined by a repeated measures ANOVA.
Given that mutation of the p40phox SH3 domain did not reduce ROS responses to physiological agonists and that deletion of the domain indeed enhanced ROS responses to some stimuli, we postulated that the SH3 domain might play an auto-inhibitory role in oxidase activation, which may be alleviated by phosphorylation on threonine 154. To test this, we created the T154A mutation in the context of a deleted SH3 domain. p40phox-ΔSH3/T154A was expressed normally in p40phox−/− BMNs and this mutant fully rescued p67phox expression (supplemental Figure 4). The defects in ROS responses in BMNs expressing p40phox-ΔSH3/T154A were generally slightly greater than the defects seen in BMNs expressing p40phox-T154A (supplemental Figure 4 and Figure 3), indicating SH3 domain deletion does not relieve the inhibition caused by a block in phosphorylation of threonine 154.
Phosphorylation of p40phox on T154 is mediated by PKC
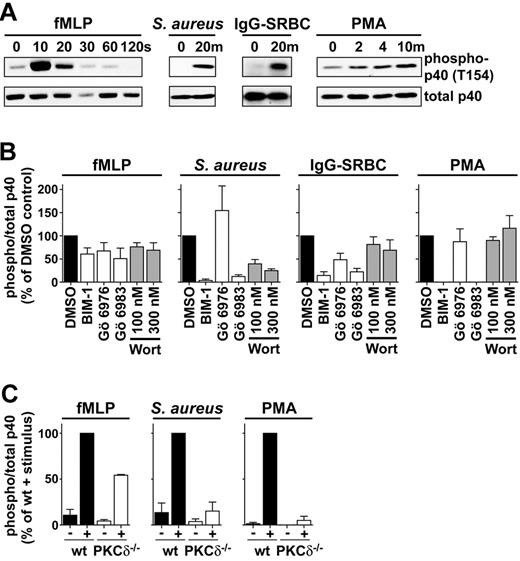
We used a phospho-specific antibody raised against phospho-threonine 154 to measure the phosphorylation of this site in BMNs stimulated with various agonists. Stimulation with PMA, fMLP, S aureus, or IgG-SRBCs resulted in substantial increases in T154 phosphorylation that were consistent with the time courses of oxidase activation to these agonists (Figure 5). T154 is located within a polybasic motif (RRLRPRTRKIK) that is a consensus site for phosphorylation by PKC enzymes (consensus-S/T-X-R/K-). Further, previous work has shown that a PKC can phosphorylate this site in vitro23,25 and that broad-range inhibitors of PKC block PMA-induced phosphorylation of p40phox in neutrophils.22,23 We therefore sought to establish whether PKCs were responsible for phosphorylation of T154 in our assays using previously well characterized inhibitors of these enzymes.
Phosphorylation of p40phox on T154 in response to fMLP and IgG-SRBCs is partially dependent on cPKCs, whereas phosphorylation of p40phox in response to PMA and S aureus is mediated by PKCδ. (A) Stimulus-elicited phosphorylation of p40phox. BMNs from wt C57Bl/6J mice (5 × 105/sample) were incubated with fMLP, S aureus, IgG-SRBCs, or PMA. Assays were terminated at the indicated time points by placing samples on ice with immediate addition of an excess of ice-cold buffer. Cells were pelleted and sonicated into SDS sample buffer. Proteins were resolved by SDS-PAGE, after which immunoblotting was performed using an antibody directed against p40phox phosphorylated on T154 or total p40phox as a loading control. A representative blot from 2 independent experiments is shown for each condition. (B) BMNs from wt C57Bl/6J mice were pretreated with vehicle control (0.1% dimethyl sulfoxide), BIM-1, Gö 6976, or Gö 6983 at a concentration of 1μM or with wortmannin (Wort) at a concentration of 100 nM or 300 nM. Cells were stimulated with fMLP for 10 seconds, S aureus or IgG-SRBCs for 20 minutes, PMA for 10 minutes. Assays were stopped, and cells were immediately processed as described in panel A. Quantification of levels of phospho-p40phox and total p40phox was performed using Aida software. Histograms represent ratios of phospho-p40phox:total p40phox as a percentage of vehicle control. Data are means ± SEM (n = 3 independent experiments for each agonist, performed in duplicate). (C) wt or PKCδ−/− neutrophils were stimulated with fMLP for 10 seconds, S aureus for 20 minutes, PMA for 10 minutes. Assays were stopped, and cells were processed as described in A. Quantification of levels of phospho-p40phox and total p40phox was performed using Aida software. Histograms represent ratios of phospho-p40phox:total p40phox as a percentage of wt stimulated BMNs. Data are means ± range (n = 2) for fMLP and means ± SEM (n = 3) for S aureus and PMA.
Phosphorylation of p40phox on T154 in response to fMLP and IgG-SRBCs is partially dependent on cPKCs, whereas phosphorylation of p40phox in response to PMA and S aureus is mediated by PKCδ. (A) Stimulus-elicited phosphorylation of p40phox. BMNs from wt C57Bl/6J mice (5 × 105/sample) were incubated with fMLP, S aureus, IgG-SRBCs, or PMA. Assays were terminated at the indicated time points by placing samples on ice with immediate addition of an excess of ice-cold buffer. Cells were pelleted and sonicated into SDS sample buffer. Proteins were resolved by SDS-PAGE, after which immunoblotting was performed using an antibody directed against p40phox phosphorylated on T154 or total p40phox as a loading control. A representative blot from 2 independent experiments is shown for each condition. (B) BMNs from wt C57Bl/6J mice were pretreated with vehicle control (0.1% dimethyl sulfoxide), BIM-1, Gö 6976, or Gö 6983 at a concentration of 1μM or with wortmannin (Wort) at a concentration of 100 nM or 300 nM. Cells were stimulated with fMLP for 10 seconds, S aureus or IgG-SRBCs for 20 minutes, PMA for 10 minutes. Assays were stopped, and cells were immediately processed as described in panel A. Quantification of levels of phospho-p40phox and total p40phox was performed using Aida software. Histograms represent ratios of phospho-p40phox:total p40phox as a percentage of vehicle control. Data are means ± SEM (n = 3 independent experiments for each agonist, performed in duplicate). (C) wt or PKCδ−/− neutrophils were stimulated with fMLP for 10 seconds, S aureus for 20 minutes, PMA for 10 minutes. Assays were stopped, and cells were processed as described in A. Quantification of levels of phospho-p40phox and total p40phox was performed using Aida software. Histograms represent ratios of phospho-p40phox:total p40phox as a percentage of wt stimulated BMNs. Data are means ± range (n = 2) for fMLP and means ± SEM (n = 3) for S aureus and PMA.
The PKC family of enzymes comprises 10 different isoforms identified to date, which are classified according to their primary structure and mode of regulation (Table 1).35 The PKC inhibitor Gö 6976 inhibits conventional (c) PKCs over other PKCs and is generally accepted to be a selective cPKC inhibitor (Table 1). BIM-1 inhibits cPKCs and novel (n) PKCs with similar potency, and Gö 6983 inhibits cPKCs, nPKCs, and atypical (a) PKCs with similar potency (Table 1). We used all 3 PKC inhibitors at a concentration of 1μM, which falls within the range of concentrations characterized in the literature.36 Phosphorylations on T154 induced by PMA and S aureus were substantially and selectively inhibited by BIM-1 and Gö 6983 (Figure 5B). fMLP-induced phosphorylation of T154 was only modestly inhibited by each of the inhibitors tested and IgG-SRBC–induced phosphorylation was more potently inhibited by each of these inhibitors (Figure 5B). The most parsimonious explanation of these results is that the T154 phosphorylation induced by PMA or S aureus depends heavily on nPKC activation but that more redundancy is likely to exist between PKC isoforms, including cPKC and possibly other protein kinases, in the responses to fMLP or IgG-SRBCs. Further, the effects of these PKC inhibitors on ROS responses to these agonists were equivalent to, or greater than, would be predicted on the basis of their effects on T154 phosphorylation (supplemental Figure 5), consistent with previous data indicating PKC enzymes are also important in the activation of the oxidase through phosphorylation of p47phox, particularly with respect to fMLP.37-39
The PKC family of enzymes, their expression in mouse neutrophils, their activators, and their inhibitors
| Subfamily . | Isoforms . | Direct activators . | Agonists . | |||
|---|---|---|---|---|---|---|
| cPKC | α* | βI† | βII* | γ† | DAG + Ca2+ | fMLP,37 PMA (β), Fc (β)47 |
| nPKC | δI-III* | ϵ‡ | η‡ | θI-III‡ | DAG | fMLP (δ)37,44 |
| aPKC | ζ* | ι/λ† | fMLP (ζ)38 | |||
| Subfamily . | Isoforms . | Direct activators . | Agonists . | |||
|---|---|---|---|---|---|---|
| cPKC | α* | βI† | βII* | γ† | DAG + Ca2+ | fMLP,37 PMA (β), Fc (β)47 |
| nPKC | δI-III* | ϵ‡ | η‡ | θI-III‡ | DAG | fMLP (δ)37,44 |
| aPKC | ζ* | ι/λ† | fMLP (ζ)38 | |||
| Inhibitor . | IC50 for PKC isoforms, μM . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| α* . | βI† . | βII* . | γ† . | δ* . | ϵ‡ . | ζ* . | η‡ . | μ† . | ι/λ† . | |
| BIM-I48-50 | 0.008 | 0.017 | 0.016 | 0.020 | 0.21 | 0.132 | 5.8 | § | 2 | § |
| Gö 697648,49,51 | 0.0023 | 0.006 | 0.002-0.0079 | 0.002-0.006 | — | — | — | § | 0.02 | § |
| Gö 698348 | 0.007 | 0.007 | 0.007 | 0.006 | 0.01 | § | 0.06 | § | 20 | § |
| Inhibitor . | IC50 for PKC isoforms, μM . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| α* . | βI† . | βII* . | γ† . | δ* . | ϵ‡ . | ζ* . | η‡ . | μ† . | ι/λ† . | |
| BIM-I48-50 | 0.008 | 0.017 | 0.016 | 0.020 | 0.21 | 0.132 | 5.8 | § | 2 | § |
| Gö 697648,49,51 | 0.0023 | 0.006 | 0.002-0.0079 | 0.002-0.006 | — | — | — | § | 0.02 | § |
| Gö 698348 | 0.007 | 0.007 | 0.007 | 0.006 | 0.01 | § | 0.06 | § | 20 | § |
Expressed in mouse neutrophils.
Unknown.
Not expressed in mouse neutrophils.
Effect of inhibitor not tested.
To date, the only nPKC that is known to be expressed in BMNs is PKCδ.40 We therefore sought to confirm a role for this PKC isoform in the phosphorylation of T154 in p40phox stimulated by PMA and S aureus using BMNs isolated from mice lacking this protein (PKCδ−/−). Phosphorylation of T154 was almost completely abolished in response to PMA and S aureus in PKCδ−/− neutrophils (Figure 5C), whereas loss of PKCδ had a much smaller effect on this response in fMLP-stimulated cells (Figure 5C), consistent with the inhibitor studies described in the previous paragraph. In addition, the ROS response to S aureus was reduced to approximately 50% of wt in PKCδ−/− BMNs, which is equivalent to the reduction in the ROS response caused by the T154A mutation (supplemental Figure 5). The ROS response to PMA, however, was almost completely abolished by loss of PKCδ (supplemental Figure 5), suggesting this enzyme is also required for other critical inputs into the oxidase downstream of PMA, such as the phosphorylation of other oxidase components.
Phosphorylation of p40phox on T154 is partially sensitive to the PI3K inhibitor wortmannin
A previous study has suggested that T154 phosphorylation induced by IgG targets is partially inhibited by the general PI3K inhibitor wortmannin.24 We found that 100 nM and 300 nM wortmannin substantially inhibited T154 phosphorylation in response to S aureus (approximately 60%-75%), partially inhibited T154 phosphorylation in response to fMLP and IgG-SRBCs (approximately 20%-30%), and had no effect on T154 phosphorylation in response to PMA (Figure 5B). These results suggest PI3Ks may be differentially involved upstream of PKC-mediated T154 phosphorylation.
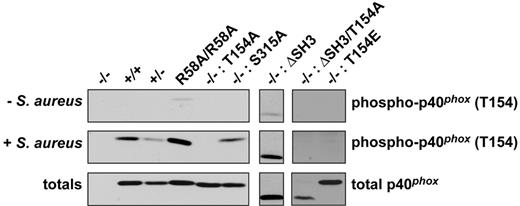
Phosphorylation of p40phox on T154 does not depend on a functional PX or SH3 domain or upon phosphorylation of S315
T154 phosphorylation induced by S aureus was normal in BMNs isolated from p40phoxR58A/R58A mice and p40phox−/− BMNs reconstituted with p40phox-S315A or p40phox-ΔSH3 (Figure 6). p40phox−/− BMNs reconstituted with p40phox-T154A, -ΔSH3/T154A, or -T154E displayed no detectable phosphorylation, which served to confirm the specificity of the antibody used (Figure 6). The lack of effect of the R58A mutation on T154 phosphorylation suggests the wortmannin sensitivity identified above is not mediated via a requirement for PtdIns3P-binding to p40phox.
Phosphorylation of p40phox on T154 is unaffected in p40phox-R58A, -S315A, or -ΔSH3 neutrophils. p40phox−/− (−/−), wt (+/+), p40phox+/− (+/−), p40phoxR58A/R58A (R58A/R58A) BMNs, or p40phox−/− BMNs reconstituted with mutated versions of p40phox as indicated were incubated without or with S aureus (ratio S aureus/BMN of 20:1) for 20 minutes at 37°C. Cell lysates were subjected to SDS-PAGE, after which immunoblotting was performed using an antibody directed against p40phox phosphorylated on T154 or total p40phox as a loading control. Representative blots from 2-5 independent experiments are shown.
Phosphorylation of p40phox on T154 is unaffected in p40phox-R58A, -S315A, or -ΔSH3 neutrophils. p40phox−/− (−/−), wt (+/+), p40phox+/− (+/−), p40phoxR58A/R58A (R58A/R58A) BMNs, or p40phox−/− BMNs reconstituted with mutated versions of p40phox as indicated were incubated without or with S aureus (ratio S aureus/BMN of 20:1) for 20 minutes at 37°C. Cell lysates were subjected to SDS-PAGE, after which immunoblotting was performed using an antibody directed against p40phox phosphorylated on T154 or total p40phox as a loading control. Representative blots from 2-5 independent experiments are shown.
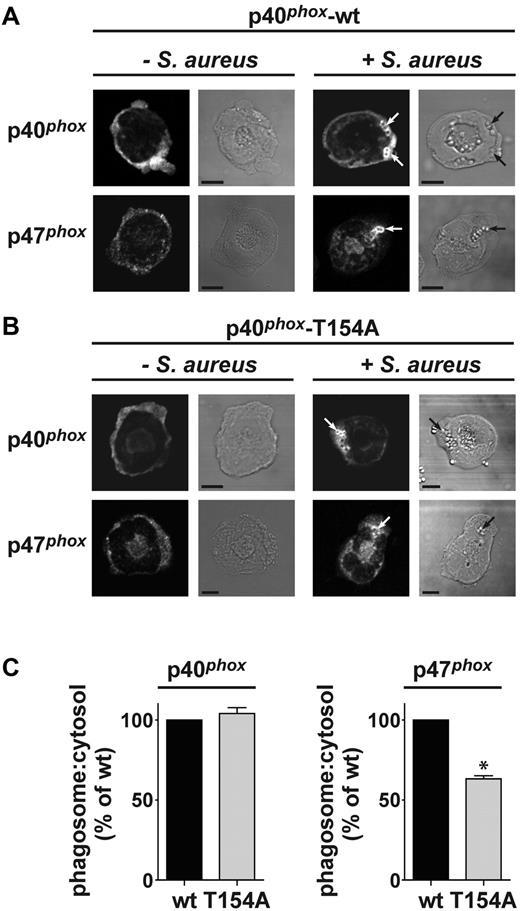
Phosphorylation of p40phox on T154 is required for translocation of p47phox to S aureus phagosomes
We had noted previously that translocation of p47phox to S aureus–containing phagosomes is highly dependent on the presence of p40phox (supplemental Figure 6), and so we used this system to investigate whether the T154A mutation might exert its effects on ROS production through impaired assembly of the oxidase complex. After ingestion of S aureus, wt p40phox and p40phox-T154A localized to phagosomal membranes to the same relative extents (Figure 7A-C). However, there was a marked decrease in the translocation of endogenous p47phox in neutrophils expressing p40phox-T154A (Figure 7B-C). This decrease was very similar in scale to the decrease in ROS effected by the T154A mutation, suggesting they may be causally linked.
p47phox accumulation is decreased around S aureus phagosomes in p40phox-T154A neutrophils. A quantity of 5 × 104 wt or p40phox-T154A neutrophils were incubated without or with 1 × 106 serum-opsonized S aureus for 7 minutes at 37°C. Samples were cytospun onto glass coverslips, fixed, and immunostained for p40phox or p47phox as described in “Methods.” Mounted samples were visualized on a Zeiss LSM 510 META point-scanning microscope using fluorescence and DIC optics, as described in the Methods. Shown are representative fluorescence and DIC images for conditions tested. The position of internalized bacteria is indicated by arrows. Phagosomal accumulation of phox components was quantified using LSM 510 Image browser software. The average phagosomal fluorescence intensity per cell was calculated for n ≥ 71 cells, in 2-3 experiments, and was expressed as a fold increase in fluorescence intensity over cytosolic fluorescence intensity. Data were combined and are presented as fold increase in fluorescence intensity around the phagosome (% of wt; mean ± SEM). Where indicated, difference between means of wt and T154A is statistically significant. *P = .013 as determined by 2-way ANOVA. p40phox: n = 87 cells for wt; n = 71 cells for T154A (3 experiments). p47phox: n = 74 cells for wt; n = 84 cells for T154A (2 experiments).
p47phox accumulation is decreased around S aureus phagosomes in p40phox-T154A neutrophils. A quantity of 5 × 104 wt or p40phox-T154A neutrophils were incubated without or with 1 × 106 serum-opsonized S aureus for 7 minutes at 37°C. Samples were cytospun onto glass coverslips, fixed, and immunostained for p40phox or p47phox as described in “Methods.” Mounted samples were visualized on a Zeiss LSM 510 META point-scanning microscope using fluorescence and DIC optics, as described in the Methods. Shown are representative fluorescence and DIC images for conditions tested. The position of internalized bacteria is indicated by arrows. Phagosomal accumulation of phox components was quantified using LSM 510 Image browser software. The average phagosomal fluorescence intensity per cell was calculated for n ≥ 71 cells, in 2-3 experiments, and was expressed as a fold increase in fluorescence intensity over cytosolic fluorescence intensity. Data were combined and are presented as fold increase in fluorescence intensity around the phagosome (% of wt; mean ± SEM). Where indicated, difference between means of wt and T154A is statistically significant. *P = .013 as determined by 2-way ANOVA. p40phox: n = 87 cells for wt; n = 71 cells for T154A (3 experiments). p47phox: n = 74 cells for wt; n = 84 cells for T154A (2 experiments).
Discussion
We were able to successfully express p40phox in p40phox−/− mouse neutrophils through retroviral transduction of FL cells and repopulation of the hematopoietic compartment in lethally irradiated mice. Remarkably, heterologous expression of p40phox driven by the murine stem cell virus promoter in pMIG R1-IRES-GFP reconstituted normal levels of p40phox expression and normal p40phox-dependent ROS responses in BMNs isolated from these chimeric mice. This provided us with an opportunity to investigate the function of conserved domains in p40phox by reconstituting p40phox−/− BMNs with p40phox mutants. It is possible that the expression levels of p40phox and its direct binding partner p67phox are mutually self-regulating,5,41-43, and thus it remains to be established how applicable this approach might be for probing the function of other oxidase subunits for which the appropriate knockout mice are available, such as gp91phox or p47phox. In any event, this represents a powerful methodology for expressing desired proteins in fully differentiated mouse neutrophils.
Our data suggest that the SH3 domain of p40phox does not play an essential role in neutrophil oxidase activation. The introduction of a W207Y mutation predicted to disrupt SH3 domain binding to PP motifs caused a decrease in the ROS response to PMA but had no effect on ROS responses to physiological stimuli. This is consistent with a recent study that reported addition of p40phox-W207R to permeabilized neutrophils could only partially restore a p40phox-dependent ROS response to PMA.27 It contrasts, however, with observations that the W207R mutation also reduces the ability of p40phox to fully support IgG-SRBC–stimulated production of ROS in the model COSphox cell system.28 We also report that deletion of the entire SH3 domain causes elevated ROS production in response to fMLP, S aureus, and IgG-SRBCs, suggesting that any role for the SH3 domain in oxidase activation is likely to be that of an “inhibitory gate.” A previous study conducted by Sathyamoorthy and coworkers18 also reported an inhibitory role for the SH3 domain of p40phox in whole-cell oxidase activity in transfecto, although in their assay system, full-length p40phox was also found to inhibit oxidase activity. It is currently difficult to reconcile all of this data, suggesting we still do not have a clear understanding of the physiological role of this domain.
Our studies pinpoint PKC-mediated phosphorylation of p40phox on T154 as a key regulatory step in the activation of the NADPH oxidase. T154A mutation resulted in substantial defects in ROS responses to fMLP, S aureus, and IgG-SRBCs but did not clearly affect the ROS response to PMA. Mutation of the other documented phosphorylation site on p40phox, S315, only resulted in a modest defect in the ROS response to PMA. Using a combination of pharmacological and genetic inhibition of PKC isoforms, we were able to establish that phosphorylation of T154 in response to PMA or S aureus is entirely dependent on PKCδ but that probably a combination of both PKCδs and cPKCs are responsible for T154 phosphorylation in response to fMLP or IgG-SRBCs. Further, we identified wortmannin-sensitive pathways that lie upstream of PKC-mediated phosphorylation of T154, particularly in response to S aureus. cPKCs and PKCδs have previously been implicated in phosphorylation of p47phox and subsequent activation of the NADPH oxidase, and in some cases PKCδ has been placed downstream of PI3K activation,37,44 and thus our data extend the known influence of these enzymes in pathways regulating oxidase activation.
Our discovery of an important, positive role for T154 phosphorylation in the activation of the oxidase is in apparent disagreement with a study by Lopes and coworkers,25 which demonstrates that in a cell-free system using membranes and recombinant proteins, p40phox phosphorylated on T154 but not unphosphorylated p40phox inhibits activation of the NADPH oxidase. This discrepancy is likely to reflect a difference in the experimental settings used. Lopes and coworkers use p47phox phosphorylated in vitro by PKC as the activator of the NADPH oxidase. It is possible that in their in vitro recombinant assay system, the regulatory mechanism that depends on T154 phosphorylation is absent, lies upstream of p47phox phosphorylation, or is bypassed by addition of excess amounts of phosphorylated p47phox. In this regard, we also note that T154 phosphorylation is not essential for PKCδ-mediated ROS formation in PMA-stimulated BMNs.
In an attempt to further elucidate the mechanism by which T154 phosphorylation regulates the oxidase, we sought to establish if this event is linked to the function of other domains in p40phox. R58A mutation in the PX domain of p40phox stops p40phox binding to PtdIns3P and causes significant defects in ROS responses to phagocytosis of both S aureus and IgG-opsonized targets.20,32 p40phox-R58A was phosphorylated normally on T154 in response to S aureus, indicating PtdIns3P-binding to p40phox is not a prerequisite for phosphorylation of this site. Further, p40phox-ΔSH3 or p40phox-S315A were also phosphorylated normally in response to S aureus, suggesting T154 phosphorylation is also independent of the SH3 domain or phosphorylation on S315. Further, deletion of the SH3 domain did not overcome the inhibition of ROS responses caused by the T154A mutation, suggesting this phosphorylation event does not mediate its effects through derepression of an SH3 domain interaction. It remained possible that T154 phosphorylation is required for binding of p40phox to PtdIns3P. Indeed, phosphorylation of p40phox on T154 could be a trigger that alleviates the closed conformation of p40phox in which PX-PB1 domain interaction masks the PX domain from binding to its target.45 Although we have not directly tested this hypothesis, the normal translocation of p40phox-T154A to S aureus phagosomes, as opposed to the approximate 40% decrease in translocation of p40phox-R58A,46 argue against phosphorylation of p40phox being upstream of its binding to PtdIns3P. In agreement with these observations, p40phox-T154A translocates to early endosomes after stimulation of the macrophage-like cell line RAW 264.7 with arachidonic acid, indicating that a p40phox unable to be phosphorylated on T154 can still bind to PtdIns3P-rich structures.16
Our observation that T154 phosphorylation plays a partial but important role in oxidase activation by stimuli that direct oxidase assembly on the plasma membrane (fMLP), internalized phagosomes (S aureus), or a combination of the 2 (IgG-SRBCs) suggests this event lies downstream of the differential signaling webs engaged by these stimuli (emanating from activation of G protein coupled receptors, toll-like receptors, integrins, or Fcγ receptors). This in turn suggests T154 phosphorylation may directly participate in the efficient assembly of the soluble oxidase subunits around cytochrome b558. In support of this concept, we found that T154A mutation in p40phox significantly reduced the accumulation of p47phox on S aureus phagosomes to a similar degree as it affected ROS formation. Given that p47phox is accepted to be a critical regulator of oxidase assembly and activity,8,10 it is possible that T154 phosphorylation on p40phox induces a more effective interaction between p47phox and the other subunits of the oxidase complex and, through this, regulates oxidase activity. Unfortunately, T154 resides in a flexible linker between the PX and SH3 domains of p40phox that is invisible in the existing crystallographic structure of the full-length protein,45 and thus it is difficult to speculate meaningfully as to how T154 phosphorylation might achieve this. Lopes and coworkers,25 as well as Bouin and coworkers,22 found that phosphorylation of p40phox on T154 conferred protection against cleavage by thrombin, suggesting that unphosphorylated and phosphorylated p40phox indeed differ in their conformation. However, we found that phosphorylation on T154 could not be mimicked by substitution with a glutamate residue, suggesting phosphorylation may provide additional contacts with other protein domains. Clearly, more work will need to be done to understand the mechanism by which T154 phosphorylation on p40phox regulates oxidase activity, but our data identify for the first time that this is an important physiological event.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Chris Ellson, Gordon John Ferguson, and Suhasini Kulkarni for helpful discussion and critical reading of the manuscript. We would also like to thank Tom Henley and Elena Vigorito for helpful advice and technical assistance, John Coadwell and Simon Andrews for bioinformatic support, and Anne Segonds-Pichon for statistical analysis. We also would like to thank Geoff Morgan and Arthur Davies for support with flow cytometry and staff in the small animal unit and small animal barrier unit at the Babraham Institute for animal husbandry.
This work was supported by the BBSRC and Wellcome Trust (WT085889MA). T.C. is a UCB BBSRC-CASE award student.
Authorship
Contribution: T.A.M.C. designed and performed experiments, analyzed and interpreted data, and wrote the manuscript; K.E.A. designed and performed experiments and analyzed and interpreted data; Y.H., Q.X., and O.R. provided valuable reagents; and L.R.S. and P.T.H. designed experiments, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no completing financial interests.
Correspondence: Phillip Hawkins, Inositide Laboratory, Babraham Institute, Babraham Research Campus, Cambridge CB22 3AT, United Kingdom; e-mail: phillip.hawkins@bbsrc.ac.uk.
References
Author notes
L.R.S. and P.T.H. contributed equally to this study.

![Figure 3. Phosphorylation of p40phox on T154, but not S315, is essential for NADPH oxidase activity in mouse neutrophils. (A) Lysates from p40phox−/− BMNs reconstituted with GFP (empty vector [ev]), wt p40phox (wt), p40phox-T154A (T154A), or p40phox-S315A (S315A) were subjected to SDS-PAGE and immunoblotted for phox components as described in “Methods.” A representative blot of 3 independent experiments is shown. Levels of p40phox and p67phox were normalized against levels of p47phox, which was used as a loading control. Histograms represent relative levels of p40phox (left) or p67phox (right) as a percentage of wt. Data are mean ± SEM (n = 3 independent experiments performed in duplicate). (B-E) p40phox−/− BMNs expressing GFP (empty vector [ev]; ○), wt p40phox (▾), p40phox-T154A (), or p40phox-S315A (■) were pre-incubated with luminol/HRP before addition to fMLP (B) or PMA (E) or with luminol before addition to S aureus (C) or IgG-SRBCs (D), as described in “Methods.” ROS responses were measured by chemiluminescence as described in “Methods.” Shown are profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for 3-6 independent experiments (mean ± SEM), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. **P = .006; ***P = .000; as determined by a paired Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/26/10.1182_blood-2010-08-300889/5/m_zh89991062730003.jpeg?Expires=1769102027&Signature=20cloFybPCDh8DRc1Y9xk~oIHTlNENqDtW6GVoEbkjlXaoI66zWpkdJfyNjaewuDjyW7vXCDcwZ0w15n0RviXBk1xR6h0E3zX57e2igq~wOAH5uuqkpPAY6OsRtoLkrNPeUEhy~5OtP2N0N-PHJmRvMelrFRtyJ2QcIhoO7b~Gc6rMtB3gVXoahTWPbpIXNEUyEBtDV~dHMoP0F80W1GhleYTW0JEhHWAexjmdl2LxzFmetK~ZNmqQroq2RMd5uPCbFB~EYYeMUiFPkMQo78nUkjRBBbB3asPJrigBHTPuu2rxVBsrXsKZzz1L4qizlG~8qPUa5Iymlczx-T9kJpDw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Mutation or deletion of the SH3 domain of p40phox does not impair ROS production in mouse neutrophils. (A) p40phox−/− BMNs reconstituted with GFP (empty vector [ev]), wt p40phox (wt), p40phox-ΔSH3 (ΔSH3), or p40phox-W207Y (W207Y) were sonicated, subjected to SDS-PAGE, and immunoblotted for phox components as described in “Methods.” A representative blot of 2 independent experiments is shown. Levels of p40phox and p67phox were normalized against levels of p47phox, which was used as a loading control. Histograms represent relative levels of p40phox (left) or p67phox (right) as a percentage of wt. Data are mean ± range (n = 2 independent experiments performed in duplicate). (B-E) p40phox−/− BMNs expressing GFP (ev; ○), wt p40phox (▾), p40phox-ΔSH3 (), or p40phox-W207Y (■) were used in assays for ROS production as described in Figure 3. Shown are through profiles of kinetics of ROS production, measured in RLU/s (mean ± range), from one representative experiment performed in duplicate, as well as total integrated ROS responses for a minimum of 3 independent experiments (mean ± SEM; ev: n = 8-9; wt: n = 9; ΔSH3: n = 9; W207Y: n = 3), expressed as a percentage of the response in p40phox−/− BMNs expressing wt p40phox. The shaded area highlights the p40phox-independent response. Where indicated, differences between means of wt and mutated versions of p40phox are statistically significant. +P = .009; as determined by a repeated measures ANOVA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/26/10.1182_blood-2010-08-300889/5/m_zh89991062730004.jpeg?Expires=1769102027&Signature=lQKyPrVS6CBXWToYKCLdyWXKgXfk69wkUhdDdS8wYZSSNWUiZdkSQVLZfEz43y170-HUNd9ZjVX1vDd6MIGktHjTzqNOOj2RzmV~Olbd1PcXe5UTq0T7-lxCIWxaW9kdSYvcKSb4kYMgzN1sGNU00TTdIvT0tqxY4wOLQKyg4NMuIcYsn7v7L6g6R~NwXwIukQ~OrLyZaDcx8QZJTDthtB9We2bXNTVexCirl-RXfpq4QjS9rwq6zsteLAtHGmw-QR5UXtOKM2J3H-M6ia~o7J18Xp3KnRgQs8WnSOx3Fnh~XLdW95f1BLEi-W~A0seLlA6N6Pp6iS09SlEprez6oA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)