Abstract
Recent studies have revealed that definitive hematopoiesis in vertebrates initiates through the formation of a non–self-renewing progenitor with limited multilineage differentiation potential termed the erythromyeloid progenitor (EMP). EMPs are specified before hematopoietic stem cells (HSCs), which self-renew and are capable of forming all mature adult blood lineages including lymphoid cells. Despite their differences, EMPs and HSCs share many phenotypic traits, making precise study of their respective functions difficult. Here, we examine whether embryonic specification of EMPs requires Notch signaling as has been shown for HSCs. In mindbomb mutants, which lack functional Notch ligands, we show that EMPs are specified normally: we detect no significant differences in cell number, gene expression, or differentiation capacity between EMPs purified from wild-type (WT) or mindbomb mutant embryos. Similarly N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT), a chemical inhibitor of Notch receptor activation, has no effect on EMP specification. These studies establish that HSCs are the only hematopoietic precursor that requires Notch signaling and help to clarify the signaling events underlying the specification of the 2 distinct waves of definitive hematopoiesis.
Introduction
In adult vertebrates, constant replenishment of mature blood cells depends upon the existence of rare hematopoietic stem cells (HSCs). The 2 hallmarks of HSCs, multilineage differentiation capacity and long-term self-renewal potential, are sufficient to sustain hematopoiesis for life.
In the mouse, embryonic HSCs emerge in close association with the major arteries of the conceptus: the vitelline arteries of the yolk sac (YS)1-3 the dorsal aorta,3-5 and the umbilical arteries of the placenta6,7 between 9 and 12 days postcoitus (dpc). HSCs subsequently migrate to the fetal liver and spleen before seeding the bone marrow,8,9 which is the main site of mammalian hematopoiesis during adulthood. Lineage tracing of embryonic HSCs showed contribution to the adult hematopoietic system,10 although additional subsequent sources of HSCs could not be ruled out as contributors to the adult HSC pool. Importantly, recent studies have implicated hemogenic endothelium from the midgestation embryo as the original source of HSCs.11,12 Similar to mammals, zebrafish possess shifting sites of hematopoiesis during embryonic development, with HSCs first appearing along the dorsal aorta between 26 and 28 hours postfertilization (hpf).13-16 Nascent HSCs migrate from this analog of the mammalian aorta-gonads-mesonephros (AGM) region to seed the caudal hematopoietic tissues, which serve a role similar to that of the mammalian fetal liver in transiently hosting multilineage hematopoiesis16 until hematopoiesis transitions to the kidney, the major site of hematopoiesis in adult teleosts (reviewed in Stachura and Traver17 ). Presumably, these migrations reflect the changing needs of HSCs to experience different signaling environments at different points in their maturation.
A major challenge in the field of developmental hematopoiesis is to determine the molecular cues that instruct each hematopoietic wave from mesoderm. In both mammals and zebrafish, signaling through the Notch pathway is important in HSC specification. Notch is a single-pass transmembrane receptor, which, upon association with its ligands Delta or Jagged, undergoes 2 sequential cleavage events. The first cleavage occurs after ligand binding, resulting in the detachment of the notch extracellular domain from its membrane-bound portion. The cleaved extracellular domain–ligand complex is endocytosed by the adjacent signal-emitting cell. Gamma secretase then mediates the secondary cleavage event, releasing the Notch intracellular domain from the Notch receptor, which translocates to the nucleus to bind Mastermind and CSL/RBPjk factors to promote transcription of Notch-dependent genes such as Hes1, Gata3, and Runx1.18-21 Mouse embryos with targeted deletions of Notch1 show no AGM hematopoiesis, demonstrating a requirement for Notch1 in intraembryonic HSC production.21,22 Studies with mindbomb mutant zebrafish embryos have confirmed the role of Notch in HSC specification. Mindbomb encodes an E3 ubiquitin ligase essential for maturation of Delta and Jagged ligands.23 Zebrafish mindbomb mutants display a complete absence of transcripts for HSC marker genes cmyb and runx1 in the ventral floor of the dorsal aorta at 36 hpf.24 Conversely, overexpression of the Notch intracellular domain resulted in an expansion of cmyb and runx1 at 36 hpf, suggesting increased HSC production.24
Despite the absence of intraembryonic hematopoiesis in murine Notch knockout animals, YS hematopoietic development appears largely unaffected. Indeed, primitive and definitive blood cell types can be obtained from the YS of Notch1- and Mindbomb-deficient embryos.22,23 These data are in agreement with the observation that Notch1-deficient embryonic stem (ES) cells contribute to primitive and definitive hematopoiesis in the YS but not to definitive hematopoiesis intraembryonically.25 Definitive hematopoiesis in the YS has been attributed to the presence of multipotent erythromyeloid progenitors (EMPs) capable of macrophage, neutrophil, and definitive erythrocyte differentiation.26 CD45−/locKit+CD41+ EMPs can be purified from YS between 8.5 and 10.5 dpc.27 Studies in the mouse embryo show that EMPs and HSCs have similar cell-surface markers, differing only in their differentiation and self-renewal potentials.27,28 Because HSC activity has been reported in the YS before or concomitant with onset of circulation, it has been difficult to distinguish between EMPs and HSCs in the murine YS. We have recently identified EMPs in the zebrafish embryo,14 where discrimination between EMPs and HSCs is possible based on anatomic location. EMPs arise between 24 and 30 hpf in the posterior blood island (PBI), from lmo2+ precursors.14 In contrast, HSCs arise between 26 and 48 hpf in the dorsal aorta, anterior to the PBI.13,14,16 These findings reflect the conservation of consecutive waves of definitive hematopoiesis from zebrafish to mouse.
To better understand the molecular signaling events underpinning differential specification of HSCs and EMPs from prehematopoietic mesoderm, we analyzed the role of Notch signaling. We used mindbomb mutant animals24,29 to determine whether Notch signaling is required for EMP formation. In mindbomb mutants carrying gata1:DsRed; lmo2:eGFP transgenes, we show that EMPs are present in the PBI, and thus do not require Notch signaling for their specification. EMPs isolated from wild-type and mindbomb mutant animals showed no significant differences in number, gene expression, or differentiation potential. These findings demonstrate that, unlike HSCs, EMPs do not require Notch signaling, providing a likely explanation for differential specification of these populations.
Methods
Zebrafish strains and maintenance
Adult fish were maintained in accordance with University of California, San Diego Institutional Animal Care and Use Committee guidelines. Zebrafish were mated, staged, and raised as previously described.30 AB* wild-type (WT) fish and transgenic lines Tg(lmo2:eGFP)zf72, Tg(lmo2:DsRed)zf73,31 Tg(cmyb:eGFP),32 and Tg(gata1:DsRed)sd2,33 were used. Hereafter, transgenic lines will be referred to without the Tg(xxx:xxx) nomenclature for clarity. Mutant mindbomb (mibta52b)29 embryos were generated from heterozygous matings and subsequently scored for brain and spinal curvature defects at 24 or 30 hours after fertilization (hpf) as previously described.29,34 Hereafter, mibta52b mutant fish lines will be referred to as mindbomb (mib) for simplicity. Adult lmo2+gata1+ animals were mated with mib heterozygotes, and lmo2+gata1+ progeny were screened, maintained, and outcrossed to known mib mutants. Embryos were dechorionated with pronase (Sigma) and kept in 0.003% 1-phenyl-2-thiourea (Sigma) to prevent pigmentation for optimal visualization as previously described.30
WISH
Dechorionated, 1-phenyl-2-thiourea–treated embryos were fixed with 4% paraformaldehyde (Sigma) at 26, 30, 36, or 48 hpf. Digoxygenin-labeled antisense and sense RNA probes were synthesized using a digoxygenin RNA Labeling Kit (SP6/T7; Roche). RNA probes were generated for HSC markers cmyb and runx1, the myeloid marker pu.1, and granulocyte marker mpx by linearizing TOPO Blunt or Bluescript vectors containing the gene sequence reverse-transcribed with SP6 or T7 polymerase to generate the desired sense and antisense probes. Whole-mount RNA in situ hybridization (WISH) was performed as described.35
Cell suspension preparation
lmo2+gata1+ cells were isolated from WT or mib mutant animals at 30 hpf as described.17 Embryos were treated with 10mM dithiothreitol (Sigma) in E3 medium,30 transferred to Hanks balanced salt solution (with calcium and magnesium; Invitrogen), and digested with Liberase Blendzyme III (Sigma-Aldrich) for 1 to 2 hours at 32°C. Cell suspensions were washed with 0.9× phosphate-buffered saline (PBS) supplemented with 1% fetal bovine serum (FBS; ATCC) and pelleted by centrifugation at 250g for 5 minutes. Supernatant was decanted, and cells were resuspended in 0.9× PBS with 1% FBS and filtered through 35-μm nylon mesh.
FACS
Cells were resuspended in 0.9× PBS, 1% FBS, with propidium iodide (Sigma) added at 1 μg/mL to exclude dead cells and debris. Fluorescence-activated cell sorting (FACS) was performed as described33 using a FACSAria flow cytometer (Becton Dickinson). Data analyses were performed using FlowJo software (TreeStar).
In vitro differentiation potential assay and cytology
Erythromyeloid precursors (EMPs) were purified by flow cytometry and plated onto confluent zebrafish kidney stroma (ZKS) cells as described14 in 1 mL of ZKS media36 at a density of 1 × 104 cells/well in a 12-well plate. For morphologic analyses, cells were gently aspirated and 500 μL was removed for cytocentrifugation on days 3 and 5 of culture. Hematopoietic cells were concentrated by cytocentrifugation at 400g for 4 minutes onto glass slides using a Shandon Cytospin 4 (Thermo Fischer Scientific). Cells were stained with May-Grünwald-Giemsa stain (Sigma-Aldrich) according to the manufacturer's protocol and o-Dianisidine as described.36 Statistical analyses were done using a 2-sample unequal variance t test with 2-tailed distribution.
Real-time qPCR analyses
For quantitative polymerase chain reaction (qPCR) analysis, lmo2+gata1+ cells were sorted from WT or mutant embryos by FACS. RNA was extracted using the RNeasy Kit (QIAGEN) according to the manufacturer's instructions. Total RNA was reverse-transcribed with oligo dT primers to generate cDNA with the Superscript III RT-PCR kit (Invitrogen). Real-time qPCR was performed with the Mx3000P system (Stratagene) following the manufacturer's protocol. In each experiment, elongation factor 1α (ef1α) expression was measured for each population and applied to normalize signals for the transcripts queried using the ΔΔ threshold cycle method. These data were normalized using whole kidney marrow (WKM) expression levels, defined as 100% for all analyzed transcripts. The following primers were used: notch1b forward primer (FP): GAATGCATCTTTTCTTCGTG, notch1b reverse primer (RP): CAGACACTTGCATTCTCCTC; notch3 FP: AATGCACAGGATAACACAGG, notch3 RP: GCTTCAACGTTATTGACTGC; her6 FP: CGT TAA TCT TGG ATG CTC TG, her6 RP: CTT CAC ATG TGG ACA GGA AC. Primers for ef1α, gata1, pu.1, mpx, cd41, and runx1 have been previously described.13
Notch inhibition assay
Embryos were collected and raised at 28°C until shield stage. At 6 hpf, embryos were dechorionated and placed in 100 μM N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT; Calbiochem) diluted in E3 as described.37 Embryos were fixed with 4% paraformaldehyde at 30 hpf and WISH was performed.
Microscopy
Fixed embryos in 3% methylcellulose (Sigma) were imaged using a Leica MZ12.5 dissecting microscope and Sony Cybershot 5.0 camera. Stained cytospins were observed with an Olympus BX51 light microscope, imaged with an Olympus DP70 camera, and processed with DP Controller software (Olympus).
Results
Notch signaling components discriminate between HSCs and EMPs
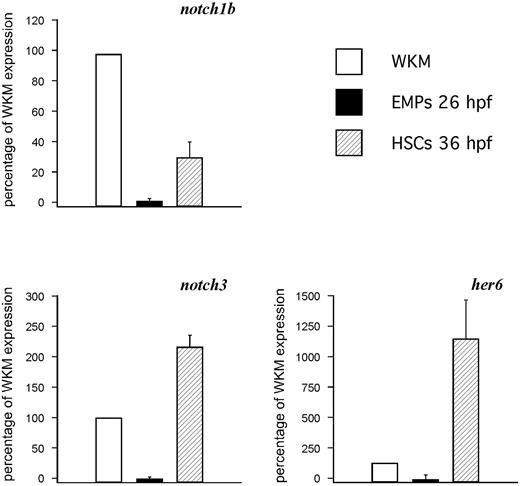
Notch signaling is required for HSC specification in the AGM region of mice and zebrafish.20,21,24 To better understand what Notch receptors control HSC specification, and to ascertain which ones might be present in the PBI where EMPs arise, we examined the expression of notch1a, notch1b, notch2, and notch3 by WISH between 24 and 36 hpf. The notch1b and notch3 receptors were expressed robustly along the aorta, whereas notch1a and notch2 showed no expression in the AGM or PBI regions (not shown). To determine whether these Notch receptors are expressed in HSCs, 36-hpf lmo2:DsRed+; cmyb:gfp+ HSCs were isolated by flow cytometry13,32 and subjected to qPCR analyses. HSCs expressed detectable levels of notch1b and notch3 transcripts (Figure 1), suggesting that one or both likely transduce the signals previously shown to be required for zebrafish HSC specification. To investigate whether notch signaling is also involved in the induction of EMPs, we isolated lmo2+gata1+ cells from 26-hpf transgenic embryos.14 Expression of notch1b and notch3 transcripts was nearly undetectable in EMPs compared with HSCs or adult WKM, suggesting that Notch signaling may not be required for EMP specification. These results are consistent with previous findings in the mouse embryo.21,22
HSCs, but not EMPs, express Notch pathway genes. qPCR analysis of notch1b (top left), notch3 (bottom left), and the Notch target her6 (bottom right) on 26-hpf EMPs and 36-hpf HSCs, with whole kidney marrow (WKM) as reference standard. The y-axis indicates percentage relative to WKM, defined as 100% in all examples. Results are the average of 3 experiments; error bars represent standard deviation.
HSCs, but not EMPs, express Notch pathway genes. qPCR analysis of notch1b (top left), notch3 (bottom left), and the Notch target her6 (bottom right) on 26-hpf EMPs and 36-hpf HSCs, with whole kidney marrow (WKM) as reference standard. The y-axis indicates percentage relative to WKM, defined as 100% in all examples. Results are the average of 3 experiments; error bars represent standard deviation.
To complement our Notch receptor studies, we investigated target genes implicated in hematopoietic development. The hairy enhancer of split (hes) family of transcriptional regulators are well-documented targets of Notch signaling,38,39 and initiation of Hes1 and Runx1 expression correlates with HSC emergence in the AGM.20 We investigated expression of her6, the zebrafish orthologue of Hes1,40 in HSCs and EMPs (Figure 1). We observed 4-fold higher her6 expression in HSCs compared with WKM. Intriguingly, no her6 expression was observed in EMPs, suggesting that Notch-induced her6 expression is specific to HSCs and can distinguish them from EMPs. Collectively, these data suggest that EMPs arise independently of Notch signaling.
Mindbomb embryos lack HSCs but retain normal myeloid transcripts in the PBI
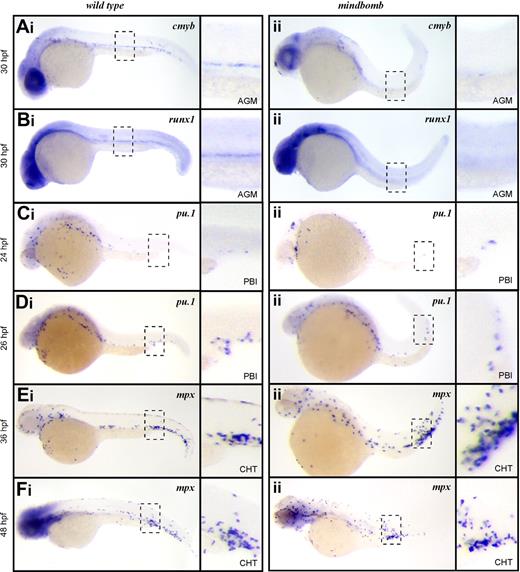
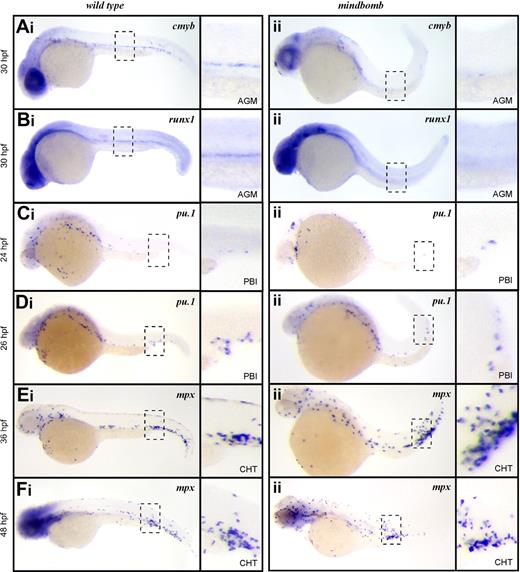
To further examine the role of Notch signaling in HSCs and EMPs, we examined the mindbomb E3 ubiquitin ligase mutant zebrafish. Mindbomb is critical for maturation of Delta-like and Jagged ligands, and disruption of this maturation abrogates Notch signaling. Studies in zebrafish show that abrogated Notch signaling in mindbomb embryos leads to an absence of HSCs in the ventral aspect of the dorsal aorta at 36 hpf.24 In accord with these findings, cmyb and runx1 transcripts were not detected in the AGM of mindbomb mutants at 30 hpf (Figure 2Aii,Bii), although they were readily observable in stage-matched WT embryos (Figure 2Ai,Bi). These findings are consistent with mammalian studies indicating that Notch signaling is required for HSC specification.21,22
EMPs, but not HSCs, are specified in mib mutant embryos. Expression of the HSC transcripts cmyb in wild-type (Ai) and mib (Aii) and runx1 in wild-type (Bi) and mib (Bii) at 30 hpf. Expression of the EMP transcript pu.1 at 24 hpf in wild-type (Ci) and mib (Cii) and 26 hpf in wild-type (Di) and mib (Dii) and mpx at 36 hpf in wild-type (Ei) and mib (Eii) and 48 hpf in wild-type (Fi) and mib (Fii). Embryos are shown with anterior to the left and dorsal up. Dashed line boxes describe the embryonic regions (AGM: Ai-ii, Bi-ii; PBI: Ci-ii, Di-ii; and CHT: Ei-ii, Fi-ii) shown in magnified images to the right of each whole embryo view. Magnification: ×100 in whole embryo images; ×300 in AGM, PBI, and CHT images.
EMPs, but not HSCs, are specified in mib mutant embryos. Expression of the HSC transcripts cmyb in wild-type (Ai) and mib (Aii) and runx1 in wild-type (Bi) and mib (Bii) at 30 hpf. Expression of the EMP transcript pu.1 at 24 hpf in wild-type (Ci) and mib (Cii) and 26 hpf in wild-type (Di) and mib (Dii) and mpx at 36 hpf in wild-type (Ei) and mib (Eii) and 48 hpf in wild-type (Fi) and mib (Fii). Embryos are shown with anterior to the left and dorsal up. Dashed line boxes describe the embryonic regions (AGM: Ai-ii, Bi-ii; PBI: Ci-ii, Di-ii; and CHT: Ei-ii, Fi-ii) shown in magnified images to the right of each whole embryo view. Magnification: ×100 in whole embryo images; ×300 in AGM, PBI, and CHT images.
We recently described the PBI as the first site of definitive hematopoiesis in the zebrafish embryo.14 We showed that the earliest myelomonocytic cells observed in this region arise from EMPs that are produced in situ from lmo2+ precursors.14 In vitro progenitor assays and in vivo transplantation experiments demonstrated that EMPs possess myelomonocyte and erythroid, but not lymphocyte, differentiation potential.14 EMPs in mouse and zebrafish are further distinguished from HSCs by their inability to self-renew.14,27,28 The first sign of myelopoietic activity in the PBI is expression of pu.1 at 24 to 26 hpf. In embryos fixed just before the onset of circulation, we observed rare cells expressing pu.1 in the PBI of both mindbomb animals and their WT siblings (Figure 2Ci-ii), and the number of pu.1+ cells increased over time in this region (Figure 2Di-ii). After 36 hpf, the PBI undergoes extensive remodeling to become the caudal hematopoietic tissue (CHT), where further myeloid differentiation and hematopoietic expansion occurs.16 It is during this phase of myelopoiesis that mpx-expressing neutrophils are first observed. In agreement with our pu.1 expression findings, mpx transcripts were detected in the CHT of mindbomb animals at 36 and 48 hpf (Figure 2Ei-ii,Fi-ii). These data suggest that although Notch signaling is essential for HSC emergence in the developing embryo, EMP-derived myeloid cells can appear normally.
To rule out the possibility that maternally provided Mindbomb allows Notch signaling to occur in mindbomb mutant embryos, we blocked Notch signaling pharmacologically with DAPT, a gamma secretase inhibitor. Gamma secretase mediates the intracellular cleavage of Notch receptors, which is required for Notch signal propagation, thus DAPT treatment can block any residual Notch signaling due to maternally loaded transcripts or protein. Exposure to DAPT from shield stage (6 hpf) to 36 hpf led to loss of cmyb+ HSCs along the dorsal aorta (supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article), whereas the pu.1+ progeny of EMPs was unaffected in the PBI (supplemental Figure 1B). Untreated embryos showed normal cmyb (n = 10) or pu.1 (n = 10) expression. Embryos treated with DAPT had ablated (n = 9) or severely reduced (n = 9) cmyb staining. All DAPT-treated embryos had normal pu.1 expression (n = 16).
Mindbomb EMPs are indistinguishable from their WT counterparts
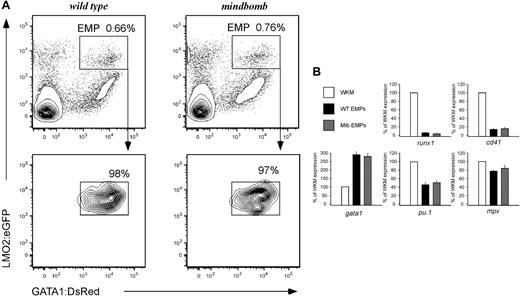
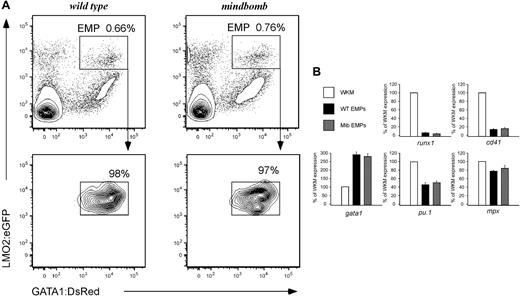
To better characterize the role of Notch signaling in EMP formation and function, we bred mib mutant animals carrying lmo2:eGFP and gatal:DsRed transgenes. EMPs can be isolated from transgenic animals based on their lmo2higata1+ phenotype. When lmo2:eGFP; gatal:DsRed; mib heterozygotes were incrossed, 14% of the progeny obtained were mib−/−; lmo2:eGFP+; gata1:DsRed+, in agreement with standard Mendelian segregation (not shown). FACS analysis and subsequent isolation of EMPs showed similar numbers of lmo2higata1+ EMPs in mutant and WT embryos at 30 hpf (Figure 3A), indicating that EMPs are present in normal numbers in animals lacking Notch signaling.
Lack of Notch signaling does not affect EMP number or gene expression. (A) lmo2higata1+ dual-positive cells (EMPs) are present in wild-type (top left) and mindbomb (top right) embryos at 30 hpf. EMPs (in boxed gate) are a fraction of the total live cells in the embryo at this time point. Resorting shows > 97% purity of isolated EMP populations (bottom row). (B) Gene expression of sorted 28-30 hpf WT and mib EMPs was examined by qPCR and compared with WKM as a reference. HSC (runx1), thrombocytic (cd41), erythroid (gata1), and myeloid (pu.1, mpx) transcript levels were determined. The y-axis indicates expression relative to WKM, defined as 100% in all examples. Levels reflect the average of 3 experiments; error bars represent standard deviation.
Lack of Notch signaling does not affect EMP number or gene expression. (A) lmo2higata1+ dual-positive cells (EMPs) are present in wild-type (top left) and mindbomb (top right) embryos at 30 hpf. EMPs (in boxed gate) are a fraction of the total live cells in the embryo at this time point. Resorting shows > 97% purity of isolated EMP populations (bottom row). (B) Gene expression of sorted 28-30 hpf WT and mib EMPs was examined by qPCR and compared with WKM as a reference. HSC (runx1), thrombocytic (cd41), erythroid (gata1), and myeloid (pu.1, mpx) transcript levels were determined. The y-axis indicates expression relative to WKM, defined as 100% in all examples. Levels reflect the average of 3 experiments; error bars represent standard deviation.
To determine whether altered Notch signaling would affect the gene expression of EMPs, we analyzed mib and WT EMPs by qPCR for the HSC-associated runx1 and cmyb genes, the thrombocyte gene cd41 (also known as itga2b), the erythroid gata1 gene, and myeloid pu.1 and mpx genes (Figure 3B and data not shown). EMPs from WT and mib mutant animals did not display significant differences in transcript abundance for any of the genes examined. Thus, loss of Notch signaling does not alter the molecular phenotype of EMPs.
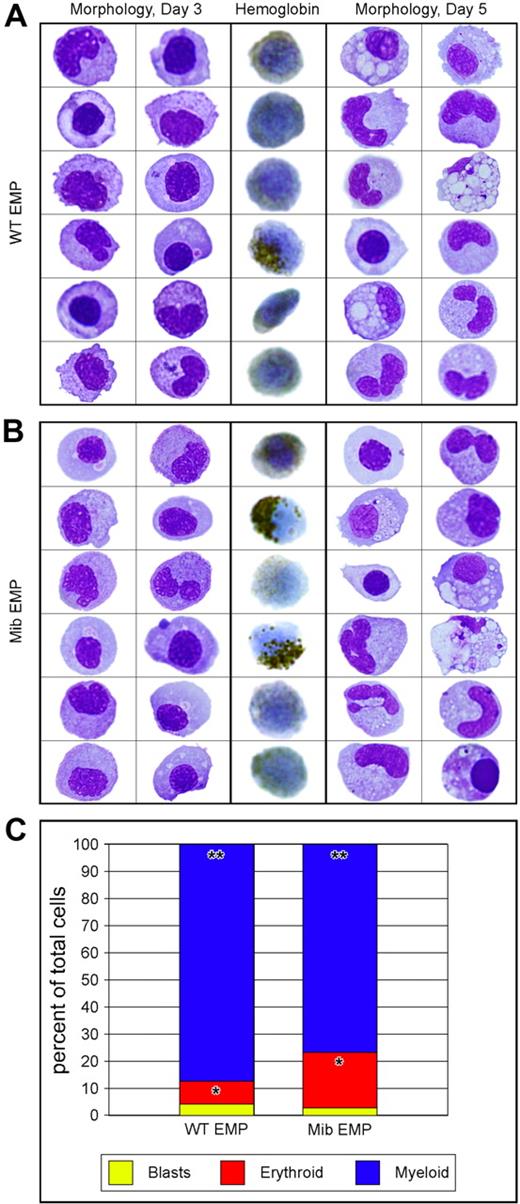
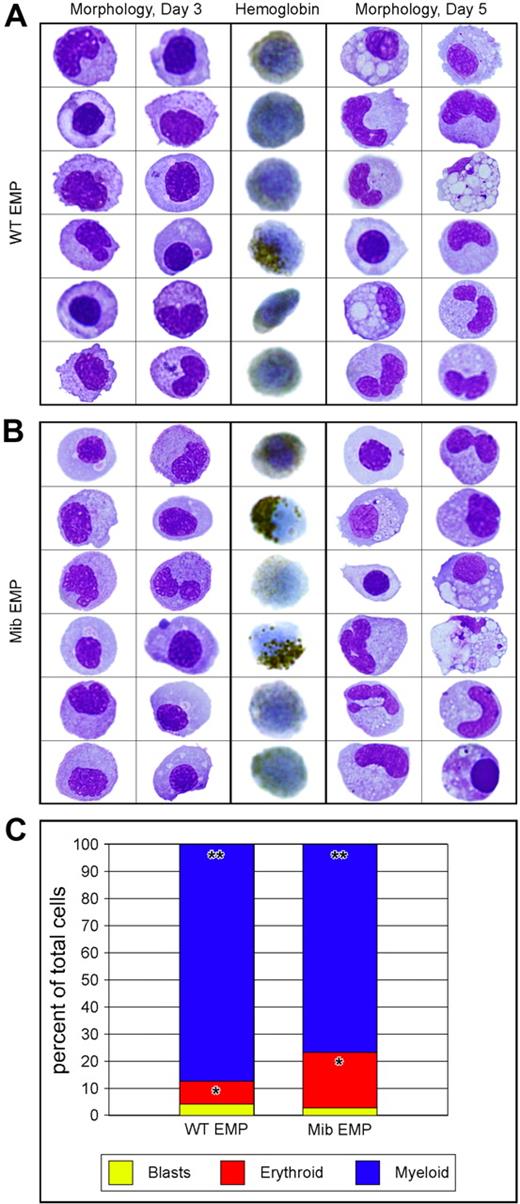
To determine whether EMPs from animals lacking Notch signaling are functional, we plated lmo2higata1+ cells on ZKS cell lines.14,36 EMPs from 28- to 30-hpf mib and WT embryos were isolated by FACS (Figure 3A top row), resorted for purity (Figure 3A bottom row), and plated on ZKS in complete medium supplemented with recombinant zebrafish erythropoietin and carp serum.36 EMP progeny were collected from the cultures at day 3, cytocentrifuged, and subjected to either an erythroid-specific hemoglobin stain or May-Grünwald/Giemsa stains to assess morphology. In addition, parallel cultures were collected at day 5 and assessed by May-Grünwald/Giemsa staining. At both time points, mib and WT EMPs differentiated into mature erythroid and myeloid lineages (Figure 4A-B). Differential cell counts for mutant and WT EMP progeny showed no statistically significant difference in erythroid (P = .678) or myeloid (P = .850) differentiation capacities (Figure 4C). These data strongly suggest that mib and WT EMPs are equivalent in gene expression and function, and thus are not affected by a lack of Notch signaling.
Similar differentiation capacity of WT and mib mutant EMPs in vitro.lmo2hi, gata1+ 30-hpf EMPs from WT and mib embryos were isolated by FACS and plated on zebrafish kidney stroma (ZKS). Samples from WT (A) and mib (B) cultured cells were cytocentrifuged and stained after 3 (3 left columns) or 5 (3 right columns) days in culture. Cells were stained for morphology with May-Grünwald/Giemsa (2 left and 2 right columns). o-Dianisidine, a chemical stain for hemoglobin, was used to assess erythroid differentiation (middle column). (C) Differential cell counts for immature blasts (yellow), erythroid (red), and myeloid (blue) cell types yielded statistically similar percentages from WT or mib EMPs (*P = .988; **P = .802). The y-axis indicates percentages of cultured cells. Levels reflect the average of 3 experiments. Statistical analyses were performed by a 2-sample, unequal-variance t test with 2-tailed distribution.
Similar differentiation capacity of WT and mib mutant EMPs in vitro.lmo2hi, gata1+ 30-hpf EMPs from WT and mib embryos were isolated by FACS and plated on zebrafish kidney stroma (ZKS). Samples from WT (A) and mib (B) cultured cells were cytocentrifuged and stained after 3 (3 left columns) or 5 (3 right columns) days in culture. Cells were stained for morphology with May-Grünwald/Giemsa (2 left and 2 right columns). o-Dianisidine, a chemical stain for hemoglobin, was used to assess erythroid differentiation (middle column). (C) Differential cell counts for immature blasts (yellow), erythroid (red), and myeloid (blue) cell types yielded statistically similar percentages from WT or mib EMPs (*P = .988; **P = .802). The y-axis indicates percentages of cultured cells. Levels reflect the average of 3 experiments. Statistical analyses were performed by a 2-sample, unequal-variance t test with 2-tailed distribution.
Discussion
The appearance of a definitive erythromyeloid progenitor that arises before HSC emergence has been established in mammals26,28 and zebrafish.14 In the mouse embryo, EMPs display the same surface phenotype (CD45locKit+CD41+) as HSCs in the AGM.27,41 The only known phenotypic difference is high Gata3 expression in HSCs.27 We recently reported a similar difference in zebrafish.13 Gata3 has been described as a target gene of the Notch signaling pathway,18,19 which is required for HSC specification in the AGM.21,22,24 Absence of Gata3 in EMPs suggests the possibility that Notch signaling is not required for their specification. Here, we show that zebrafish EMPs do not express Notch receptors, in contrast to AGM HSCs, which express notch1b and notch3, as well as the Notch target, her6. Moreover, we show that in mib mutants lacking functional Notch signaling, EMPs are present as well as phenotypically and functionally indistinguishable from their WT counterparts.
It was recently found that macrophages and neutrophils appear in mib zebrafish despite the absence of HSCs.42 Preservation of granulopoiesis was attributed to the existence of previously undescribed bipotent precursors in the rostral blood islands. Our finding that EMPs are unaffected in mindbomb mutants suggests an alternative explanation. We propose that granulocytes present in the PBI of mindbomb animals are the progeny of EMPs, whereas the rostral blood island produces primitive macrophages only. This postulate is strongly supported by recent fate-mapping studies in the zebrafish embryo.43 In our model, fish primitive macrophages are more similar to the primitive macrophage lineage in the murine yolk sac, which has been demonstrated to be unipotent.28 Because EMPs arise normally in HSC-deficient mindbomb mutants, we have been able to monitor their differentiation potential in the absence of confounding HSC-derived progeny. Our results show that this transient definitive precursor is capable of generating erythrocytes and myelomonocytes. Thus the EMP may have evolved as a means for the developing embryo to rapidly produce new erythrocytes and myelomonocytes until HSC-derived progenitors become the primary producers of all adult hematopoietic lineages.
Our data show that dependence upon Notch signaling to discriminate HSCs and EMPs in zebrafish is similar to the situation in mice. HSCs fail to form in the AGM of Notch1–deficient22 or Mindbomb1–deficient44 mouse embryos. Despite the absence of AGM HSCs, definitive hematopoiesis in the yolk sac of mutant animals appeared normal, with numbers of colony-forming units nearly equivalent to those of WT controls.22,44 In vitro, Notch1-deficient ES cells can generate normal numbers of mixed erythromyeloid colony-forming units.25 In vivo, chimeric analyses showed that Notch1-deficient ES cells could transiently produce primitive and definitive myeloid (but not lymphoid) cell lineages.25 The absence of HSCs from Notch1−/− cells strongly suggests that Notch1 is required cell autonomously for specification of HSCs. Taken together, results in both the mouse and zebrafish embryo demonstrate that, in hematopoietic ontogeny, Notch signaling is required for HSC emergence, but not for EMPs across phyla.
The prehematopoietic tissue that gives rise to EMPs remains to be determined. Lineage tracing experiments have recently revealed that murine HSCs originate from endothelial cells expressing vascular endothelial cadherin.11,12 In addition, studies using ES cells have suggested that primitive hematopoiesis is established through Flk1+Tie-2+ precursors with endothelial potential.45,46 However, it is unclear, from these studies, if the primitive wave progenitor is in fact an endothelial cell that transfates to become hematopoietic (ie, a hemogenic endothelium) or a mesodermal precursor with endothelial and hematopoietic bipotentiality (ie, a hemangioblast). We previously determined that EMPs are derived from lmo2+ cells in the caudal part of the zebrafish, but our lineage tracing experiments were performed at a stage when vasculogenesis had not yet occurred in this area.14 EMPs arise concomitant with or just after vascular tube formation in the vascular plexus of the PBI. It is therefore possible that EMPs derive from hemogenic endothelium, and that this derivation is a shared feature of definitive hematopoietic ontogeny. Because HSC development is closely linked to hemogenic endothelium along arteries,8 and because arterial fate is established by Notch signaling,34,47,48 it is unlikely that EMPs derive from aortic endothelium. In the PBI, the site of EMP emergence, the vascular plexus has venous origins.49 It is conceivable that EMPs arise from venous hemogenic endothelium in a Notch-independent manner, or they may arise from lmo2+ hemangioblasts. Further studies are required to better understand the process of EMP formation.
In summary, our findings have reinforced the importance of Notch signaling in the induction of HSCs from the aortic endothelium. We have shown that specification of the first definitive hematopoietic progenitors, EMPs, occurs independently of the Notch pathway. These studies provide important insight into the signaling environments that instruct myeloid development, and explain the ontogeny of EMPs. Moreover, they help explain the differential specification of early definitive blood precursors. A clear understanding of the molecular signaling events underlying these events is an essential precondition to deriving bona fide HSCs from pluripotent ES cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Wilson Clements for critical evaluation of the paper; Roger Rainville, Evie Wright, and Lisa Phelps for animal care; and Kerstin Richter for laboratory management.
This work was supported by a fellowship from the California Institute of Regenerative Medicine (CIRM; J.Y.B.), T32 Training Grant HL086344 from the National Institutes of Health (NIH; D.L.S.), the American Society of Hematology (D.T.), the Arnold and Mabel Beckman Foundation (D.T.), the Senyei Family Foundation (D.T.), NIH grant R01-DK074482 (D.T.), and a New Investigator Award from the CIRM (D.T.).
National Institutes of Health
Authorship
Contribution: J.Y.B. and J.L.C. performed the research and analyzed the data; D.L.S. performed aspects of the research; J.Y.B. and D.T. designed the research; and J.Y.B., J.L.C., D.L.S., and D.T. wrote the article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Traver, University of California at San Diego Division of Biological Sciences, Section of Cell and Developmental Biology, La Jolla, CA 92093-0380; e-mail: dtraver@ucsd.edu.
References
Author notes
J.Y.B. and J.L.C. contributed equally to this work.