Abstract
Expression of inhibitory killer cell immunoglobulin-like receptors (KIRs) specific for self–major histocompatibility complex (MHC) class I molecules provides an educational signal that generates functional natural killer (NK) cells. However, the effects of activating KIRs specific for self-MHC class I on NK-cell education remain elusive. Here, we provide evidence that the activating receptor KIR2DS1 tunes down the responsiveness of freshly isolated human NK cells to target cell stimulation in donors homozygous for human leukocyte antigen (HLA)–C2, the ligand of KIR2DS1. The tuning was apparent in KIR2DS1+ NK cells lacking expression of inhibitory KIRs and CD94/NKG2A, as well as in KIR2DS1+ NK cells coexpressing the inhibitory MHC class I–specific receptors CD94/NKG2A and KIR2DL3, but not KIR2DL1. However, the tuning of responsiveness was restricted to target cell recognition because KIR2DS1+ NK cells responded well to stimulation with exogenous cytokines. Our results provide the first example of human NK-cell education by an activating KIR and suggest that the education of NK cells via activating KIRs is a mechanism to secure tolerance that complements education via inhibitory KIRs.
Introduction
Natural killer (NK)–cell function is regulated by inhibitory and activating receptors that recognize cell-bound ligands. Human and murine NK cells express major histocompatibility complex (MHC) class I–specific receptors, which on ligation inhibit NK-cell effector functions.1-4 These inhibitory receptors allow NK cells to recognize cells whose expression of self-MHC class I molecules is down-regulated, for example, as a consequence of viral infection or tumor transformation, a phenomenon referred to as “missing-self” recognition.5 Most of the inhibitory MHC class I–specific receptors belong to the killer cell immunoglobulin-like receptor (KIR) family in humans and the Ly49 receptor family in mice.6 In addition, humans as well as mice express the C-type lectin-like inhibitory receptor CD94/NKG2A, which recognizes HLA-E7,8 and Qa1-b,9 respectively.
Data accumulated over the last 15 years indicate that NK cells, like T and B cells, undergo an educational process that ensures the formation of a functional yet self-tolerant NK-cell repertoire. It is now well established that the ligands for inhibitory MHC class I–specific receptors, for example, KIR2DL1/2,10 KIR3DL1,11,12 and NKG2A12-14 in humans, and Ly49 receptors in mice,15-17 must be expressed in the host in order for the NK cells expressing these receptors to be fully responsive to target cell stimulation. Conversely, NK cells lacking inhibitory receptors specific for MHC class I are hyporesponsive to target cell stimulation.10,18,19 The MHC class I–dependent education of NK cells via inhibitory receptors has been termed “licensing,”16 but the mechanisms underlying this educational process remain unknown.
In addition to inhibitory KIRs, human NK cells can also express activating KIRs. Several of the activating KIRs have at a genetic level been associated with clinical outcomes of infectious diseases,20 autoimmunity,21-23 and transplantation.24-26 Activating KIRs lack immunoreceptor tyrosine-based inhibition motifs and instead associate with adaptor proteins, such as DAP-12/KARAP, containing immunoreceptor tyrosine-based activation motifs.27,28 The extracellular domains of many activating KIRs are highly homologous to their inhibitory counterparts, probably resulting from evolution by gene duplication.29 However, despite the homology to inhibitory KIRs, it has been difficult to demonstrate interactions between activating KIRs and human leukocyte antigen (HLA) class I molecules. One exception is KIR2DS1, which has been shown to interact with group 2 HLA-C molecules (HLA-C2), both in binding studies with KIR2DS1 fusion proteins30-32 and in functional assays with in vitro expanded human KIR2DS1+ NK cells and clones.30,31,33-35
Because inhibitory KIRs have an established role in the education of NK cells, we hypothesized that their activating counterparts may also influence NK-cell education when the appropriate ligands are present. Support for a role of activating NK-cell receptors in NK-cell education can be found in recent murine studies where forced expression of virally encoded or stress-induced ligands during development rendered NK cells expressing receptors specific for these ligands hyporesponsive to stimulation.36-39 Using KIR2DS1 as a model for activating KIRs, we investigated whether the expression of activating KIRs in the presence or absence of their endogenous, ubiquitously expressed ligands could affect NK-cell education. To this end, we designed a multicolor flow cytometry strategy that allowed us to analyze KIR2DS1 expression on NK cells in conjunction with the 4 major inhibitory KIRs and NKG2A. Our results demonstrate that NK cells expressing KIR2DS1 are hyporesponsive against cellular targets from donors homozygous for HLA-C2, the ligand of KIR2DS1. The hyporesponsiveness was apparent in NK cells lacking all inhibitory KIRs and CD94/NKG2A, as well as in NK cells educated via the inhibitory receptors KIR2DL3 and CD94/NKG2A. Our results suggest that the education of human NK cells via activating KIRs is dependent on the presence of their ligands and represents a mechanism that complements education via inhibitory KIRs.
Methods
Blood samples and cell lines
Buffy coats from healthy human donors were obtained from the blood bank at the Karolinska University Hospital in Huddinge, Stockholm, Sweden. This study was approved by the regional ethical review board in Stockholm, Sweden (2005-229/31-5). The donors were screened for expression of KIR2DS1, and peripheral blood mononuclear cells (PBMCs) from 28 KIR2DS1+ donors were isolated by density centrifugation (Ficoll-Hypaque; GE Healthcare) and cryopreserved in fetal bovine serum supplemented with 10% dimethyl sulfoxide until analysis. Eleven of the donors were HLA-C1 homozygous, 7 were HLA-C1/C2 heterozygous, and 10 were HLA-C2 homozygous. Of these 28 donors, 23 lacked KIR2DL2/S2 and were thus KIR2DL3 homozygous. The 5 donors that were KIR2DL2/S2+ were excluded in the analysis of coexpression of KIR2DS1 and KIR2DL3, as well as in the analysis of KIR2DL3 single-positive (sp) NK, but were included in the analysis of KIR2DL2/3/S2− (eg, KIR2DS1sp and KIR2DL1sp). The K562 cells, P815 cells (ATCC), and the EBV− Burkitt lymphoma cell line DG75 (provided by Dr D. Donati) were cultured in complete medium (RPMI 1640 with 10% fetal bovine serum, 5mM l-glutamine, and 50mM streptomycin/penicillin).
KIR and KIR ligand genotyping
Genomic DNA was isolated from 100 μL of buffy coats using the DNeasy Blood & Tissue Kit (QIAGEN). KIR genotyping was performed using PCR-SSP technology with a KIR typing kit (Olerup-SSP). KIR ligands were determined with the KIR HLA ligand kit (Olerup-SSP), which detects the HLA-C1, HLA-C2, and Bw4 motifs.
Real-time PCR for KIR2DS1 and KIR2DL1
KIR2DS1−KIR2DL1−, KIR2DS1+KIR2DL1−, KIR2DS1+KIR2DL1+, and KIR2DS1−KIR2DL1+ NK cells were sorted to more than 99% purity using a FACSAria sorter (BD Biosciences). Total RNA was extracted from bulk or sorted NK cells, and cDNA was synthesized using a reverse-transcriptase kit (Applied Biosystems) with 1 μg of RNA. Taqman-based quantitative polymerase chain reaction (PCR) was performed on a 7500 FAST Thermocycler (Applied Biosystems) to analyze KIR2DS1 and KIR2DL1 cDNA content in each subset. Primers and probes specific for KIR2DL1 and KIR2DS1 have been published previously.18 Relative KIR mRNA was calculated using the ΔΔCt method; 18s RNA was used as an endogenous control, and the KIR− subset was used as calibrator. Data are displayed as logarithmic values of the difference between the subset of interest and the KIR− subset (2−ΔΔCt).
Flow cytometry
PBMCs were stained with anti–KIR2DL1-FITC (clone 143211; R&D Systems), anti–KIR2DL2/3/S2-PE (clone GL183), anti–KIR2DL1/S1-APC (clone EB6), anti–NKG2A-Pacific Blue (clone Z199; Coulter Biotech), anti–KIR3DL1-Alexa 700 (clone DX9; BioLegend), anti–CD56-PE-Cy7 (clone NCAM16.2; BD Biosciences), anti–CD3-Cascade Yellow (clone SK7; Dako), and either anti–KIR3DL2-biotin (clone DX31, kindly provided by Dr J. Phillips, DNAX Research Institute), or anti–KIR2DS4-biotin (clone JJC11.6; Miltenyi Biotec). Importantly, the anti-KIR2DL1/S1 monoclonal antibody (mAb) was added 15 minutes after addition of the other antibodies to allow analysis of all KIR2DS1+ NK cells. For analysis of NK-cell degranulation, PBMCs were stained with the same mAb panel, and anti–CD107a-PerCP-Cy5.5 (BioLegend) or anti–CD107a-biotin (BD Biosciences). The cells were subsequently washed and stained with streptavidin–Qdot 605 and Live/Dead Aqua (Invitrogen). For analysis of interferon-γ (IFN-γ) production, PBMCs were stained and fixed as described above in this section, except for using biotin-conjugated instead of phycoerythrin (PE)–conjugated GL183, permeabilized with 0.5% saponin, and stained with anti–IFN-γ–PE (clone 4S.B3; BD Biosciences). Details on analyses of flow cytometric data can be found in supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Functional assays
Cryopreserved PBMCs from healthy donors were thawed and rested overnight in complete medium. PBMCs (106) were cultured in medium alone or cocultured with 105 target cells (K562 or DG75) for 2 hours at 37°C. Where indicated (Figures 3B-C, 4), PBMCs were incubated overnight with 10 ng/mL interleukin-15 (IL-15; PeproTech) or 104 U/mL IFN-α (PBL InterferonSource) before stimulation with K562 cells. Antibody-redirected antibody-dependent cellular cytotoxicity was performed by coculturing 106 PBMCs with 105 P815 cells alone, together with 1 μg/mL purified anti-CD16 (clone 2G4; BD Biosciences) or with 5 μg/mL purified anti-KIR2DL1/S1 (clone 11PB6; Miltenyi Biotec) for 2 hours at 37°C. Details on the analysis of redirected antibody-dependent cellular cytotoxicity assays using 11PB6 appear in supplemental data. For analysis of IFN-γ production, 106 PBMCs were stimulated with IL-12 and IL-15 (each at 20 ng/mL) for 22 hours, and brefeldin A was added after 18 hours of stimulation, followed by staining with mAbs.
Statistical analyses
Statistical analyses were performed with GraphPad Prism software, Version 4.0. A nonparametric Kruskal-Wallis test with a Dunn multiple comparison posttest was used to test for differences between HLA-C1 homozygous, HLA-C2 homozygous, and HLA-C1/C2 heterozygous donors. Wilcoxon matched-pairs test was used for comparison between 2 groups of paired data, and the Mann-Whitney test was used for comparison of 2 groups of nonpaired data.
Results
HLA-C2 does not influence the overall frequency of KIR2DS1+ NK cells
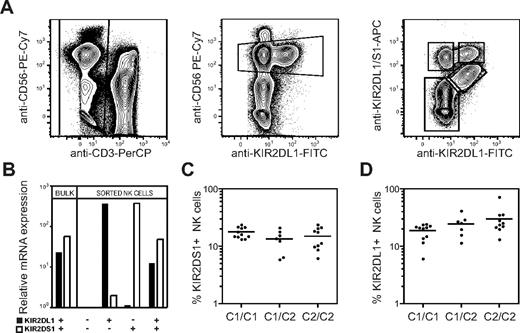
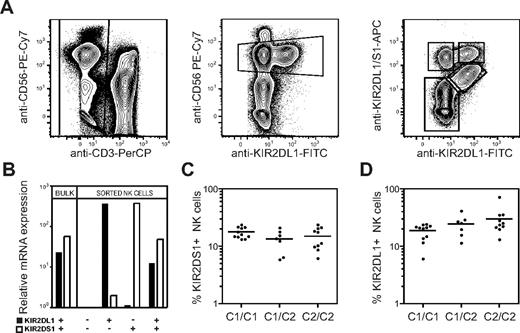
The extracellular domains of several activating KIRs, including KIR2DS1, are highly homologous to their inhibitory counterparts, making it hard to generate antibodies that specifically recognize only the activating form of a KIR. Here we used the competing properties of 2 commercial mAbs to distinguish KIR2DS1+ cells from KIR2DL1+ cells. We first incubated PBMCs with an anti-KIR2DL1–specific mAb (clone 143211), before adding a mAb cross-reactive with KIR2DL1 and KIR2DS1 (clone EB6). This staining procedure partially blocked binding of EB6 to KIR2DL1 on freshly isolated human NK cells and allowed us, for the first time, to detect expression of all combinations of KIR2DL1 and KIR2DS1 (Figure 1A). The specificity of staining by these antibodies was confirmed by real-time PCR-analysis of KIR2DL1 and KIR2DS1 mRNA expression in sorted populations of KIR2DS1+KIR2DL1−, KIR2DS1+KIR2DL1+, KIR2DS1−KIR2DL1+, and KIR2DS1−KIR2DL1− NK cells (Figure 1B).
Expression of HLA-C2 does not influence the overall frequency of KIR2DS1+ NK cells. (A) Gating strategy to identify KIR2DS1- and KIR2DL1-expressing NK cells. CD56dim NK cells were defined within CD3− single live lymphocytes in an anti-CD56 versus anti-KIR2DL1 plot. KIR2DS1+KIR2DL1−, KIR2DS1+KIR2DL1+, and KIR2DS1−KIR2DL1+ NK cells were identified based on binding of anti-KIR2DL1 and anti-KIR2DL1/S1 mAbs. (B) Bulk NK cells and sorted KIR2DS1−KIR2DL1−, KIR2DS1+KIR2DL1−, KIR2DS1+KIR2DL1+, and KIR2DS1−KIR2DL1+ NK cells were analyzed for expression of KIR2DL1 and KIR2DS1 by real-time PCR. Expression relative to KIR2DL1−KIR2DS1− NK cells is shown. (C) The frequency of KIR2DS1+ NK cells among CD56dim NK cells in HLA-C1 homozygous (C1/C1), HLA-C1/C2 heterozygous (C1/C2), and HLA-C2 homozygous donors (C2/C2). (D) The frequency of KIR2DL1+ NK cells among CD56dim NK cells in HLA-C1 homozygous, HLA-C1/C2 heterozygous, and HLA-C2 homozygous donors. Horizontal bars represent the mean frequency of KIR2DS1+ and KIR2DL1+ NK cells. All graphs depict data on a log10 scale.
Expression of HLA-C2 does not influence the overall frequency of KIR2DS1+ NK cells. (A) Gating strategy to identify KIR2DS1- and KIR2DL1-expressing NK cells. CD56dim NK cells were defined within CD3− single live lymphocytes in an anti-CD56 versus anti-KIR2DL1 plot. KIR2DS1+KIR2DL1−, KIR2DS1+KIR2DL1+, and KIR2DS1−KIR2DL1+ NK cells were identified based on binding of anti-KIR2DL1 and anti-KIR2DL1/S1 mAbs. (B) Bulk NK cells and sorted KIR2DS1−KIR2DL1−, KIR2DS1+KIR2DL1−, KIR2DS1+KIR2DL1+, and KIR2DS1−KIR2DL1+ NK cells were analyzed for expression of KIR2DL1 and KIR2DS1 by real-time PCR. Expression relative to KIR2DL1−KIR2DS1− NK cells is shown. (C) The frequency of KIR2DS1+ NK cells among CD56dim NK cells in HLA-C1 homozygous (C1/C1), HLA-C1/C2 heterozygous (C1/C2), and HLA-C2 homozygous donors (C2/C2). (D) The frequency of KIR2DL1+ NK cells among CD56dim NK cells in HLA-C1 homozygous, HLA-C1/C2 heterozygous, and HLA-C2 homozygous donors. Horizontal bars represent the mean frequency of KIR2DS1+ and KIR2DL1+ NK cells. All graphs depict data on a log10 scale.
Next, we tested whether the frequency of NK cells expressing KIR2DS1 was affected by expression of its ligand HLA-C2. In our cohort of 28 donors genotyped as KIR2DS1+, the frequency of KIR2DS1+ NK cells was on average 15% (range, 5.9%-24%) and did not significantly differ between HLA-C1 homozygous, HLA-C2 homozygous, and HLA-C1/C2 heterozygous donors (Figure 1C). Similarly, the levels of KIR2DS1 expression on the cell surface did not differ between HLA-C1 and HLA-C2 homozygous donors (data not shown). As a comparison, the average frequency of KIR2DL1+ NK cells was 24% (range, 6.0%-41%). Similar to KIR2DS1, the overall frequency of KIR2DL1+ NK cells was not significantly different between HLA-C1 and HLA-C2 homozygous donors (Figure 1D). The results suggest that the frequency of NK cells expressing activating KIRs, like inhibitory KIRs,13,40 is not greatly influenced by their ligands.
Combined expression of KIR2DS1 and HLA-C2 renders NK cells hyporesponsive to target cell stimulation
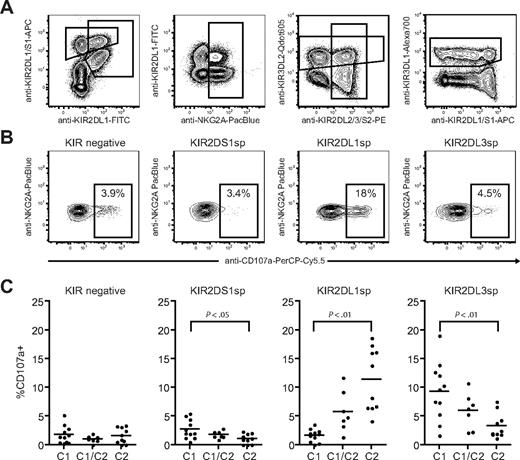
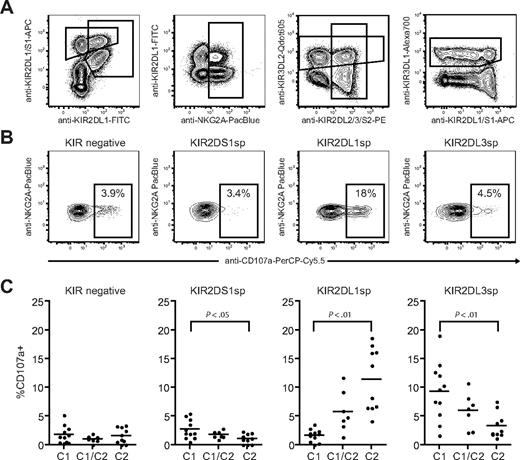
Next, we investigated how NK-cell responsiveness to stimulation with the HLA class I–negative target cell K562 was affected by expression of KIR2DS1 and its ligand HLA-C2. Degranulation by freshly isolated resting NK cells was measured using a 9-color antibody panel, which allowed simultaneous analyses of NKG2A, KIR2DS1, KIR2DL1, KIR2DL2/3/S2, KIR3DL1, and KIR3DL2 (Figure 2A). NKG2A−KIR− NK cells were hyporesponsive when stimulated with K562 cells (Figure 2B-C left panels), in line with recent findings.10 Interestingly, freshly isolated NKG2A−KIR2DS1 sp NK cells (ie, NKG2A−KIR2DS1+KIR2DL1−KIR2DL3−KIR3DL1− cells) were also hyporesponsive when stimulated with K562 cells and degranulated at similar low levels as NKG2A−KIR− NK cells (Figure 2B-C middle left panels). Corroborating previous data on education of NK cells via inhibitory KIRs,10-12,14 NKG2A−KIR2DL1sp NK cells from HLA-C2 homozygous donors responded much better than NKG2A−KIR2DL1sp NK cells from HLA-C1 homozygous donors (Figure 2B-C middle right panels). Conversely, NKG2A−KIR2DL3sp NK cells from HLA-C1 homozygous donors responded much better than NKG2A−KIR2DL3sp NK cells from HLA-C2 homozygous donors (Figure 2C right panel). Furthermore, the responses of NKG2A−KIR2DL1sp and NKG2A−KIR2DL3sp NK cells were intermediate in HLA-C1/C2 heterozygous donors (Figure 2C right panels), indicating a dose effect of HLA-C1 and HLA-C2 in the education of these NK cells, similar to that recently observed for KIR3DL1 and HLA-Bw4.11 In line with previous reports demonstrating that KIR3DL2 does not educate NK cells,12,14 expression of KIR3DL2 did not affect the NK-cell responsiveness (data not shown). These results indicate that resting NKG2A−KIR2DS1sp NK cells were hyporesponsive to stimulation with K562 cells.
Resting KIR2DS1+ NK cells lacking expression of inhibitory KIRs and CD94/NKG2A are hyporesponsive to stimulation with MHC class I–negative target cells. (A) Gating strategy to identify expression of KIR2DS1, KIR2DL1, KIR2DL3, KIR3DL1, KIR3DL2, and NKG2A on CD56dim NK cells simultaneously by flow cytometry. CD56dim NK cells were identified within single live CD3− lymphocytes based on CD56 expression. (B) Representative example of degranulation by NK cells from a HLA-C2 homozygous donor, measured as CD107a cell-surface expression after stimulation with K562 cells. Degranulation by NKG2A−KIR− NK cells, NKG2A−KIR2DS1sp NK cells, NKG2A−KIR2DL1sp NK cells, and NKG2A−KIR2DL3sp NK cells is shown. (C) Summary of NK-cell degranulation measured as CD107a cell-surface expression after stimulation with K562 cells within NKG2A−KIR−, NKG2A−KIR2DS1sp, NKG2A−KIR2DL1sp, and NKG2A−KIR2DL3sp NK cells. In each graph, HLA-C1 homozygous (C1), HLA-C1/C2 heterozygous (C1/C2), and HLA-C2 homozygous (C2/C2) donors are compared. Horizontal bars represent the mean frequency of CD107a+ NK cells within each subset. P values indicate a statistically significant difference between the groups, tested by a nonparametric Kruskal-Wallis test with Dunn posttest.
Resting KIR2DS1+ NK cells lacking expression of inhibitory KIRs and CD94/NKG2A are hyporesponsive to stimulation with MHC class I–negative target cells. (A) Gating strategy to identify expression of KIR2DS1, KIR2DL1, KIR2DL3, KIR3DL1, KIR3DL2, and NKG2A on CD56dim NK cells simultaneously by flow cytometry. CD56dim NK cells were identified within single live CD3− lymphocytes based on CD56 expression. (B) Representative example of degranulation by NK cells from a HLA-C2 homozygous donor, measured as CD107a cell-surface expression after stimulation with K562 cells. Degranulation by NKG2A−KIR− NK cells, NKG2A−KIR2DS1sp NK cells, NKG2A−KIR2DL1sp NK cells, and NKG2A−KIR2DL3sp NK cells is shown. (C) Summary of NK-cell degranulation measured as CD107a cell-surface expression after stimulation with K562 cells within NKG2A−KIR−, NKG2A−KIR2DS1sp, NKG2A−KIR2DL1sp, and NKG2A−KIR2DL3sp NK cells. In each graph, HLA-C1 homozygous (C1), HLA-C1/C2 heterozygous (C1/C2), and HLA-C2 homozygous (C2/C2) donors are compared. Horizontal bars represent the mean frequency of CD107a+ NK cells within each subset. P values indicate a statistically significant difference between the groups, tested by a nonparametric Kruskal-Wallis test with Dunn posttest.
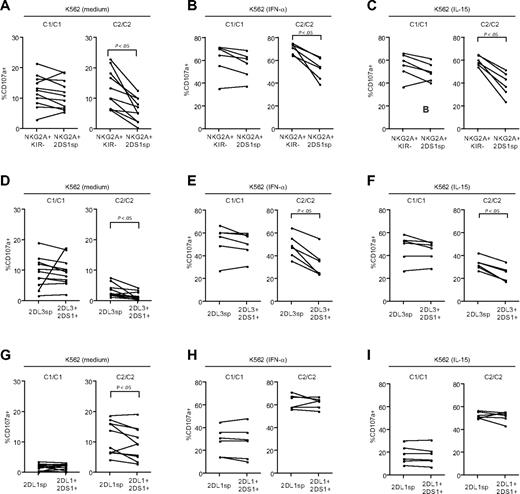
Viral infections can result in increased levels of IL-15 and IFN-α, both of which can stimulate NK cells,41 and potentially reverse the hyporesponsiveness of NK cells lacking education via inhibitory KIRs or CD94/NKG2A. Therefore, we tested how expression of KIR2DS1, in the absence of inhibitory KIRs and CD94/NKG2A, affected the responsiveness to K562 cells after overnight incubation with IL-15 or IFN-α. Compared with NK cells cultured in medium alone (Figure 3A), incubation with either of these cytokines greatly increased the degranulation of NKG2A−KIR− NK cells in response to K562 cells, both in HLA-C1 and HLA-C2 homozygous donors (Figure 3B-C). Interestingly, NKG2A−KIR2DS1sp NK cells from HLA-C2 homozygous donors had a clearly decreased response after overnight incubation with IL-15 or IFN-α compared with NKG2A−KIR− NK cells from the same donors (Figure 3B-C). In contrast, NKG2A−KIR2DS1sp NK cells from HLA-C1 homozygous donors responded at levels similar to those of NKG2A−KIR− NK cells from the same donors (Figure 3B-C). Of note, recognition of K562 cells is largely dependent on expression of ligands for non–MHC-specific activating NK-cell receptors, for example, ligands for NKp30 and NKG2D. Importantly, there was no difference in expression of NKp30 or NKG2D between NKG2A−KIR2DS1sp NK cells from HLA-C1 homozygous and HLA-C2 homozygous donors (supplemental Figure 1).
Combined expression of KIR2DS1 and HLA-C2 renders NK cells hyporesponsive to target cell stimulation. Paired analysis of CD107a cell-surface expression after stimulation with K562 cells within NKG2A−KIR− and NKG2A−KIR2DS1sp NK cells cultured overnight in (A) complete medium, (B) 104 U/mL IFN-α, or (C) 10 ng/mL IL-15. (D) Paired analysis of CD107a cell-surface expression after stimulation with P815 cells and anti-CD16 mAb by NKG2A−KIR− and NKG2A−KIR2DS1sp NK cells. (A-D) Left panels represent responses in HLA-C1 homozygous (C1/C1) donors, and right panels represent responses in HLA-C2 homozygous (C2/C2) donors. P values indicate a statistically significant difference between the groups, tested by a Wilcoxon matched-pairs test.
Combined expression of KIR2DS1 and HLA-C2 renders NK cells hyporesponsive to target cell stimulation. Paired analysis of CD107a cell-surface expression after stimulation with K562 cells within NKG2A−KIR− and NKG2A−KIR2DS1sp NK cells cultured overnight in (A) complete medium, (B) 104 U/mL IFN-α, or (C) 10 ng/mL IL-15. (D) Paired analysis of CD107a cell-surface expression after stimulation with P815 cells and anti-CD16 mAb by NKG2A−KIR− and NKG2A−KIR2DS1sp NK cells. (A-D) Left panels represent responses in HLA-C1 homozygous (C1/C1) donors, and right panels represent responses in HLA-C2 homozygous (C2/C2) donors. P values indicate a statistically significant difference between the groups, tested by a Wilcoxon matched-pairs test.
CD16 (FcγRIII) is an activating receptor expressed by virtually all CD56dim NK cells. In contrast to stimulation of NK cells with K562 cells, which is dependent on the cooperation between different activating NK-cell receptors, stimulation via CD16 alone is sufficient to trigger strong NK-cell responses.42 Therefore, we tested whether the observed hyporesponsiveness of NKG2A−KIR2DS1sp NK cells from HLA-C2 homozygous donors also extended to stimulation via CD16 (Figure 3D). Indeed, NKG2A−KIR2DS1sp NK cells from HLA-C2 homozygous donors had a clearly decreased response after stimulation via CD16, compared with NKG2A−KIR− NK cells from the same donors, whereas no such effect was observed in HLA-C1 homozygous donors. Altogether, the results reveal that expression of KIR2DS1 in the presence of HLA-C2 renders NK cells hyporesponsive to target cell stimulation.
Expression of KIR2DS1 in the presence of HLA-C2 tunes down the responsiveness of NK cells educated via inhibitory HLA class I–specific NK-cell receptors
Because the expression of KIR2DS1 together with HLA-C2 resulted in hyporesponsiveness by NK cells lacking expression of inhibitory KIRs and CD94/NKG2A (Figure 3B-D), we investigated whether KIR2DS1 could also affect the responsiveness of NK cells educated via inhibitory KIRs or CD94/NKG2A. To this end, we measured responses to stimulation with K562 cells by NKG2A+KIR− NK cells, NKG2A−KIR2DL3sp, and NKG2A−KIR2DL1sp NK cells in the presence or absence of KIR2DS1. Interestingly, in HLA-C2 homozygous donors, there was a striking decrease in degranulation by resting, as well as by IFN-α– or IL-15–primed NKG2A+KIR2DS1sp NK cells, compared with NKG2A+KIR− NK cells from the same donors (Figure 4A-C right panels), whereas no such effect was observed in HLA-C1 homozygous donors (Figure 4A-C left panels) or HLA-C1/C2 heterozygous donors (data not shown). Similarly, in HLA-C2 homozygous donors, resting as well as IFN-α– and IL-15–primed NKG2A−KIR2DL3sp NK cells coexpressing KIR2DS1 also showed a decreased response compared with NKG2A−KIR2DL3sp NK cells lacking KIR2DS1 expression (Figure 4D-F right panels), whereas no significant effect was noted in HLA-C1 homozygous donors (Figure 4D-F left panels). Of note, and in contrast to NKG2A+KIR− and KIR2DL3sp NK cells, degranulation by NKG2A−KIR2DL1sp NK cells coexpressing KIR2DS1 was not, or only very modestly, reduced compared with NKG2A−KIR2DL1sp NK cells lacking expression of KIR2DS1 in HLA-C2 homozygous donors (Figure 4G-I right panel), and the responsiveness was not affected in HLA-C1 homozygous donors (Figure 4G-I left panels). Similar results were obtained when analyzing IFN-γ responses by resting NK cells stimulated with K562 cells (data not shown). Taken together, these results indicate that expression of KIR2DS1 in combination with HLA-C2 can tune down the responsiveness of resting, as well as IL-15– or IFN-α–primed NK cells educated via CD94/NKG2A and KIR2DL3, but not KIR2DL1.
Expression of KIR2DS1 in the presence of HLA-C2 tunes down the responsiveness of NK cells educated via inhibitory HLA class I–specific NK-cell receptors. (A-C) Paired analyses of NK-cell degranulation after stimulation with K562 cells by NKG2A+KIR− and NKG2A+KIR2DS1sp NK cells cultured overnight in (A) medium alone, (B) 104 U IFN-α/mL, or (C) 10 ng/mL IL-15. (D-F) Paired analyses of NK-cell degranulation after stimulation with K562 cells by NKG2A−KIR2DL3sp and NKG2A−KIR2DL3sp NK cells coexpressing KIR2DS1 cultured overnight in (D) medium alone, (E) 104 U IFN-α/mL, or (F) 10 ng/mL IL-15. (G-I) Paired analyses of NK-cell degranulation after stimulation with K562 cells by NKG2A−KIR2DL1sp and NKG2A−KIR2DL1sp NK cells coexpressing KIR2DS1 cultured overnight in (G) medium alone,(H) 104 U IFN-α/mL, or (I) 10 ng/mL IL-15. P values indicate a statistically significant difference between groups, tested by a Wilcoxon matched-pairs test.
Expression of KIR2DS1 in the presence of HLA-C2 tunes down the responsiveness of NK cells educated via inhibitory HLA class I–specific NK-cell receptors. (A-C) Paired analyses of NK-cell degranulation after stimulation with K562 cells by NKG2A+KIR− and NKG2A+KIR2DS1sp NK cells cultured overnight in (A) medium alone, (B) 104 U IFN-α/mL, or (C) 10 ng/mL IL-15. (D-F) Paired analyses of NK-cell degranulation after stimulation with K562 cells by NKG2A−KIR2DL3sp and NKG2A−KIR2DL3sp NK cells coexpressing KIR2DS1 cultured overnight in (D) medium alone, (E) 104 U IFN-α/mL, or (F) 10 ng/mL IL-15. (G-I) Paired analyses of NK-cell degranulation after stimulation with K562 cells by NKG2A−KIR2DL1sp and NKG2A−KIR2DL1sp NK cells coexpressing KIR2DS1 cultured overnight in (G) medium alone,(H) 104 U IFN-α/mL, or (I) 10 ng/mL IL-15. P values indicate a statistically significant difference between groups, tested by a Wilcoxon matched-pairs test.
Human NK cells can also express KIR2DS4, an activating KIR that interacts weakly with HLA-C2.43 To test whether KIR2DS4 in combination with HLA-C2 could tune down the responsiveness of NKG2A+KIR− NK cells, we analyzed the effect of KIR2DS4 in the subset of our donor cohort that expressed KIR2DS4 on the cell surface. However, in contrast to NKG2A+KIR2DS1sp NK cells from HLA-C2 homozygous donors, no difference was found in the responses by NKG2A+KIR− NK cells and NKG2A+KIR2DS4sp NK cells (supplemental Figure 2).
KIR2DS1sp NK cells from HLA-C2 homozygous donors are hyporesponsive to stimulation with HLA-C2+ target cells
Next, we set out to investigate whether donor HLA-C2 homozygosity also influenced the responsiveness of KIR2DS1+ NK cells to stimulation with HLA-C2–expressing target cells. To this end, we measured NK-cell degranulation after stimulation with a HLA-C2 homozygous cell line, DG75, that expresses ligands for activating receptors (eg, CD48 and ULBP4) and is targeted by NK cells, albeit at a lower level than K562 cells (data not shown). As expected, NKG2A−KIR− NK cells responded poorly to stimulation with DG75, whether the cells were from HLA-C1 or HLA-C2 homozygous donors (Figure 5A). Interestingly, NKG2A−KIR2DS1sp NK cells from HLA-C2 homozygous donors responded as poorly as NKG2A−KIR− NK cells, whereas NKG2A−KIR2DS1sp NK cells from HLA-C1 homozygous donors clearly responded (Figure 5B), in line with recent conclusions from studies of responses to HLA-C2–expressing targets by long-term in vitro activated polyclonal KIR2DS1+ NK cells derived from HLA-C2–negative persons.33-35 Furthermore, the responses by NKG2A−KIR2DL3sp NK cells coexpressing KIR2DS1 in HLA-C2 homozygous donors were much lower than those observed in HLA-C1 homozygous donors (Figure 5C). Importantly, in HLA-C1 homozygous donors, the responses by NKG2A−KIR2DL3sp NK cells coexpressing KIR2DS1 (Figure 5C) were significantly higher than responses by NKG2A−KIR2DL3sp NK cells lacking expression of KIR2DS1 (Figure 5D), suggesting that “missing-self” recognition via inhibitory KIRs can work additively with activating KIRs. Finally, NKG2A−KIR2DL1sp NK cells did not respond to stimulation with DG75, irrespective of coexpression of KIR2DS1 and the donors' HLA-C (Figure 5E-F). Together, these results indicate that freshly isolated KIR2DS1+ NK cells from HLA-C2 homozygous donors are hyporesponsive also to stimulation via KIR2DS1 by HLA-C2+ target cells.
Freshly isolated KIR2DS1sp NK cells from HLA-C2 homozygous donors are hyporesponsive to stimulation with HLA-C2+ target cells. Degranulation in response to stimulation with HLA-C2 homozygous DG75 cells in (A) KIR− NK cells, (B) KIR2DS1sp NK cells, (C) KIR2DL3sp NK cells coexpressing KIR2DS1, (D) KIR2DL3sp NK cells, (E) KIR2DL1sp NK cells coexpressing KIR2DS1, and (F) KIR2DL1sp NK cells. In each graph, the responses in HLA-C1 homozygous and HLA-C2 homozygous donors are compared. All subsets were gated on NKG2A− NK cells. Horizontal bars represent the mean expression of CD107a on the cell surface. P values indicate a statistically significant difference between groups, tested by a 2-tailed Mann-Whitney test.
Freshly isolated KIR2DS1sp NK cells from HLA-C2 homozygous donors are hyporesponsive to stimulation with HLA-C2+ target cells. Degranulation in response to stimulation with HLA-C2 homozygous DG75 cells in (A) KIR− NK cells, (B) KIR2DS1sp NK cells, (C) KIR2DL3sp NK cells coexpressing KIR2DS1, (D) KIR2DL3sp NK cells, (E) KIR2DL1sp NK cells coexpressing KIR2DS1, and (F) KIR2DL1sp NK cells. In each graph, the responses in HLA-C1 homozygous and HLA-C2 homozygous donors are compared. All subsets were gated on NKG2A− NK cells. Horizontal bars represent the mean expression of CD107a on the cell surface. P values indicate a statistically significant difference between groups, tested by a 2-tailed Mann-Whitney test.
KIR2DS1+ NK cells from HLA-C2 homozygous donors are fully responsive to antibody-mediated cross-linking of KIR2DS1
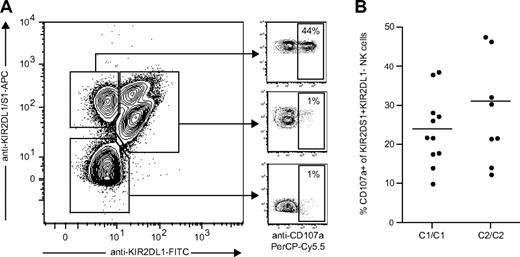
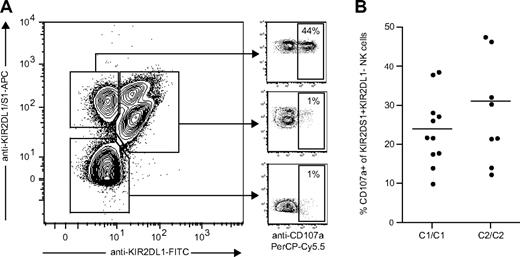
Although KIR2DS1 can bind to HLA-C2, the affinity between HLA-C2 and KIR2DS1 is 3- to 4-fold lower than that between HLA-C2 and KIR2DL1.32 Still not clear, though, is whether KIR2DS1 has additional ligands or if the affinity between KIR2DS1 and HLA-C2 can be increased by modification of HLA-C2 (eg, by viral peptides or proteins). To test whether the hyporesponsiveness of KIR2DS1+ NK cells from HLA-C2 homozygous donors could be overcome by stimulation with a higher-affinity ligand, we used antibody cross-linking of KIR2DS1 as a model. KIR2DS1+KIR2DL1− NK cells responded readily to cross-linking with the anti-KIR2DL1/S1 mAb 11PB6, whereas KIR2DL1+KIR2DS1+ and KIR2DL1+KIR2DS1− NK cells did not (Figure 6A). However, we did not detect any significant difference in the response of KIR2DS1+KIR2DL1− NK cells from HLA-C2 homozygous donors compared with HLA-C1 homozygous donors (Figure 6B), indicating that ligation of KIR2DS1 with a high-affinity ligand could indeed override the hyporesponsiveness induced by expression of HLA-C2.
KIR2DS1+KIR2DL1− NK cells from HLA-C2 homozygous donors are fully responsive to antibody-mediated cross-linking of KIR2DS1. (A) Example of KIR2DS1 and KIR2DL1 expression and degranulation measured by CD107a cell surface after coculture with P815 cells and anti-KIR2DL1/S1 mAb (clone 11PB6). The cells were stained with anti-KIR2DL1-FITC and anti-KIR2DL1/S1-APC (clone EB6) before stimulation to verify that only KIR2DS1+KIR2DL1− NK cells responded above background to stimulation with 11PB6. (B) Summary of responses to cross-linking with 11PB6, normalized to the frequency of KIR2DS1+KIR2DL1− NK cells in each of the donors.
KIR2DS1+KIR2DL1− NK cells from HLA-C2 homozygous donors are fully responsive to antibody-mediated cross-linking of KIR2DS1. (A) Example of KIR2DS1 and KIR2DL1 expression and degranulation measured by CD107a cell surface after coculture with P815 cells and anti-KIR2DL1/S1 mAb (clone 11PB6). The cells were stained with anti-KIR2DL1-FITC and anti-KIR2DL1/S1-APC (clone EB6) before stimulation to verify that only KIR2DS1+KIR2DL1− NK cells responded above background to stimulation with 11PB6. (B) Summary of responses to cross-linking with 11PB6, normalized to the frequency of KIR2DS1+KIR2DL1− NK cells in each of the donors.
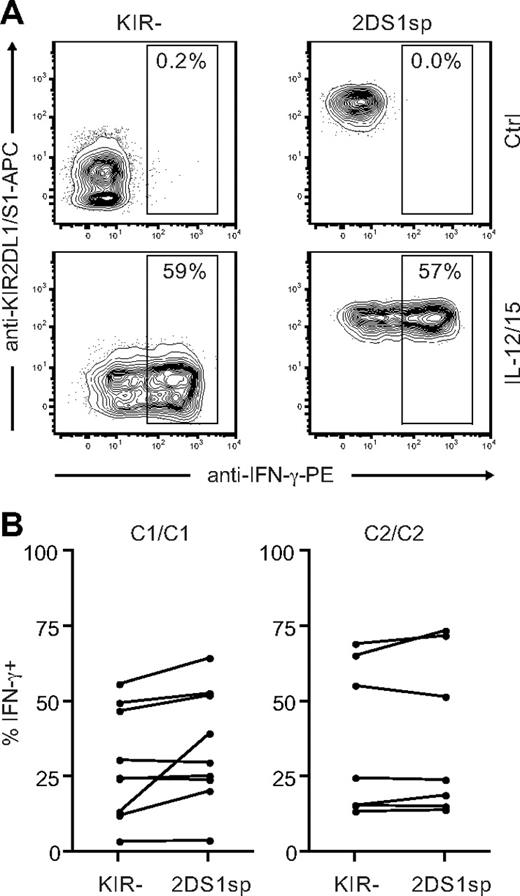
KIR2DS1sp NK cells are not hyporesponsive to stimulation with IL-12 and IL-15
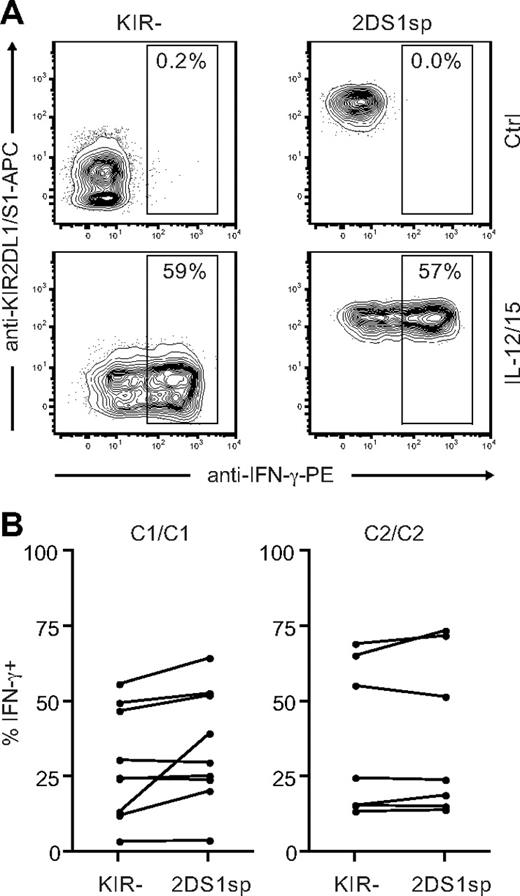
So far, our results indicated that expression of KIR2DS1 in combination with HLA-C2 tunes the responsiveness of NK cells after stimulation with cellular targets. In addition to stimulation via cell-bound ligands, NK cells can be stimulated by cytokines.44 Cytokine receptors use signaling pathways that are quite distinct from those used by activating NK-cell receptors to recognize cell-bound ligands.6 To test whether the hyporesponsiveness of KIR2DS1sp NK cells in HLA-C2 homozygous donors was limited to stimulation with cellular targets, we stimulated NK cells with IL-12 and IL-15 in combination and measured IFN-γ intracellularly. Surprisingly, NKG2A−KIR2DS1sp NK cells from both HLA-C1 and HLA-C2 homozygous donors responded equally well as NKG2A−KIR− NK cells from the same donors (Figure 7), indicating that the hyporesponsiveness induced by KIR2DS1 is limited to stimulation via cell-bound ligands.
KIR2DS1sp NK cells in HLA-C2 homozygous donors are not hyporesponsive to stimulation with IL-12 and IL-15. (A) Representative example of IFN-γ production by NKG2A−KIR− (left panels) and NKG2A−KIR2DS1sp (right panels) NK cells from a HLA-C2 homozygous donor after culture in medium alone (top panels), and after stimulation with IL-12 and IL-15 in combination (bottom panels). (B) Paired comparison of the frequency of IFN-γ–producing cells within NKG2A−KIR− and NKG2A−KIR2DS1sp NK cells from HLA-C1 homozygous (left panel) and HLA-C2 homozygous (right panel) donors.
KIR2DS1sp NK cells in HLA-C2 homozygous donors are not hyporesponsive to stimulation with IL-12 and IL-15. (A) Representative example of IFN-γ production by NKG2A−KIR− (left panels) and NKG2A−KIR2DS1sp (right panels) NK cells from a HLA-C2 homozygous donor after culture in medium alone (top panels), and after stimulation with IL-12 and IL-15 in combination (bottom panels). (B) Paired comparison of the frequency of IFN-γ–producing cells within NKG2A−KIR− and NKG2A−KIR2DS1sp NK cells from HLA-C1 homozygous (left panel) and HLA-C2 homozygous (right panel) donors.
Discussion
In this study, we set out to investigate the impact of activating KIRs on human NK-cell education. We demonstrate that expression of the activating receptor KIR2DS1 together with its MHC class I ligand, HLA-C2, tunes down the responsiveness of NK cells to stimulation with cellular targets, but not with cytokines alone. Our study provides the first example of human NK-cell education by an activating KIR and self-MHC class I, and suggests that education of NK cells via activating KIRs represents a mechanism to secure tolerance that complements education via inhibitory KIRs.
It is now well established that interactions between inhibitory receptors and their self-MHC class I ligands affect NK-cell education and endow NK cells with a capacity to recognize MHC class I–negative target cells.10-12,14,16,18,19 In contrast, the effect of activating KIRs on human NK-cell education has remained largely elusive. The findings presented here suggest that interactions between activating KIRs and their corresponding MHC class I ligands indeed affect human NK-cell education. However, the outcome of education via activating KIRs is distinct from education via inhibitory KIRs; interactions between a self-MHC class I and an activating KIR decreased NK-cell responsiveness, whereas interactions between a self-MHC class I and an inhibitory KIR increased NK-cell responsiveness. The present results are consistent with recent studies in mice, where forced expression of viral and stress-induced ligands for the activating receptors Ly49H and NKG2D during development rendered NK cells hyporesponsive to cellular stimulation.36-39 Whereas the previous studies involved the introduction of ligands that are normally not expressed during development, our study provides the first example of education via an activating NK-cell receptor specific for a ubiquitously expressed, endogenous ligand.
Furthermore, the use of 9-color flow cytometry enabled us to analyze the expression of KIR2DS1 in conjunction with the 4 major inhibitory KIRs and CD94/NKG2A, and could thus test the impact of activating and inhibitory KIRs on NK-cell education simultaneously. Our data indicate that education via activating and inhibitory KIRs operates independently of each other, as evident by the hyporesponsiveness of KIR2DS1+ NK cells in HLA-C2 homozygous donors in the absence of the major inhibitory KIRs and CD94/NKG2A. At the same time, education of NK cells via activating KIRs seems to have an impact on NK cells educated via inhibitory KIRs, and vice versa, because expression of KIR2DS1 on NK cells educated via CD94/NKG2A and KIR2DL3 tuned down, but did not abolish, the responsiveness of these cells in HLA-C2 homozygous persons.
Despite numerous studies of NK-cell education via inhibitory receptors, the underlying mechanisms remain unknown. Previous data indicate that the hyporesponsive state of NK cells lacking education via inhibitory KIRs or CD94/NKG2A cannot be explained simply by reduced expression of non–MHC-specific activating receptors.10 Similarly, the hyporesponsiveness of KIRDS1+ NK cells observed in this study was independent of expression levels of non–MHC-specific activating receptors involved in recognition of K562 cells. Although the mechanism underlying education of NK cells remains unknown, 2 distinct models have been proposed.45 The arming model proposes that NK cells are hypofunctional by default and that the interactions between self-MHC class I and the inhibitory receptors set NK cells in a responsive state, independently of signals via activating receptors. In contrast, the disarming model suggests that NK cells are active by default and those that do not engage self-MHC class I specific inhibitory receptors become hyporesponsive resulting from excessive stimulation via activating receptors.45 The finding that expression of KIR2DS1 in the presence of HLA-C2 renders NK cells hyporesponsive, with or without inhibitory KIRs and CD94/NKG2A, at a first glance perhaps best fits within the disarming model. However, our data do not exclude the arming model because it is equally possible that the observed hyporesponsiveness of KIR2DS1+ NK cells is mediated via a mechanism that works in parallel with arming by inhibitory receptors.
The effect of education via inhibitory MHC class I–specific receptors has commonly been described as an on-off switch of NK-cell functionality. However, as has become increasingly clear, NK-cell functionality seems to operate along a spectrum that depends on the strength of the educational signals, as proposed in the rheostat model.46,47 According to this model's prediction, NK-cell education is influenced by the expression levels of MHC class I in the host, the affinity between inhibitory receptors and their ligands, and the number of inhibitory self-MHC classI–specific receptors expressed by a given NK cell. The rheostat model is compatible with both the disarming and arming models of NK-cell education and is, perhaps, best used to describe the continuum of education mediated via inhibitory MHC class I–specific receptors and their ligands. In the present study, the HLA-C1/C2 heterozygous donors probably have a reduced expression of HLA-C1 and HLA-C2, compared with HLA-C1 and HLA-C2 homozygous donors, respectively, as is the case for H-2Dd heterozygous mice.47 The finding that the responsiveness of KIR2DL1sp and KIR2DL3sp NK cells was intermediate in HLA-C1/C2 heterozygous donors, compared with HLA-C1 and HLA-C2 homozygous donors, is thus in line with the rheostat model. Although the rheostat model has been used mainly to describe the outcome of education via inhibitory receptors, it does not exclude a role for activating receptors in the education of NK cells. Indeed, several of our findings indicate that education via activating KIRs tunes the NK-cell responsiveness. First, coexpression of KIR2DS1 on NKG2A+KIR− and KIR2DL3sp NK cells decreased the responsiveness (or alternatively, coexpression of NKG2A or KIR2DL3 on KIR2DS1+ NK cells increased the responsiveness), whereas coexpression of KIR2DS1 on KIR2DL1sp NK cells did not affect the responsiveness. The lack of tuning of KIR2DL1sp NK cells could possibly be the result of competition between KIR2DL1 and KIR2DS1 for binding to HLA-C2. Second, education via KIR2DS1 and HLA-C2 was observed only in NK cells from HLA-C2 homozygous donors, but not from HLA-C1/C2 heterozygous donors, suggesting that the expression level of HLA-C2 in the host influences the education via KIR2DS1. Finally, no tuning of the response was observed in HLA-C2 homozygous donors expressing the activating receptor KIR2DS4, a receptor that probably has a lower affinity for HLA-C2 than does KIR2DS1. Our results are thus in agreement with the rheostat model and suggest that the education via both activating and inhibitory receptors acts in a continuum, where the integrated signals in these systems will determine the responsiveness of NK cells.
A fundamental question in NK-cell education is why hyporesponsive NK cells are not deleted during the education process. After all, it is probably a considerable cost for the host to maintain these cells. One possible explanation is that the hyporesponsive NK cells (resulting from either education by activating KIRs or lack of education by inhibitory KIRs) can still respond to stimulation via other signaling pathways, for example, via cytokine stimulation. In support of this notion, KIR2DS1+ NK cells were highly responsive to stimulation with IL-12 and IL-15 in HLA-C2 homozygous donors, despite their hyporesponsiveness to stimulation with cellular targets. Therefore, CD56dim NK cells that are hypofunctional to stimulation with cellular targets may still be useful as cytokine-producing immunoregulators, as are the CD56bright NK cells.
The hyporesponsiveness of KIR2DS1+ NK cells from HLA-C2 homozygous donors after stimulation with K562 cells and anti–CD16-coated target cells was also extended to stimulation with HLA-C2+ target cells. The observed hyporesponsiveness to HLA-C2+ target cells is in line with a recent study of KIR2DS1+ NK cells expanded during long-term culture, where these cells had a low response to stimulation with HLA-C2+ target cells when they were derived from HLA-C2+ donors.35 However, although it remained possible that the decreased responsiveness to HLA-C2+ targets by in vitro expanded KIR2DS1+ NK cells was a consequence of the in vitro culture conditions, the use of freshly isolated NK cells in the present study indicate that these cells are indeed probably tolerant to stimulation via KIR2DS1 and HLA-C2 also in vivo. The results raise the question of why we have activating receptors specific for self-MHC class I if the NK cells that express them are rendered hyporesponsive. It makes little sense to have a functional activating receptor if the host is never going to encounter the ligand, as is the case is for KIR2DS1 in HLA-C1 homozygous donors. Although these receptors are probably important in allogeneic stem cell transplantation, it is questionable whether they have evolved to recognize allogeneic cells in a natural setting. An alternative possibility is that they recognize pathogen-encoded, -induced, or -altered ligands, and that the specificity for self-MHC class I merely reflects a cross-reactivity. If so, tolerance induction by education via activating KIRs and their endogenous ligands may have evolved to avoid autoimmunity but could still allow these cells to respond to a higher-affinity ligand. Indeed, our findings support this possibility because cross-linking of KIR2DS1 with antibodies induced equally good responses in KIR2DS1+ NK cells from HLA-C2 homozygous and HLA-C1 homozygous donors.
In conclusion, we have demonstrated that expression of KIR2DS1 in the presence of HLA-C2 renders NK cells hyporesponsive to stimulation with cellular targets. Our results indicate that education via activating KIRs in the presence of their ligands provides an alternative and complementary mechanism to education via inhibitory KIRs and CD94/NKG2A. The findings should have bearing on the understanding of NK-cell education and function in general, as well as on the interpretation of data on activating KIRs and HLA class I in transplantation, infectious diseases, and autoimmunity.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank N. Björkström for helpful discussions, H. Concha for help with cell sorting, and J. Fink and A. T. Nguyen Hoang for help with quantitative PCR analyses.
This work was supported by the Swedish Research Council (C.F., J.M.), the Swedish International Development Cooperation Agency (J.M.), the Swedish Foundation for Strategic Research, and the Swedish Cancer Society (C.F., K.-J.M., and H.-G.L.).
Authorship
Contribution: C.F., M.A.I., and J.M. performed research and analyzed data; C.F. and J.M. designed research; and C.F., H.-G.L., K.-J.M., and J.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jakob Michaëlsson, Center for Infectious Medicine, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, Stockholm, 141 86, Sweden; e-mail: jakob.michaelsson@ki.se.