Abstract
The present study investigated the function of p13, a mitochondrial protein of human T-cell leukemia virus type 1 (HTLV-1). Although necessary for viral propagation in vivo, the mechanism of function of p13 is incompletely understood. Drawing from studies in isolated mitochondria, we analyzed the effects of p13 on mitochondrial reactive oxygen species (ROS) in transformed and primary T cells. In transformed cells (Jurkat, HeLa), p13 did not affect ROS unless the cells were subjected to glucose deprivation, which led to a p13-dependent increase in ROS and cell death. Using RNA interference we confirmed that expression of p13 also influences glucose starvation-induced cell death in the context of HTLV-1–infected cells. ROS measurements showed an increasing gradient from resting to mitogen-activated primary T cells to transformed T cells (Jurkat). Expression of p13 in primary T cells resulted in their activation, an effect that was abrogated by ROS scavengers. These findings suggest that p13 may have a distinct impact on cell turnover depending on the inherent ROS levels; in the context of the HTLV-1 propagation strategy, p13 could increase the pool of “normal” infected cells while culling cells acquiring a transformed phenotype, thus favoring lifelong persistence of the virus in the host.
Introduction
Human T-cell leukemia virus type 1 (HTLV-1) is a complex retrovirus that infects an estimated 20 million people worldwide. HTLV-1 is the causative agent of adult T-cell leukemia/lymphoma (ATLL), an aggressive neoplasm of mature CD4+ T cells that is refractory to current therapies. ATLL arises in approximately 3% of HTLV-1–infected persons and is preceded by a decades-long clinical latency.
Many aspects of HTLV-1 replication, persistence, and pathogenesis remain incompletely understood (reviewed in Verdonck et al1 ). Studies so far have been focused primarily on the transcriptional activator Tax, which is essential for expression from the viral promoter, and the posttranscriptional factor Rex, which is required for expression of incompletely spliced viral transcripts. Tax also plays a critical role in T-cell transformation through its ability to deregulate the expression of a vast array of cellular genes and interfere with cell-cycle checkpoints, producing major alterations in cell proliferation and survival and promoting genetic instability (reviewed in Lairmore et al2 ). Indeed, expression of Tax in mouse thymocytes is sufficient for induction of T-cell leukemia/lymphoma.3 However, the contrast between the powerful oncogenic properties of Tax and the low prevalence and long latency of ATLL suggests the existence of mechanisms that limit the transforming potential of the virus and favor its lifelong persistence in the host in the absence of disease.
Recent studies indicate that the viral accessory proteins p12, p21, p30, HBZ, and p13 may also contribute to HTLV-1 replication and pathogenesis (reviewed in Nicot et al4 ). The present study is focused on p13, an 87-amino acid accessory protein that accumulates in mitochondria (reviewed in D'Agostino et al5 ). Studies of a p13-knockout virus showed that although the protein is dispensable for viral replication in cultured cells,6,7 it is required for establishing a persistent infection in a rabbit experimental model.8
Functional studies of p13 revealed that it induces mitochondrial fragmentation,9 inhibits proliferation of HeLa cells and Jurkat T cells,10,11 sensitizes Jurkat T cells to apoptosis triggered by ceramide and Fas ligand,10,11 and interferes with tumor growth in experimental models using HeLa cells and rat embryo fibroblasts transformed by the myc and ras oncogenes.11 Biophysical and biochemical analyses of a synthetic peptide spanning residues 9-41 (p139-41) indicated that this region folds into an amphipathic α-helix12 upon exposure to membrane-mimetic solutions and changes mitochondrial membrane permeability to K+.13 Additional in vitro assays carried out with full-length synthetic p13 and isolated mitochondria demonstrated that it triggers an inward K+ current that induces depolarization and activation of the electron transport chain; these changes are accompanied by increased mitochondrial ROS production, which along with membrane depolarization, lowers the opening threshold of the permeability transition pore.14
In the present study, we show that p13 increases ROS production in living cells. We also provide evidence that this change may have a distinct impact on cell survival and proliferation depending on the cell's inherent ROS levels, with activation predominating in normal resting T-cells and death-promoting effects in transformed cells. In the context of the HTLV-1 propagation strategy, p13 would provide a mechanism to increase the pool of “normal” infected cells while promoting the elimination of cells acquiring a transformed phenotype, thus favoring lifelong persistence of the virus in the host.
Methods
Cell culture
HeLa and 293T cells were maintained in Dulbecco modified Eagle medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS; Invitrogen), 100 units/mL penicillin, and 20 units/mL streptomycin. Jurkat T cells and the HTLV-1–infected cell line C91PL15 were maintained in RPMI 1640 containing 2 g/L D-glucose (Sigma-Aldrich) supplemented with 10% FCS and penicillin/streptomycin. For glucose deprivation experiments in HeLa cells the standard DMEM containing 4.5 g/L D-glucose was replaced by DMEM containing 1 g/L D-glucose (GIBCO) supplemented with 10% FCS and penicillin/streptomycin. Glucose deprivation experiments in Jurkat T cells and C91PL cells were carried out in glucose-free DMEM (GIBCO). All media contained or were supplemented with 2mM l-glutamine. Cell death assays in Jurkat cells were carried out in the presence of 10% FCS. DMEM formulations lacking phenol red were used in time-lapse confocal microscopy experiments.
Plasmids and lentiviral vectors
Eukaryotic expression plasmid pSGp13, derived from pSG5 (Stratagene), contained the SV40 promoter, rabbit β-globin intron, p13 coding sequence and SV40 polyadenylation signal/site.13 pcDNAp13-GFP, derived from pcDNA3.1 (Invitrogen) expressed p13 fused to enhanced green fluorescent protein (GFP) under the control of the human cytomegalovirus (HCMV) promoter. pcDNAp13mut-GFP contained mutations in the 4 critical arginine residues of the α-helical domain of p13 (R-Q). Plasmid mt-GFP-HA1, derived from pcDNA1, contained the HCMV promoter and expressed GFP modified with the mitochondrial targeting sequence of cytochrome oxidase subunit VIII and the HA1 epitope at its N terminus16 (referred to as mt-GFP in the text). The p13-expressing lentiviral vector pRRLp13 was constructed from pRRLsin.PPTs.hCMV.GFP.Wpre17 by substituting the HCMV promoter and GFP gene with the SV40 promoter, rabbit β-globin intron, and p13 coding sequence obtained from plasmid pSGp13. Lentiviral vector pRRLp13-mut contained substitutions of the 4 arginines in the amphipathic α-helical domain (R to A and L).13 The control vector pRRLΔ contained the SV40 promoter and β-globin intron obtained from pSG5. Restriction enzymes were purchased from New England Biolabs and Roche. Plasmids were purified by chromatography (QIAGEN).
Lentiviral stocks were prepared from transfected 293 T cells as described,17 using the protocol discussed here. Cells were seeded into 10 cm2 culture dishes at 2 to 3 × 106 cells/well and cotransfected 1 day later by calcium phosphate coprecipitation with pCMVΔR 8.74, which coded for the HIV-1 Gag and Pol proteins,18 pHIT-VSVG, coding for vesicular stomatitis virus glycoprotein,19 and either pRRLp13, pRRLp13-mut or pRRLΔ. After 24 hours, the cells were washed in DMEM and refed with complete DMEM. After an additional 24 hours, the culture supernatants were collected and centrifuged at 260g for 10 minutes to eliminate cells and debris. The supernatants were then filtered through a 0.45-μm diameter filter and ultracentrifuged for 2 hours at 76 220g in a Beckman Opti L-90 Ultracentrifuge. The pellet was resuspended in 1 mL of complete RPMI 1640 medium and used to infect 1 × 106 Jurkat cells seeded in 6-well plates; transduced cultures were expanded after 2 days. Experiments were carried out on 2 independently transduced bulk cultures to minimize possible effects resulting from clonal variants in the cell population. The presence of the transduced gene (p13 wild-type, p13-mut, or empty vector) was periodically tested by PCR of genomic DNA using primers specific for the SV40 promoter. Total RNA was treated with DNAase (Invitrogen) and reverse transcribed with Superscript II (Invitrogen) following the supplier's recommendations. p13 expression was assessed by real-time reverse transcriptase (RT)-PCR of total RNA as described previously using primer/probe sets spanning the exon 1-C splice junction (infected cells) or exon C (transduced cells).11 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control and was amplified with a kit from Applied Biosystems. Amplified products were quantified using standard curves constructed from 5-fold serial dilutions of the p13 and GAPDH amplicons.
Analysis of ROS production in living cells
ROS were measured using MitoTracker Red CM-H2XRos (Molecular Probes), referred to in this study as H2-MTR, which is a derivative of dihydro-X-rosamine that accumulates in mitochondria and produces a red fluorescent signal upon oxidation to MitoTracker Red CMXRos (MTR) by H2O2. Monoamine oxidases (MAO) were inhibited by using 2mM pargyline hydrochloride.
To measure mitochondrial ROS production in Jurkat cells transduced with lentiviral vectors, cultures were washed once with phosphate-buffered saline (PBS), resuspended at 600 000 cells/mL in DMEM with or without glucose, pretreated with 2mM pargyline for 15 minutes, and then labeled with 25nM H2-MTR for 15 minutes in the presence of pargyline.
Cells were then analyzed for ROS production using a Zeiss LSM510 confocal laser microscope equipped with a 37°C, 5% CO2 incubator using Helium Neon (543 nm) and Argon (488 nm) lasers. Laser intensity, pinhole aperture, and photomultiplier parameters were standardized to allow comparison of signals obtained in different samples. The mean fluorescent signal of MTR in mitochondria of individual cells was quantitated with the Zeiss “Histogram” software tool. The rate of ROS accumulation (shown in Figure 2) was calculated using the ΔF/F0 formula, where F0 is the fluorescent signal measured at beginning of the experiment, and ΔF is the difference in fluorescent signal measured at each individual time point subtracted of the F0 value. Mean and SE of ΔF/F0 recorded in at least 60 cells per time point per sample in 3 independent experiments were calculated. End-point fluorescence recordings collected after 30 minutes (shown in Figure 2D) represent mean and SE of at least 30 cells in randomly selected fields, from a total of 3 experiments.
To measure mitochondrial ROS production in HeLa cells, cultures transfected with plasmids expressing p13-GFP or mt-GFP were grown overnight in complete DMEM (standard or low glucose), pretreated with 2mM pargyline for 15 minutes and then incubated with 25nM H2-MTR for an additional 15 minutes. The MTR signal in transfected cells (identified by GFP fluorescence) was analyzed by laser scanning microscopy. Data accumulated from 3 experiments were used to plot mean fluorescence intensity (shown in Figure 3).
‘Blue death’ assay
The “blue death” assay to detect cell death was performed as previously described.20 Briefly, HeLa cells were transfected with plasmids expressing p13-GFP or mt-GFP using the calcium phosphate coprecipitation technique; the β-galactosidase (β-gal) expression vector CMV-β-gal was added as a control to identify transfected cells. To test for possible effects of GFP, additional transfections were carried out using pSGp13 (coding for untagged p13) or pBluescript (empty plasmid control; Stratagene), together with CMV-β-gal. After 8 hours, cells were washed to remove excess calcium phosphate precipitate and cultured for an additional 18 hours in standard or low-glucose DMEM, with or without 1M N-acetyl cysteine (NAC). To detect β-gal–expressing cells, cultures were then fixed with 0.05% glutaraldehyde in PBS for 5 minutes at room temperature, washed twice with PBS, and stained with X-gal solution (1 mg/mL 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside, 5mM K3Fe(CN)6, 5mM K4Fe(CN)6, 2mM MgCl2 prepared in PBS) at 37°C for at least 5 hours. Stained cultures were examined using an Olympus IX70 inverted microscope to count cells positive for β-gal (β-gal+ cells) that showed normal morphology versus those showing signs of death (ie, plasma membrane blebbing, shrinkage, chromatin condensation). The percentage of dead cells was calculated by dividing the number of dead β-gal+ cells by the total number of β-gal+ cells counted. Results from transfections using GFP-tagged versus untagged proteins were similar; therefore we combined these data to arrive at approximately 500 to 600 cells counted for each transfection combination. Specific cell death was calculated by the following formula: (percentage of cell death in low glucose − percentage of cell death in high glucose) / (100 − the percentage of cell death in high glucose) × 100.
Annexin V cell death assay
Jurkat T cells (6 × 105) were seeded into 12-well plates in 1.0 mL per well of complete DMEM with or without glucose and cultured for 24 hours. Where indicated, the cells were preincubated with 100μM NAC overnight. The annexin V assay was performed using an annexin V–Alexa Fluor 647 conjugate (Molecular Probes) according to the manufacturer's instructions. In brief, 300 000 cells were collected, centrifuged, and then resuspended in 200 μL of Annexin-binding buffer (10mM HEPES/RPMI 1640), followed by incubation with 1 μL of annexin V–Alexa Fluor 647 for 10 minutes at room temperature. Cells positive for annexin V were detected by flow cytometry using a FACSCalibur apparatus and CellQuest software (both BD Biosciences), with 30 000 ungated events examined for each sample. Specific cell death was calculated using the formula specified in “‘Blue death’ assay.”
Silencing of p13 expression
C91PL cells were washed in PBS and resuspended in aliquots of 4 × 106 cells in 120 μL buffer R (Digitalbio) supplemented with 3μM of a small interfering RNA (siRNA) against p13 (CCGGAGGACCUGCUAGACGGCGGAC), or a control siRNA (CCGGCGGAAGCCGUUCGACAGGGAC) alone or in combination with 3 μg of a plasmid expressing wild-type p13-GFP. Cells were electroporated with one 1410-V, 30-msec pulse using a Microporator (Digitalbio). Cells were then diluted in 4 mL of complete RPMI 1640 and cultured in 6-well plates. After 24 hours, 1 × 106 cells were lysed and total RNA was extracted and subjected to real-time RT-PCR as detailed in “Plasmids and lentiviral vectors” to verify silencing of p13. The remaining cells were counted and seeded in aliquots of 6 × 105 cells into 12-well plates in a total volume of 1 mL per well of DMEM with or without glucose. After 16 hours cells were collected, and cell death was analyzed by staining with propidium iodide (0.5 μg/mL) followed by flow cytometry. Specific cell death was calculated as specified in “‘Blue death’ assay.”
Analysis of primary peripheral blood T cells
Peripheral blood mononuclear cells (PBMCs) from healthy donors were isolated by Ficoll-Paque gradient centrifugation using a standard protocol. ROS levels were measured in PBMCs (stimulated or unstimulated) and Jurkat T cells by loading 500 000 cells with 25nM H2-MTR for 1 hour at 37°C in the presence of 20 ng/mL verapamil to inhibit multidrug resistance activity. MTR fluorescence was then analyzed by flow cytometry. For electroporation, PBMC were washed once in PBS and then resuspended in aliquots of 2 × 107 cells for each electroporation in 120 μL of buffer R (Digitalbio) and mixed with 5 μg of plasmids expressing wild-type p13-GFP, mutant p13-GFP, or mt-GFP. Cells were electroporated with one 1800-V, 30-msec pulse using a Microporator and then diluted in 4 mL of complete RPMI 1640 and cultured overnight. The next day, half of the cells were treated with Allophycocyanin (PHA, 10 μg/μL) for 48 hours and then stimulated with interleukin-2 (IL-2, 100 U/mL) for 24 hours. After this period, 5 × 105 cells from the stimulated and nonstimulated cultures were labeled with peridinin chlorophyll protein–conjugated anti–CD3 and antigen-presenting cell–conjugated anti–CD38 antibodies (Biolegend) according to the supplier's recommendations and analyzed by flow cytometry.
Results
Measuring mitochondrial ROS production in living cells
ROS levels in living cells were measured using H2-MTR, a probe that is converted to fluorescent MTR upon oxidation by H2O2. The fact that the probe is targeted to mitochondria makes the assay specific for mitochondrial ROS. To increase the specificity of the assay for ROS produced by the respiratory chain, we added pargyline, an inhibitor of the MAO, which are an additional source of mitochondrial ROS.14 The assay was initially setup in Jurkat T cells, a cell line derived from a CD4+ T-cell lymphoblastic leukemia21 extensively used to study receptor signaling and T-cell activation/death. The change in MTR fluorescence within regions of interest (ROI) over time was measured in situ by laser scanning microscopy. Results were expressed as MTR ΔF/F0 ratios, calculated as described in “Methods.” To test the specificity of the probe for ROS in our system we measured MTR fluorescence in response to treatments known to affect mitochondrial ROS production. As shown in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article), the MTR fluorescent signal increased over time; this effect was enhanced upon addition of exogenous H2O2 and was abrogated by the ROS scavenger Trolox. Oligomycin, which is known to increase ROS production by the electron transport chain, resulted in an increase in MTR fluorescence, whereas FCCP (protonophore carbonyl cyanide p-trifluoromethoxy-phenylhydrazone), which is expected to dampen ROS production, resulted in a decrease in MTR fluorescence.
Effect of p13 on ROS in Jurkat T cells
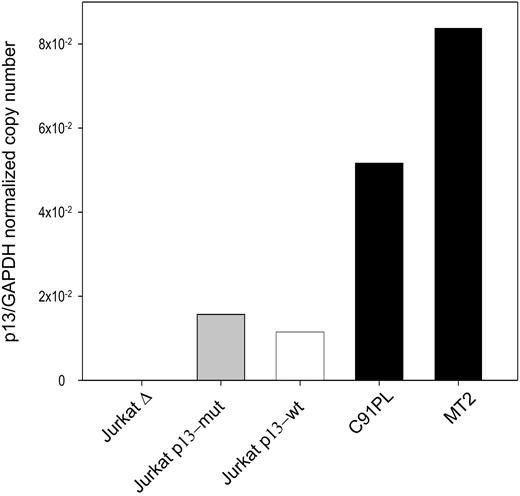
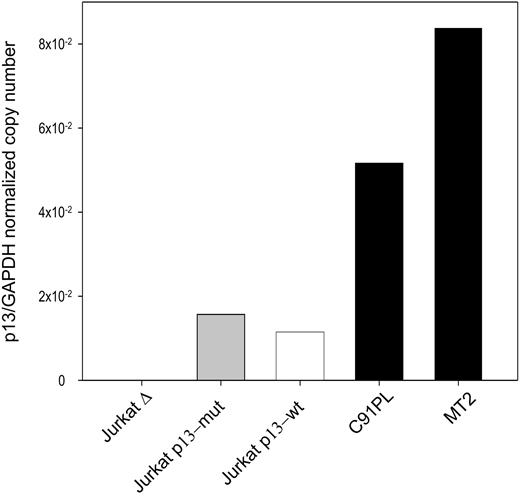
The effects of p13 on ROS in Jurkat T cells were tested in cells transduced with an “empty” lentiviral vector (Δ), a lentiviral vector expressing wild-type p13, or a lentiviral vector expressing a p13 mutant carrying substitutions in the critical arginine residues previously shown to be required to induce K+ influx and ROS production in the context of isolated mitochondria.14 Expression of the p13 transgene was assessed by quantitative real-time RT-PCR. Results showed that the expression levels obtained with this approach were lower than those measured in the HTLV-1 chronically infected cell lines C91PL and MT2 (Figure 1), thus indicating that p13 expression in this system did not exceed the levels achieved by the virus in chronically infected cells.
Real-time RT-PCR quantitation of p13 expression in Jurkat T cells transduced with lentiviral vectors expressing p13. Values on the y-axis represent the copy number of the p13 mRNA divided by the copy number of GAPDH (normalized copy number). The graph shows a comparison of the expression levels detected in HTLV-1–infected cell lines (MT2, C91PL) and Jurkat T-cell lines transduced with a lentiviral vector expressing wild-type p13, mutant p13, or with the empty vector (Δ).
Real-time RT-PCR quantitation of p13 expression in Jurkat T cells transduced with lentiviral vectors expressing p13. Values on the y-axis represent the copy number of the p13 mRNA divided by the copy number of GAPDH (normalized copy number). The graph shows a comparison of the expression levels detected in HTLV-1–infected cell lines (MT2, C91PL) and Jurkat T-cell lines transduced with a lentiviral vector expressing wild-type p13, mutant p13, or with the empty vector (Δ).
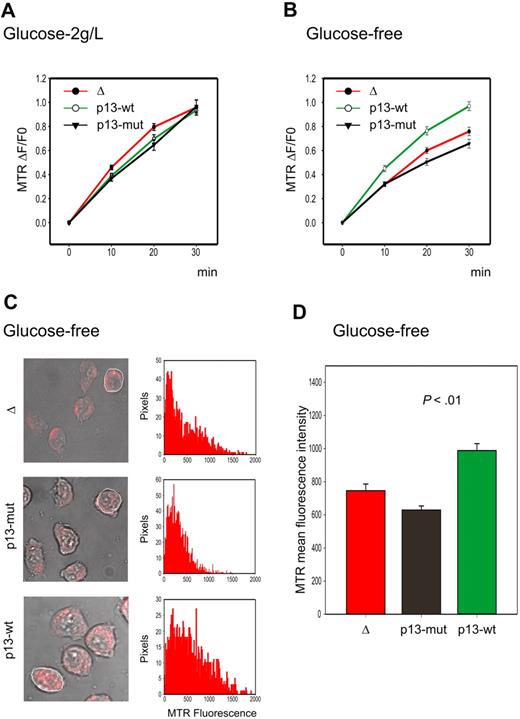
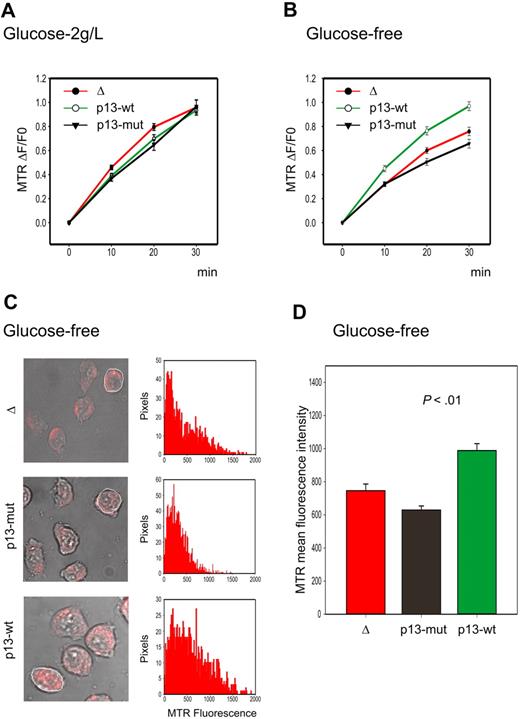
As shown in Figure 2A, Jurkat T cells expressing p13 did not produce more ROS than control cells when cultured under standard conditions. Although this result was unexpected based on previous results obtained in isolated mitochondria,14 it could be explained by recent studies indicating that high levels of ROS production may be counteracted, at least in part, by an up-regulation of the ROS scavenging enzymes. This situation is frequently found in tumor cells, which exhibit increased ROS production compared with normal cells derived from the same tissues. This change is accompanied by increased glucose consumption required to fuel the pentose phosphate pathway that reduces NADP+ to NADPH, which is required to regenerate ROS scavenging enzymes.22 We therefore reasoned that possible differences in ROS production in p13-expressing cells might be unmasked by glucose deprivation, resulting in increased ROS levels and possibly cell death. Indeed, glucose deprivation revealed a significant increase in the rate of ROS production in p13-expressing cells (Figure 2B), which resulted in increased ROS at the end-point of the experiment (P < .01, Student t test; Figure 2C-D). This difference was markedly reduced by the ROS scavenger NAC (data not shown).
Effects of p13 on ROS production in Jurkat cells. Jurkat T-cell lines transduced with a lentiviral vector expressing wild-type p13, mutant p13, or empty vector (Δ) were cultured in standard medium containing 2 g/L glucose or in glucose-free medium and loaded with the H2O2 probe MitoTracker Red H2-MTR. MTR signals were analyzed by confocal microscopy and quantitated in regions of interest (ROIs) using the Histogram software in a time-lapse experiment of 30 minutes. (A-B) Plots show the fractional rate of accumulation of MTR (ΔF/F0, as described in “Analysis of ROS production in living cells”) over time. Data represent mean and SE of ΔF/F0 recorded in at least 60 cells per time point per sample in 3 independent experiments. (C) Representative end-point fluorescence recordings and an example of the quantitation of the MTR signals over the indicated ROI. This analysis was carried out on at least 30 cells in randomly selected fields, with data collected after 30 minutes from a total of 3 experiments. (D) Presents the resulting mean MTR fluorescence values with SE bars. The effects of p13 on ROS were specifically triggered in response to glucose deprivation (p13-wt compared with either Δ or p13-mut, P < .01 by Student t test).
Effects of p13 on ROS production in Jurkat cells. Jurkat T-cell lines transduced with a lentiviral vector expressing wild-type p13, mutant p13, or empty vector (Δ) were cultured in standard medium containing 2 g/L glucose or in glucose-free medium and loaded with the H2O2 probe MitoTracker Red H2-MTR. MTR signals were analyzed by confocal microscopy and quantitated in regions of interest (ROIs) using the Histogram software in a time-lapse experiment of 30 minutes. (A-B) Plots show the fractional rate of accumulation of MTR (ΔF/F0, as described in “Analysis of ROS production in living cells”) over time. Data represent mean and SE of ΔF/F0 recorded in at least 60 cells per time point per sample in 3 independent experiments. (C) Representative end-point fluorescence recordings and an example of the quantitation of the MTR signals over the indicated ROI. This analysis was carried out on at least 30 cells in randomly selected fields, with data collected after 30 minutes from a total of 3 experiments. (D) Presents the resulting mean MTR fluorescence values with SE bars. The effects of p13 on ROS were specifically triggered in response to glucose deprivation (p13-wt compared with either Δ or p13-mut, P < .01 by Student t test).
Effect of p13 on ROS in HeLa cells
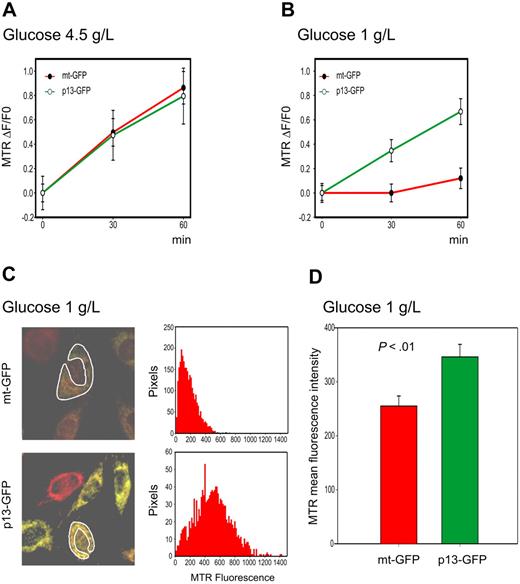
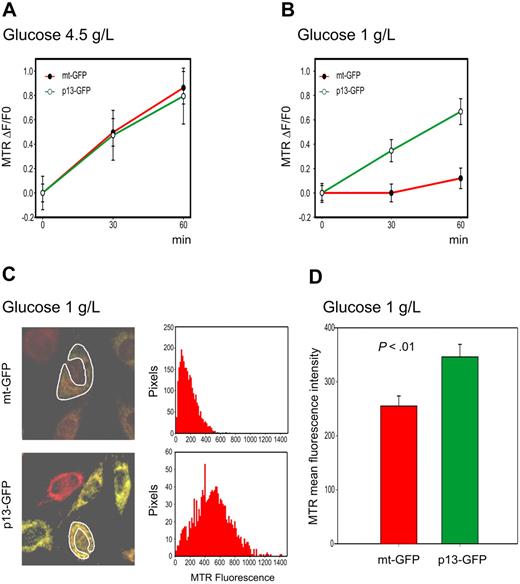
We also used the H2-MTR assay to follow the kinetics of ROS production using time-lapse microscopy in HeLa cells transiently transfected with p13-GFP or with the control mitochondrial-targeted GFP fusion protein mt-GFP (Figure 3). Similar to observations made in Jurkat T cells, results revealed no significant difference in the rate of ROS accumulation in p13-expressing HeLa cells versus mt-GFP controls (Figure 3A). However, the rate of H2O2 accumulation in HeLa cells subjected to glucose deprivation was significantly higher in p13-GFP–expressing cells than in control cells expressing mt-GFP (P < .01, Student t test; Figure 3B). As expected, this difference in the rate of ROS production in p13-expressing cells after glucose deprivation resulted in a significant increase in ROS levels at the 30-minute end-point measurement (Figure 3C-D). The addition of the ROS scavenger NAC produced a marked reduction in MTR fluorescence (data not shown).
Effects of p13 on ROS production in HeLa cells. HeLa cells were transfected with plasmids expressing p13-GFP or mt-GFP and cultured in standard (4.5 g/L) or low (1 g/L) glucose medium and loaded with MitoTracker Red H2-MTR. MTR signals were analyzed by confocal microscopy and quantitated in regions of interest (ROI) using the Histogram software tool in a time-lapse experiment of 30 minutes. (A-B) Plots show the fractional rate of accumulation of MTR (ΔF/F0, as described in “Analysis of ROS production in living cells”) over time. Data represent mean and SE of ΔF/F0 recorded in at least 60 cells per time point per sample in 3 independent experiments. (C) Representative end-point fluorescence recordings and an example of the quantitation of the MTR signals over the indicated ROI. This analysis was carried out on at least 30 cells in randomly selected fields, with data collected after 30 minutes from a total of 3 experiments. (D) Presents the resulting mean MTR fluorescence values and SE bars. The effects of p13 on ROS were specifically triggered in response to glucose deprivation (P < .01 by Student t test).
Effects of p13 on ROS production in HeLa cells. HeLa cells were transfected with plasmids expressing p13-GFP or mt-GFP and cultured in standard (4.5 g/L) or low (1 g/L) glucose medium and loaded with MitoTracker Red H2-MTR. MTR signals were analyzed by confocal microscopy and quantitated in regions of interest (ROI) using the Histogram software tool in a time-lapse experiment of 30 minutes. (A-B) Plots show the fractional rate of accumulation of MTR (ΔF/F0, as described in “Analysis of ROS production in living cells”) over time. Data represent mean and SE of ΔF/F0 recorded in at least 60 cells per time point per sample in 3 independent experiments. (C) Representative end-point fluorescence recordings and an example of the quantitation of the MTR signals over the indicated ROI. This analysis was carried out on at least 30 cells in randomly selected fields, with data collected after 30 minutes from a total of 3 experiments. (D) Presents the resulting mean MTR fluorescence values and SE bars. The effects of p13 on ROS were specifically triggered in response to glucose deprivation (P < .01 by Student t test).
Taken together, these results demonstrated that p13 increased ROS levels in response to glucose deprivation in both Jurkat and HeLa cells.
Increased death of p13-expressing cells in response to glucose deprivation
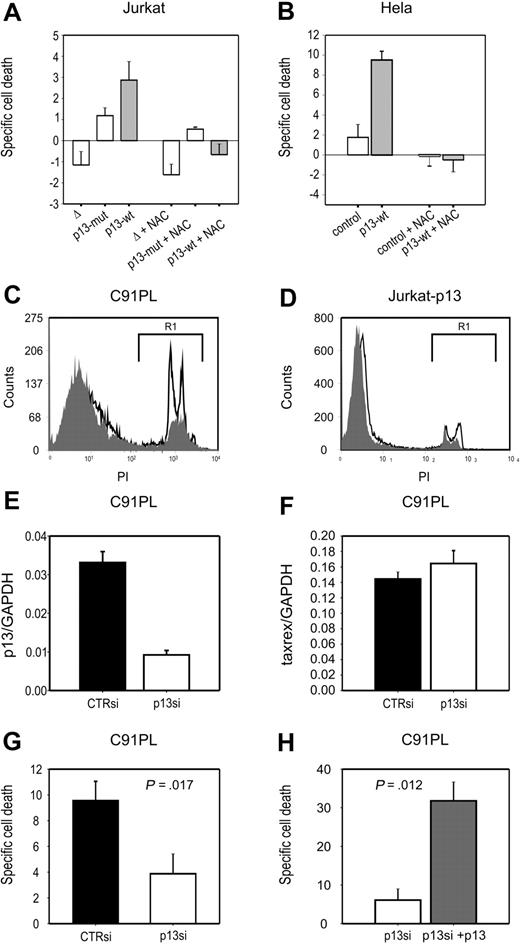
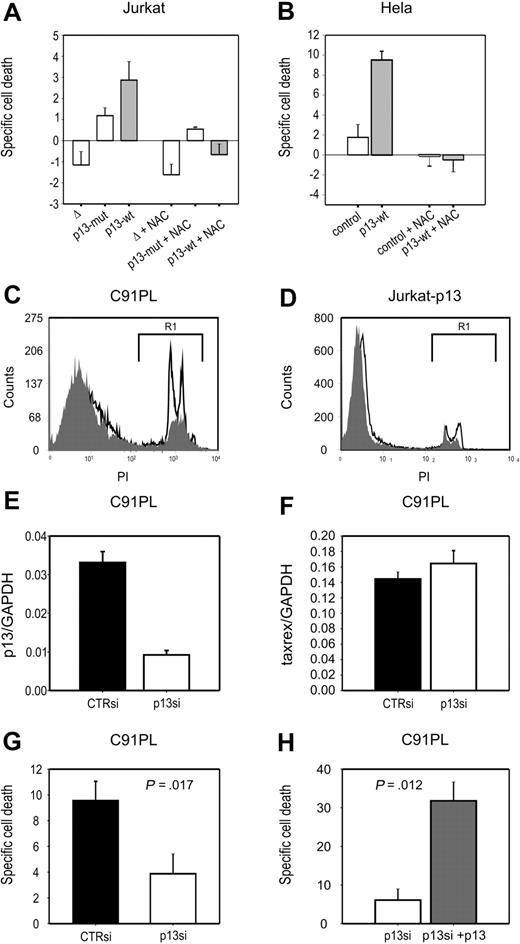
We next tested whether the effects of p13 on ROS in cells in response to glucose deprivation might result in cell death. To this end, we assessed cell viability after short-term (24 hours) culture in standard versus glucose deprivation conditions. Death specifically triggered by glucose deprivation (specific cell death) was calculated as described in “Methods.” Results showed that p13 expression induced a significant increase (P < .01) in specific cell death both in Jurkat T cells (∼ 3-fold; Figure 4A) and HeLa cells (∼ 5-fold; Figure 4B).
p13 increases cell death of tumor cell lines in response to glucose deprivation. Specific death of Jurkat (A) and HeLa (B) cells in response to glucose deprivation was calculated as described in “Analysis of ROS production in living cells.” In both cell types, p13 expression significantly enhanced cell death induced by glucose deprivation. Specific cell death triggered by p13 in response to glucose depletion was abrogated by treatment with NAC. (C-D) Comparison of the levels of cell death detected in the HTLV-1–infected cell line C91PL (C) and in p13-expressing Jurkat cells (D) in the presence (gray histogram) or absence (open histogram) of glucose. Dead cells measured in the gate areas R1 were 16.84% in the presence of glucose and 37.83% in the absence of glucose for C91PL cells, and 10.01% in the presence of glucose, and 17.10% in the absence of glucose for Jurkat-p13 cells. (E-F) Results of real-time RT-PCR to quantitate p13 (E) and tax/rex (F) mRNA copy number (normalized for GAPDH), in response to treatment with p13 siRNA. (G) The effects of p13 silencing on glucose deprivation–induced C91PL cell death. (H) The reversion of the effects of p13 siRNA upon cotransfection of a plasmid expressing p13-GFP. Indicated P values were calculated with the Mann-Whitney test.
p13 increases cell death of tumor cell lines in response to glucose deprivation. Specific death of Jurkat (A) and HeLa (B) cells in response to glucose deprivation was calculated as described in “Analysis of ROS production in living cells.” In both cell types, p13 expression significantly enhanced cell death induced by glucose deprivation. Specific cell death triggered by p13 in response to glucose depletion was abrogated by treatment with NAC. (C-D) Comparison of the levels of cell death detected in the HTLV-1–infected cell line C91PL (C) and in p13-expressing Jurkat cells (D) in the presence (gray histogram) or absence (open histogram) of glucose. Dead cells measured in the gate areas R1 were 16.84% in the presence of glucose and 37.83% in the absence of glucose for C91PL cells, and 10.01% in the presence of glucose, and 17.10% in the absence of glucose for Jurkat-p13 cells. (E-F) Results of real-time RT-PCR to quantitate p13 (E) and tax/rex (F) mRNA copy number (normalized for GAPDH), in response to treatment with p13 siRNA. (G) The effects of p13 silencing on glucose deprivation–induced C91PL cell death. (H) The reversion of the effects of p13 siRNA upon cotransfection of a plasmid expressing p13-GFP. Indicated P values were calculated with the Mann-Whitney test.
Treatment with the ROS scavenger NAC produced a marked general decrease in the levels of glucose deprivation–induced specific cell death in both cell lines. Interestingly, in p13-expressing cells the antioxidant reduced death to levels below those observed under standard culture conditions, as indicated by the negative specific cell death values.
Effects of p13 silencing on cell death in chronically infected cells
To test whether p13 affected cell death in the context of the complete viral genome, we carried out glucose starvation experiments with the HTLV-1-infected cell line C91PL. Figure 4C and D showed C91PL cells were more sensitive to glucose starvation–induced cell death (Figure 4C) compared with Jurkat-p13 cells (Figure 4D); this effect was ROS dependent, as it was completely abrogated by the ROS scavenger NAC (data not shown). The increased sensitivity of the infected cells to this treatment may reflect the higher levels of p13 expression with respect to the p13-transduced Jurkat cells (Figure 1).
We next tested the link between p13 expression and glucose starvation–induced cell death by knocking down p13 expression in C91PL cells with siRNAs. However, these experiments were complicated by the fact that the open reading frame coding for p13 overlaps with those of Tax, Rex, and HBZ. To achieve specific silencing for p13 while minimizing possible off-target effects on other viral transcripts, we designed siRNAs spanning the splice junction of the mRNA encoding p13 (singly spliced mRNA 1-C). Results of electroporation and real-time RT-PCR assays showed that one of the siRNAs inhibited p13 expression in C91PL cells (∼ 3.5-fold reduction; Figure 4E). As shown in Figure 4G, silencing of p13 significantly reduced cell death of C91PL cells in response to glucose deprivation (P = .017). The p13 siRNA had no effect on the expression of the tax/rex mRNA (Figure 4F). This is important because Tax is the major factor controlling overall viral expression and apoptosis, whereas Rex controls the balance of expression among the different viral genes. Conversely, we observed some off-target inhibition of the env and HBZ transcripts (data not shown). This inhibition would not be expected to influence the results of the experiment as neither Env nor HBZ is reported to affect cell death. Nevertheless, as a further control for the specificity of the effect for p13, we reintroduced the protein in p13-silenced C91PL, and observed restored sensitivity to glucose deprivation–induced cell death (Figure 4H). This result strongly suggests that p13 is responsible for the sensitivity of the infected cells to glucose deprivation.
Measurement of mitochondrial ROS in primary versus transformed T cells
ROS are key second messengers that may act as a “rheostat”23 with respect to cell turnover. According to this model, low levels of ROS are found in resting normal cells and moderate levels are found in activated/cycling cells, whereas high ROS levels trigger cell death. Interestingly, many tumor cell types exhibit higher levels of ROS compared with their normal counterparts.22 Application of this model to T-cell turnover and transformation leads to the prediction that p13 might exert distinct effects in normal T cells versus transformed T cells, as these cells would be expected to have different basal levels of ROS.
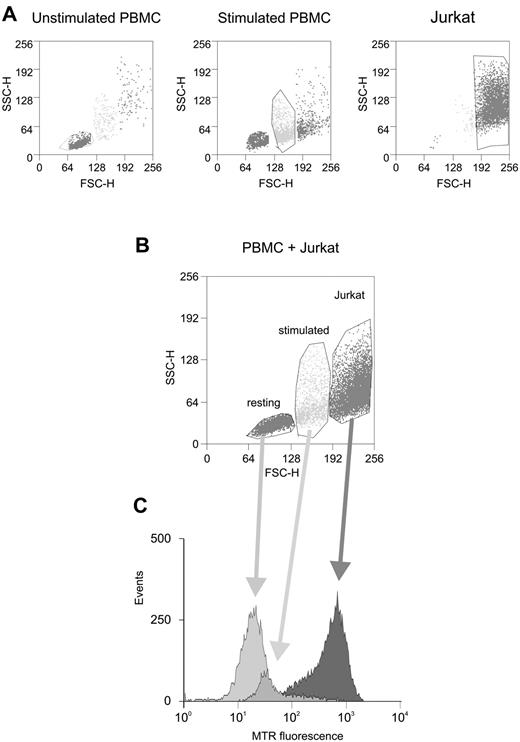
Figure 5 shows an experiment comparing ROS levels in Jurkat cells and in PBMC that were cultured for 3 days in the absence or presence of PHA and IL-2 to activate T cells. Control labeling assays with the T cell–specific marker CD3 confirmed that our PBMC preparations contained 70% to 80% T cells (data not shown). Flow cytometric analysis of the cells' forward scatter (FSC) and side scatter (SSC) properties revealed that the PBMC contained a single major population whose morphology shifted upon stimulation (Figure 5A left and middle panels). The Jurkat culture also contained a predominant population with distinct light scattering properties (Figure 5A right panel). Given that the 3 cell populations could be easily distinguished morphologically, we chose to mix them immediately before labeling with H2-MTR and added verapamil to inhibit the multidrug resistance pump, whose activity is expected to be increased in tumor cells. Analysis of the MTR signals in the 3 cell populations (Figure 5B-C) revealed that the stimulated cells showed an increase in ROS compared with unstimulated controls. Furthermore, ROS levels were substantially higher in Jurkat cells compared with either of the primary cell populations. These observations support the “rheostat” model in the context of primary T cells and Jurkat T cells.
Comparison of ROS levels in resting and activated primary PBMC versus Jurkat cells. (A) Morphologic properties of primary resting PBMC (left), mitogen-activated PBMC (middle), and Jurkat cells (right) analyzed by flow cytometry (FSC-SSC scatter plots); indicated are the gate areas containing most of the cell populations. (B) A combined FSC-SSC scatter plot of these cell populations. (C) ROS levels (measured with H2-MTR) in the 3 gate areas corresponding to primary PBMC (resting or mitogen-activated) and Jurkat cells. Results indicated that mitogen-induced activation was associated with increasing levels of ROS and that Jurkat cells exhibited markedly increased ROS levels compared with primary cells.
Comparison of ROS levels in resting and activated primary PBMC versus Jurkat cells. (A) Morphologic properties of primary resting PBMC (left), mitogen-activated PBMC (middle), and Jurkat cells (right) analyzed by flow cytometry (FSC-SSC scatter plots); indicated are the gate areas containing most of the cell populations. (B) A combined FSC-SSC scatter plot of these cell populations. (C) ROS levels (measured with H2-MTR) in the 3 gate areas corresponding to primary PBMC (resting or mitogen-activated) and Jurkat cells. Results indicated that mitogen-induced activation was associated with increasing levels of ROS and that Jurkat cells exhibited markedly increased ROS levels compared with primary cells.
Effect of p13 on activation of primary T cells
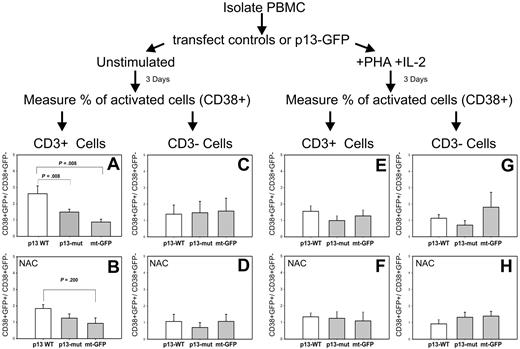
We next transfected PBMC with mt-GFP, wild-type p13-GFP, or mutant p13-GFP. Cells were cultured in the absence or presence of PHA and IL-2 for 3 days. T-cell activation was assessed by flow cytometry by measuring the percentage of activated T cells in the transfected (GFP-positive) and untransfected (GFP-negative) populations, using CD38 as an activation marker and CD3 as a marker for T cells. The possible involvement of ROS in the system was assessed by treating cultures with the ROS scavenger NAC; direct measurement of ROS using H2-MTR in this experimental setup was precluded because of the partial overlap of the GFP and MTR signals.
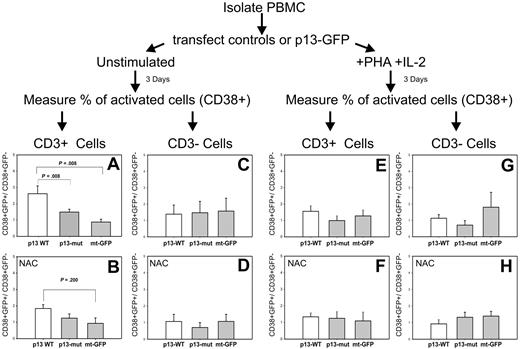
Figure 6 shows the ratio of activated (CD38+) cells in the transfected (GFP+) versus untransfected (GFP−) cells. Results showed that expression of WT-p13 resulted in a significant activation of T cells in the unstimulated PBMC culture (Figure 6A); in contrast, no effect of p13 was observed in T cells in the culture exposed to PHA and IL-2 (Figure 6E). Furthermore, mutant p13 did not activate T cells (Figure 6A). Addition of the ROS scavenger NAC abolished the ability of p13 to activate T cells (Figure 6B). Taken together, these observations demonstrated that p13 activated primary T cells through a mechanism linked with its effects on mitochondrial K+ influx and ROS. No effect on cell activation was seen in CD3-negative cells (Figure 6C), suggesting that the effects of p13 were highly specific for resting T cells.
Effects of p13 in primary T cells. Freshly isolated PBMCs were electroporated with plasmids expressing mt-GFP, wild-type p13-GFP, or mutant p13-GFP. Resting or mitogen-activated PBMCs were cultured for 3 days and then analyzed by flow cytometry to assess activation by measuring the percentage of activated (CD38+) in the transfected (GFP-positive) and untransfected populations. T cells were identified by staining with anti–CD3 antibody. (A) Expression of WT-p13 resulted in a highly significant activation of resting T cells. This effect was lost with mutant p13 (A) and in the presence of ROS scavengers (B). In contrast no effect of p13 was observed in mitogen-activated T cells (E-F), or in non-T cells (C,D,G,H). Reported are mean values and error bars from 5 independent experiments (in the absence of NAC) and 3 independent experiments (in the presence of NAC). Indicated P values were calculated with the Mann-Whitney test.
Effects of p13 in primary T cells. Freshly isolated PBMCs were electroporated with plasmids expressing mt-GFP, wild-type p13-GFP, or mutant p13-GFP. Resting or mitogen-activated PBMCs were cultured for 3 days and then analyzed by flow cytometry to assess activation by measuring the percentage of activated (CD38+) in the transfected (GFP-positive) and untransfected populations. T cells were identified by staining with anti–CD3 antibody. (A) Expression of WT-p13 resulted in a highly significant activation of resting T cells. This effect was lost with mutant p13 (A) and in the presence of ROS scavengers (B). In contrast no effect of p13 was observed in mitogen-activated T cells (E-F), or in non-T cells (C,D,G,H). Reported are mean values and error bars from 5 independent experiments (in the absence of NAC) and 3 independent experiments (in the presence of NAC). Indicated P values were calculated with the Mann-Whitney test.
Discussion
Mitochondria exert powerful control on cell proliferation and death through multiple pathways. Among these, the fine-tuning of mitochondrial ROS production has been proposed to act as a “rheostat” controlling cell fate in different lineages (reviewed in Rustin23 ). In the context of the immune system, life and death of T cells is determined by engagement of the T cell–receptor complex (reviewed by Krammer et al,24 Hildeman et al,25 and Bouillet and O'Reilly26 ), which through the generation of ROS, controls distinct redox-sensitive pathways, favoring apoptosis (mainly through Fas ligand and Bim) or proliferation (through extracellular signal-regulated kinases).27 The observation that the HTLV-1 p13 protein induced ROS production in the context of isolated mitochondria14 led us to test the effects of the protein on the redox state of transformed and primary T cells.
Our measurements of mitochondrial ROS in primary T cells versus transformed Jurkat T cells revealed a gradient of ROS, with very low levels in resting T cells, higher levels in stimulated T cells, and substantially higher levels in Jurkat T cells (Figure 5). Expression of p13 in these populations had distinct effects. In Jurkat T cells, p13 increased both ROS and cell death when cultures were subjected to glucose deprivation (Figures 2,4). In the context of primary T cells, p13 exhibited an activating effect on resting cells, whereas no effect was observed in mitogen-activated cells (Figure 6). Although technical limitations precluded direct measurement of ROS in the primary T cells expressing p13, the observation that p13-induced activation was abrogated in the presence of ROS scavengers demonstrates that this effect was ROS-dependent. By raising ROS levels in the context of resting T cells, p13 thus can be seen as mimicking receptor-mediated activation.
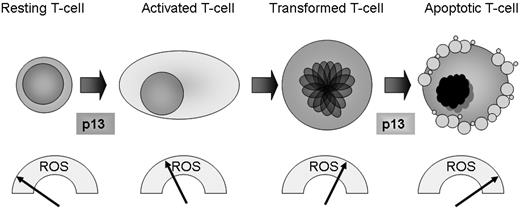
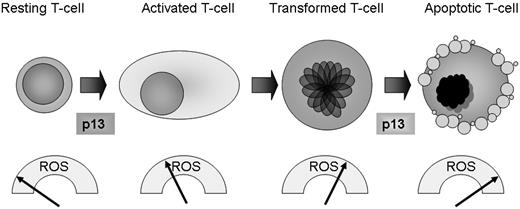
These observations were incorporated into the working model shown in Figure 7. Translating these findings in the context of the HTLV-1 life cycle, it is tempting to speculate that, by raising ROS levels in primary T cells, p13 might exert a positive role during the initial stages of the HTLV-1 life cycle by triggering activation and expansion of the infected cell pool (thus cooperating with the Tax oncoprotein). In contrast, in the context of transformed T cells that already have elevated ROS levels, p13 might have a death-promoting effect that could counteract the action of Tax, which is known to protect cells from apoptosis induced by mitochondria-mediated death stimuli.20,28 Our observations made in glucose-starved Jurkat cells suggest that the death-promoting activity of p13 might emerge in response to metabolic stress, for example when HTLV-1–infected cells are confined in ischemic environments such as solid lymphomatous masses in the context of ATLL and/or when trafficking through avascular areas of lymphoid organs. This hypothesis is consistent with previous findings that p13 inhibits the growth of experimental tumors in mice11 and increases the sensitivity of Jurkat and HeLa cells to apoptosis induced by ceramide,10,11 which induces mitochondrial generation of H2O2.29
A working model for redox regulation of T-cell turnover by p13. Data in the present study revealed a gradient of ROS in primary versus transformed T cells, with very low levels in resting cells, higher levels in stimulated cells, and substantially higher levels in transformed Jurkat cells. By increasing mitochondrial ROS production, p13 favors activation of primary resting T cells while promoting death of transformed T cells. These findings suggest that p13 may have a distinct impact on cell turnover depending on the inherent ROS levels, and that in the context of the HTLV-1 propagation strategy, p13 could increase the pool of “normal” infected cells while culling transformed T cells, thus favoring lifelong persistence of the virus in the host.
A working model for redox regulation of T-cell turnover by p13. Data in the present study revealed a gradient of ROS in primary versus transformed T cells, with very low levels in resting cells, higher levels in stimulated cells, and substantially higher levels in transformed Jurkat cells. By increasing mitochondrial ROS production, p13 favors activation of primary resting T cells while promoting death of transformed T cells. These findings suggest that p13 may have a distinct impact on cell turnover depending on the inherent ROS levels, and that in the context of the HTLV-1 propagation strategy, p13 could increase the pool of “normal” infected cells while culling transformed T cells, thus favoring lifelong persistence of the virus in the host.
Results of p13 knock-down experiments in the chronically infected, transformed T-cell line C91PL indicate that the effects of p13 on glucose starvation-induced cell death are relevant in the context of the complete provirus (ie, with “natural” expression levels and expression of other viral genes).
When integrated together, the activities of p13 described could favor lifelong persistence of the virus in the host in the absence of disease. This hypothesis is consistent with the low frequency and long clinical latency of ATLL and with results of experiments carried out in a rabbit experimental model showing that p13 is required to establish a persistent infection in vivo.8
The fact that p13 did not activate primary CD3-negative cells is interesting and suggests that p13 might also play a role in restricting HTLV-1 tropism to T cells in vivo, a poorly understood hallmark of HTLV-1 biology that is in apparent contrast with the ability of HTLV-1 to infect many cell types in tissue culture experiments.
Recent studies have revealed expression of mitochondrial proteins by many viruses, including all of the human tumor viruses.30,31 Among these, the HBx protein of hepatitis B virus is of particular interest in the context of the present study. This multifunctional protein shows partial targeting to mitochondria, where it disrupts mitochondrial morphology,32-34 reduces membrane potential,35 alters calcium homeostasis,33 reduces the levels of enzymes involved in oxidative phosphorylation and electron transport, increases ROS,36 and renders cells more sensitive to apoptosis.33,36,37 These effects of HBx on cell death are in apparent contrast with the oncogenic properties of the protein revealed by studies both in transgenic mice38,39 and in in vitro transformation models. Indeed, in other studies, HBx was reported to exert antitumor effects as a result of the induction of apoptosis.40,41 These apparently conflicting data suggest that, in analogy with the findings described in this study for p13, the effects of HBx on cell turnover and tumor transformation might be linked to a rheostat-like modulation of ROS or depend on additional factors such as cell type and expression of other genes.
The accumulated data describing the biologic properties of p13 and the analogies with proteins coded by other tumor viruses reinforce the emerging concept that manipulation of mitochondrial functions and ROS signaling is a key point of convergence in tumor virus replication strategies. Further investigations of p13 and its interactions with mitochondria may therefore provide important insights into the mechanisms of HTLV-1 replication and pathogenesis and could aid in the discovery of new targets for antitumor therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Enrica Cannizzaro, Paolo Bernardi, and Stefano Indraccolo for discussions and reagents.
This work was supported by grants from the European Union (“The role of chronic infections in the development of cancer,” contract no. 2005-018704), the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Ministero per l'Università e la Ricerca Scientifica, e Tecnologica Progetti di Ricerca di Interesse Nazionale (PRIN), the Ministero della Salute (project RFPS-2006-2-342-010), and the University of Padova. N.V. was supported by a fellowship from AIRC.
Authorship
Contribution: M.S.-B. carried out all experiments in primary PBMCs and Jurkat cells. N.V. and D.S. carried out experiments in HeLa cells. I.C. quantitated p13 expression in Jurkat cells and HTLV-1–infected cell lines. S.V. provided technical assistance for the assays to measure ROS in living cells. F.D.L. helped design the assays to measure ROS in living cells. L.C.-B., D.M.D. and V.C. designed the experiments and prepared the manuscript. All authors contributed to the analysis and interpretation of the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vincenzo Ciminale, Dipartimento di Scienze, Oncologiche e Chirurgiche, Università di Padova, Via Gattamelata 64, 35128 Padova, Italy; e-mail: v.ciminale@unipd.it.