Abstract
The directed differentiation of human pluripotent stem cells offers the unique opportunity to generate a broad spectrum of human cell types and tissues for transplantation, drug discovery, and studying disease mechanisms. Here, we report the stepwise generation of bone-resorbing osteoclasts from human embryonic and induced pluripotent stem cells. Generation of a primitive streak-like population in embryoid bodies, followed by specification to hematopoiesis and myelopoiesis by vascular endothelial growth factor and hematopoietic cytokines in serum-free media, yielded a precursor population enriched for cells expressing the monocyte-macrophage lineage markers CD14, CD18, CD11b, and CD115. When plated in monolayer culture in the presence of macrophage colony-stimulating factor and receptor activator of nuclear factor-κB ligand (RANKL), these precursors formed large, multinucleated osteoclasts that expressed tartrate-resistant acid phosphatase and were capable of resorption. No tartrate-resistant acid phosphatase-positive multinucleated cells or resorption pits were observed in the absence of RANKL. Molecular analyses confirmed the expression of the osteoclast marker genes NFATc1, cathepsin K, and calcitonin receptor in a RANKL-dependent manner, and confocal microscopy demonstrated the coexpression of the αvβ3 integrin, cathepsin K and F-actin rings characteristic of active osteoclasts. Generating hematopoietic and osteoclast populations from human embryonic and induced pluripotent stem cells will be invaluable for understanding embryonic bone development and postnatal bone disease.
Introduction
The embryonic development of the mammalian skeleton and the maintenance of skeletal and calcium homeostasis during adult life is dependent on the continuous interaction between osteoclasts, which degrade or resorb bone, and osteoblasts, which form bone.1 The origins of these cells are well established: whereas osteoblasts are derived from mesenchymal stem cells, osteoclasts are highly specialized, multinucleated cells that are derived from hematopoietic stem cells, specifically from cells of the monocyte-macrophage lineage.2,3 A large proportion of diseases that affect bone metabolism, such as osteoporosis, rheumatoid arthritis, multiple myeloma, and metastatic bone disease, is characterized by pathologic osteoclastic bone resorption. These cells are therefore very important targets for therapeutic intervention of bone diseases characterized by bone loss.
The bulk of what is known about the transcription factors, signaling molecules, and genes that control osteoclast differentiation and activation has come largely from studying mammalian osteopetroses with the use of natural and targeted mutations in mice.2-5 The breakthroughs in the past 10 years after the discovery of the tumor necrosis factor (TNF) family member, receptor activator of nuclear factor-κB ligand (RANKL), which binds to its receptor RANK and stimulates osteoclast differentiation from macrophage colony-stimulating factor (M-CSF)–dependent precursors,2,3,6,7 have made it relatively straightforward to generate osteoclasts efficiently from murine hematopoietic precursors.
However, the differentiation of human osteoclasts from circulating precursors present in peripheral blood mononuclear cells (PBMCs) is less efficient. Although it is known that osteoclast precursors reside in the CD14+ monocyte fraction from PBMCs, high donor variability, sample heterogeneity, low precursor frequency, and unpredictable responsiveness to different batches of serum for in vitro culture have provided the biggest obstacles to establishing defined, efficient, and reproducible systems for studying osteoclast differentiation and for screening novel antiresorptive compounds.8-10 Moreover, the currently available approaches for studying osteoclasts are not suitable for studying the ontogeny of osteoclasts during human embryonic development or for understanding the pathophysiology of genetic diseases affecting osteoclasts or changes in bone metabolism during aging.
The differentiation of both murine and human embryonic stem cells (ESCs) into multiple hematopoietic lineages is now well established as a powerful tool for studying early embryonic hematopoiesis and lineage restriction and for generating unlimited numbers of cell populations for transplantation and in vitro study.11-16 The exciting recent discoveries in generating induced pluripotent stem cells (iPSCs) by reprogramming somatic cells to an ESC-like state also has enabled further understanding of the ontogeny of specific embryonic lineages and has opened the door to generating patient-specific stem cells.17,18 The ability to differentiate ESCs and iPSCs in vitro to specific lineages efficiently and reproducibly has been achieved in part by the recent shift to the use of chemically defined, serum-free culture systems,12,19,20 and this has been particularly evident in the directed differentiation of ESCs to specific hematopoietic lineages.21-23
Because osteoclasts are derived from hematopoietic stem cells, we sought to establish the requirements for osteoclast precursor cell generation and differentiation. Although the potential of murine ESCs to differentiate into osteoclasts has been demonstrated previously,24,25 the generation of osteoclasts from human ESCs (hESCs), or indeed from human iPSCs (hiPSCs), is not well established. In this report, we demonstrate the directed differentiation of hESCs and hiPSCs to multipotential myeloid precursors and further differentiation to multinucleated osteoclasts that exhibit high resorptive capacity.
Methods
Maintenance and differentiation of hESCs and hiPSCs
Two hESC lines (H1, HES2)26,27 and 1 reprogrammed hiPSC line (MSC-iPS1)28 were used in this study. Undifferentiated cells were maintained in hESC media consisting of Dulbecco modified Eagle medium/F12 (50:50; Mediatech) supplemented with 20% knockout serum replacement, 100μM nonessential amino acids, 50 U/mL penicillin, 50 μg/mL streptomycin (Invitrogen), 10−4 M β-mercaptoethanol (Sigma-Aldrich), and 20 ng/mL human basic fibroblast growth factor (hbFGF) on irradiated mouse embryonic feeder cells as described previously.23 Before differentiation, the hESC and hiPSC populations were depleted of feeder cells by culturing on plates coated with Matrigel (BD Biosciences) in hESC medium for 24 to 48 hours.
To induce differentiation, embryoid bodies (EBs) were generated by culturing small aggregates (10-20 cells) of feeder-depleted hESCs or hiPSCs in 6-well low cluster plates (Corning Incorporated) in aggregation medium consisting of StemPro-34 (Invitrogen) supplemented with penicillin/streptomycin, 10 ng/mL human bone morphogenetic protein-4 (BMP-4), 2mM glutamine, 4 × 10−4 M monothioglycerol, and 50 μg/mL ascorbic acid (Sigma-Aldrich) at 37°C in an environment of 5% CO2, 5% O2, and 90% N2 for 24 hours. At this stage, the EBs were harvested, washed, and cultured in the aforementioned media supplemented with 5 ng/mL hbFGF for an additional 3 days to induce primitive streak/mesoderm formation.23
At day 4 the EBs were harvested again and recultured in supplemented StemPro-34 containing human vascular endothelial growth factor (hVEGF; 10 ng/mL), hbFGF (1 ng/mL), human interleukin-6 (hIL-6; 10 ng/mL), hIL-3 (40 ng/mL), hIL-11 (5 ng/mL), and human stem cell factor (100 ng/mL) for 4 days to promote hematopoietic specification and development. At day 8 the EBs were transferred to a 5% CO2/air environment and cultured in supplemented StemPro-34, hVEGF (10 ng/mL), human erythropoietin (4 U/mL), human thrombopoietin (50 ng/mL), and human stem cell factor, hIL-6, hIL-11, and hIL-3 for a further 10 to 14 days for hematopoietic cell maturation and expansion (see also Figure 1). Primitive streak/mesoderm induction and specification to hematopoiesis were all performed in the absence of fetal calf serum (FCS). Enrichment of myeloid precursors from day 8 was performed in both the presence and absence of FCS. All recombinant factors were purchased from R&D Systems.
Specification of hematopoietic and osteoclast lineages from human hESCs and hiPSCs. A schematic of the stepwise protocol used for the differentiation of H1, HES2, and MSC-iPS cells to the osteoclast lineage (see “Commitment of hESCs and hiPSCs to the monocyte-macrophage lineage” for details).
Specification of hematopoietic and osteoclast lineages from human hESCs and hiPSCs. A schematic of the stepwise protocol used for the differentiation of H1, HES2, and MSC-iPS cells to the osteoclast lineage (see “Commitment of hESCs and hiPSCs to the monocyte-macrophage lineage” for details).
Colony assay and flow cytometry
Analysis of hematopoietic colony potential was performed at different time points as indicated during specification and enrichment of hematopoiesis and myelopoiesis by plating 5 × 104 cells from dissociated EBs in 1% methylcellulose containing specific cytokines as described previously.23 In brief, day 8 EBs were dissociated by a 5-minute treatment with trypsin/ethylenediaminetetraacetic acid (EDTA; 0.25%) at 37°C, and cells were dispersed by passing 6 times through a 20-gauge needle. Later-stage EBs were dissociated by incubation in collagenase (0.2% phosphate-buffered saline/20% FCS; Sigma-Aldrich C-030) for 1 hour at 37°C before trypsin treatment. Colony types were determined by shape, cell size, and extent of visible erythroid content. Lineage assignment was confirmed by morphologic analyses. Flow cytometric analyses were performed at the times indicated from dissociated EBs. Cells were stained in phosphate-buffered saline/5% FCS/10% human serum with primary antibodies against CD45 (phycoerythrin conjugated), CD14 (fluorescein isothiocyanate conjugated), CD18 (allophycocyanin conjugated; all from Becton Dickinson), CD11b (allophycocyanin conjugated; Caltag), and CD115 (fluorescein isothiocyanate conjugated; R&D Systems), and analyzed on a FACSCalibur flow cytometer (Becton Dickinson) as described previously.23 Independent experiments for colony assays and flow cytometry with all 3 cell lines were performed a minimum of 3 to 4 times.
Osteoclast differentiation and confocal microscopy
Myeloid precursors were cultured on dentine slices at a density of 105 cells/6-mm well in IMDM containing human M-CSF (10 ng/mL) and the absence or presence of mouse RANKL (10 ng/mL) and 10% FCS for 10 to 14 days as indicated. Osteoclasts were identified in the first instance after fixation and staining for tartrate-resistant acid phosphatase (TRAP) activity (Sigma-Aldrich) in 50mM Na-Tartrate. Multinucleated TRAP-positive cells containing at least 3 nuclei were identified as osteoclast-like cells. Filamentous (F)–actin rings representing active osteoclasts were visualized after staining of the actin cytoskeleton with the toxin phalloidin conjugated to rhodamine (tetramethylrhodamine isothiocyanate; TRITC), and immunohistochemistry for the vitronectin receptor (VNR; αvβ3 integrin) with the 23C6 monoclonal antibody was performed as described previously.29 For confocal microscopy, resorbing osteoclasts were identified by the coexpression of F-actin rings with TRITC-phalloidin, immunostaining with an anti-β3 polyclonal antibody (Telios Pharmaceuticals Inc), and staining for the secreted form of the cysteine protease cathepsin K with an anti–cathepsin K (10C6) monoclonal antibody, as described.29
Primary antibodies were incubated for 30 minutes at room temperature, and bound antibodies were detected with Alexa-488– or Cy5-labeled goat anti–mouse secondary antibodies as indicated. Fluorescent marker and antibody distribution were monitored with a Leica TCS-NT confocal laser-scanning microscope (Leica) and imaged with the Leica TCS-NT software package as described.29 The specificities of all antibodies have been confirmed previously (data not shown, see also Stenbeck and Horton29 ). Resorption pits were visualized either by observing dentine slices under reflected light after TRAP staining or after removal of the cells and staining the slices with 1% toluidine blue or by scanning electron microscopy (Center for Ultrastructural Imaging, King's College London). Resorption area was quantified with National Institutes of Health ImageJ software.
Quantitative polymerase chain reaction analysis
Real-time quantitative polymerase chain reaction (PCR) was performed according to Bozec et al30 with SYBR Green (Molecular Probes) and an Opticon2 Monitor Fluorescence Thermocycler (MJ Research). Specific primers were as follows: cathepsin K, forward: CAGTGAAGAGGTGGTTCAGA; cathepsin K, reverse: AGAGTCTTGGGGCTCTACCTT; calcitonin receptor, forward: TCTCAGGAGTGAAAGCATTGCACATA; calcitonin receptor, reverse: AATGCTATGACCGAATGCAGCAGTTA; and NFATc1, forward: AGAATTCGGCTTGCACAGG, NFATc1, reverse: CTCTGGTGGAGAAGCAGAGC.
Statistical analysis
All statistical analyses were performed with the use of the Student t test and considered significant if P was less than .05.
Results
Commitment of hESCs and hiPSCs to the monocyte-macrophage lineage
To direct the differentiation of hESCs and hiPSCs to the osteoclast lineage, we used a staged protocol constructed on the basis of our previous induction strategy23,31 that involves primitive streak and mesoderm induction (stage 1), hematopoietic specification (stage 2), hematopoietic expansion and maturation (stage 3), and differentiation to functional osteoclasts (stage 4; Figure 1). With this approach H1, HES2, and MSC-iPS1 cells were cultured for 4 days as EBs in the presence of BMP4 and bFGF to induce a primitive-streak-like/mesoderm population, characterized by expression of the markers T (brachyury) and WNT3A as previously described.23,31 Commitment to hematopoiesis was achieved in stage 2 in serum-free media containing VEGF, bFGF, IL-6, IL-3, IL-11, and stem cell factor. At stage 3, bFGF was omitted, and erythropoietin and thrombopoietin were added to the combination of cytokines for an additional 10 to 14 days to further enrich hematopoiesis and expand multiple myeloid lineages. Although suitable enrichment was observed under serum-free conditions, the addition of FCS at the later stages resulted in greater myelopoiesis (not shown).
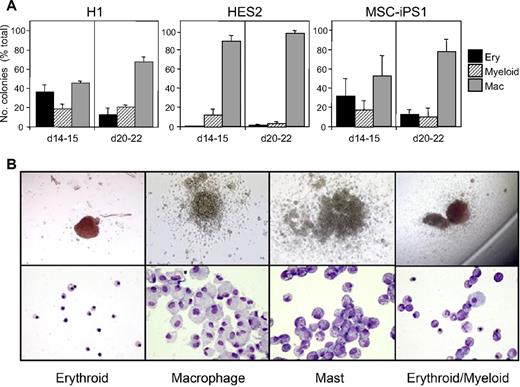
Hematopoietic specification and myeloid maturation of the EB-derived populations was monitored by analyses of hematopoietic progenitors in methylcellulose colony assays and by tracking the expression of the pan-hematopoietic surface marker CD45 and the myeloid/macrophage markers CD18, CD11b, CD115, and CD14 by flow cytometry.23 EBs from H1 and MSC-iPS1 cells showed a similar pattern of hematopoietic development characterized by the presence of erythroid, myeloid, and macrophage progenitors at 2 weeks of culture and an increase in the proportion of macrophage progenitors by 3 weeks (Figure 2). The HES2 hESC line generated fewer erythroid and myeloid progenitors, and as a consequence, macrophage progenitors predominated at both time points analyzed (Figure 2).
Colony-forming potential of hESCs and hiPSCs. EBs from hESCs (H1, HES2) and hiPSCs (MSC-iPS1) were harvested at the indicated times during hematopoietic cell commitment and myeloid cell expansion and colony forming potential was measured in methylcellulose cultures. (A) Quantification of the number of erythroid (Ery), macrophage (Mac), and myeloid colonies. Erythroid includes both pure erythroid colonies (> 90%) and mixed erythroid/myeloid colonies (< 10%). The majority of the myeloid colonies are mast cell colonies. Data represent the mean ± SEM of 6 to 8 cultures from 3 independent experiments. Values are presented as a percentage of total colonies, to demonstrate the emergence of macrophage progenitors with time during EB differentiation. We typically observe between 320 and 850 total colonies per 5 × 104 cells at 2 weeks for all 3 cell lines, which increases to 490 to 1475 colonies per 5 × 104 cells by 3 weeks. (B) Representative photographs of typical H1-derived colonies and cells within colonies after cytospin preparations. Images were acquired with a Zeiss Axioskop2 plus microscope using plan-Neofluar objectives and an Axiocam camera.
Colony-forming potential of hESCs and hiPSCs. EBs from hESCs (H1, HES2) and hiPSCs (MSC-iPS1) were harvested at the indicated times during hematopoietic cell commitment and myeloid cell expansion and colony forming potential was measured in methylcellulose cultures. (A) Quantification of the number of erythroid (Ery), macrophage (Mac), and myeloid colonies. Erythroid includes both pure erythroid colonies (> 90%) and mixed erythroid/myeloid colonies (< 10%). The majority of the myeloid colonies are mast cell colonies. Data represent the mean ± SEM of 6 to 8 cultures from 3 independent experiments. Values are presented as a percentage of total colonies, to demonstrate the emergence of macrophage progenitors with time during EB differentiation. We typically observe between 320 and 850 total colonies per 5 × 104 cells at 2 weeks for all 3 cell lines, which increases to 490 to 1475 colonies per 5 × 104 cells by 3 weeks. (B) Representative photographs of typical H1-derived colonies and cells within colonies after cytospin preparations. Images were acquired with a Zeiss Axioskop2 plus microscope using plan-Neofluar objectives and an Axiocam camera.
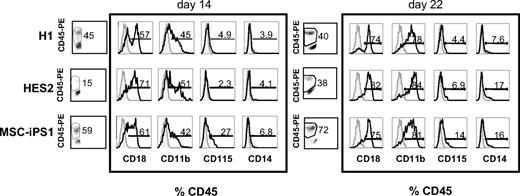
We next investigated whether the induction and enrichment protocols used resulted in the time-dependent up-regulation of specific cell-surface markers characteristic of this lineage. Flow cytometric analysis of dissociated EBs revealed the presence of large CD45+ hematopoietic populations in day 14 EBs derived from H1 ESCs and MSC-iPSCs. The size of the H1-derived population did not change significantly during the next week, whereas the iPSC-derived population increased to more than 70% of the entire culture during this time. As expected from the progenitor analyses, the emergence of the CD45+ population within the HES2-derived EBs appeared to be delayed compared with the other 2 cell lines, because only 15% of the EB cells expressed the marker at day 14. The population increased to almost 40% of the EBs at day 22 (Figure 3). These large CD45+ populations indicate that the differentiation protocol established in this study efficiently induces a hematopoietic fate from both hESC and hiPSC lines. The expression of the monocyte marker CD14 and the macrophage markers CD18 and CD11b increased with time in EBs derived from all 3 cell lines (Figure 3). CD115, representing the M-CSF receptor c-FMS, also was expressed in all 3 EB populations (Figure 3). Taken together, these data establish the stepwise generation of hematopoietic progenitors and monocyte-macrophage lineage cells from 2 independent hESC lines and 1 hiPSC line.
Monocyte-macrophage surface marker expression on developing EBs. Flow cytometric analysis of EBs from hESCs (H1, HES2) and hiPSCs (MSC-iPS1) harvested at the indicated times during hematopoietic differentiation, showing expression patterns of CD18, CD11b, CD115, and CD14, expressed as a percent of CD45+ hematopoietic progenitors. Open histograms represent populations stained with the indicated antibodies and shaded histograms represent unstained control samples.
Monocyte-macrophage surface marker expression on developing EBs. Flow cytometric analysis of EBs from hESCs (H1, HES2) and hiPSCs (MSC-iPS1) harvested at the indicated times during hematopoietic differentiation, showing expression patterns of CD18, CD11b, CD115, and CD14, expressed as a percent of CD45+ hematopoietic progenitors. Open histograms represent populations stained with the indicated antibodies and shaded histograms represent unstained control samples.
Differentiation of functional osteoclasts from hESC- and iPSC-derived myeloid precursors
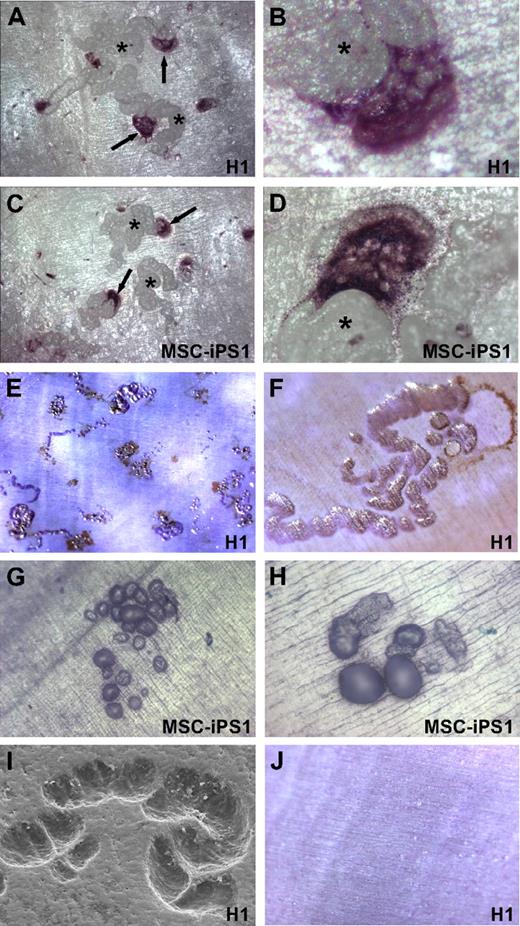
We next investigated whether the monocyte-macrophage precursors derived from H1, HES2, and MSC-iPS1 cells harbored the ability to differentiate further into functional osteoclasts. To this end, EBs at 18 to 22 days of differentiation (stage 4; Figure 1) were dissociated and the cells cultured on a mineralized dentine substrate in the presence of M-CSF and RANKL, cytokines that are essential for osteoclast differentiation, survival, and function. As shown in Figure 4, large multinucleated cells formed after 10 to 14 days in culture from both H1 (Figure 4A,B) and MSC-iPS1 (Figure 4C,D) cells. These multinucleated cells stained positively for TRAP, an enzyme highly expressed in mature osteoclasts and prefusion mononuclear precursors. HES2 cell–derived precursors also were able to form TRAP-positive cells but at a lower frequency (data not shown).
Differentiation of functional osteoclasts from hESC and hiPSCs. Representative micrographs of multinucleated osteoclasts derived from H1-derived (A-B) and MSC-iPS1–derived (C-D) hematopoietic precursors. Cells from day 20 cultures were seeded onto dentine slices in the presence of M-CSF and RANKL, and osteoclasts were stained 2 weeks later for TRAP activity (↑). Clear multinucleation is evident (B,D) and prominent resorption trails (*) are associated with all osteoclasts as viewed under reflected light. Resorption lacunae created by H1-derived (E,F,I) and MSC-iPS1–derived (G-H) osteoclasts were identified after removal of cells and staining with toluidine blue or by scanning electron microscopy (I). Cells cultured in the absence of RANKL did not differentiate into TRAP-positive cells, and resorption pits were never observed (J). Images were acquired with a Leica MZ FLIII stereomicroscope using a DFC300 FX camera. Auto adjustments were performed with Adobe Photoshop software. Original magnifications: ×4 (E); ×10 (A,C,G,J); ×20 (F,H); and ×40 (B,D).
Differentiation of functional osteoclasts from hESC and hiPSCs. Representative micrographs of multinucleated osteoclasts derived from H1-derived (A-B) and MSC-iPS1–derived (C-D) hematopoietic precursors. Cells from day 20 cultures were seeded onto dentine slices in the presence of M-CSF and RANKL, and osteoclasts were stained 2 weeks later for TRAP activity (↑). Clear multinucleation is evident (B,D) and prominent resorption trails (*) are associated with all osteoclasts as viewed under reflected light. Resorption lacunae created by H1-derived (E,F,I) and MSC-iPS1–derived (G-H) osteoclasts were identified after removal of cells and staining with toluidine blue or by scanning electron microscopy (I). Cells cultured in the absence of RANKL did not differentiate into TRAP-positive cells, and resorption pits were never observed (J). Images were acquired with a Leica MZ FLIII stereomicroscope using a DFC300 FX camera. Auto adjustments were performed with Adobe Photoshop software. Original magnifications: ×4 (E); ×10 (A,C,G,J); ×20 (F,H); and ×40 (B,D).
Reflected light microscopy after TRAP staining clearly showed osteoclasts adjacent to resorption pits. Analysis of the pits after cell removal and Toluidine blue staining demonstrated that osteoclasts derived from both H1 ESCs and MSC-iPSCs were very active and migratory, forming extensive resorption lacunae and trails (Figure 4E-H). Scanning electron microscopic analysis further confirmed the excavation of the dentine surface (Figure 4I). Precursors from H1, HES2, and MSC-iPS1 cells cultured in the absence of RANKL did not differentiate into TRAP-positive cells, and resorption pits were never observed under these conditions (Figure 4J).
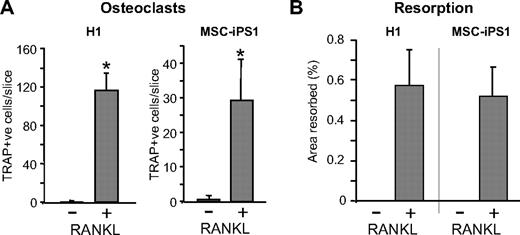
Quantification of hESC- and hiPSC-derived TRAP-positive multinucleated cells differentiated on a mineralized dentine substrate revealed that their development was RANKL dependent because comparable cells were not observed in cultures containing M-CSF only (Figure 5A). Likewise, analysis of the surface area resorbed by osteoclasts confirmed that resorption was only observed in the presence of RANKL during a period of 14 days (Figure 5B). Increasing the plating density of hESC- and hiPSC-derived myeloid precursors by 3-fold (3 × 105 cells/well) resulted in less-efficient osteoclast differentiation and resorption (data not shown).
RANKL-dependent differentiation of osteoclasts. Quantification of the number of multinucleated osteoclasts and their resorptive activity, generated by H1- and MSC-iPS1–derived osteoclast precursors. Cells from day 21 cultures were seeded at 105 cells/96-mm well on dentine slices in the absence or presence of RANKL as indicated and harvested 14 days later for TRAP staining (A) and resorption analysis (B). The data represent the mean ± SD of 4 to 6 wells. *P < .05.
RANKL-dependent differentiation of osteoclasts. Quantification of the number of multinucleated osteoclasts and their resorptive activity, generated by H1- and MSC-iPS1–derived osteoclast precursors. Cells from day 21 cultures were seeded at 105 cells/96-mm well on dentine slices in the absence or presence of RANKL as indicated and harvested 14 days later for TRAP staining (A) and resorption analysis (B). The data represent the mean ± SD of 4 to 6 wells. *P < .05.
We further examined the formation of F-actin rings, which are specific cytoskeletal structures characteristic of polarized, actively resorbing osteoclasts. Visualization of the actin cytoskeleton by phalloidin staining of hESC-derived cells cultured on a mineralized dentine substrate demonstrated clear F-actin ring formation as well as ruffled borders (Figure 6A-C). F-actin ring-positive cells were not detected in populations cultured in the absence of RANKL (Figure 6D). We next investigated the expression of the αvβ3 integrin (VNR), which is highly expressed in osteoclasts and is essential for their attachment to bone, migration, cytoskeletal maintenance, and resorption. Immunostaining of cells cultured on dentine demonstrated high VNR expression in large osteoclasts with ruffled borders (Figure 6E-G). These VNR-positive cells were highly migratory, active, and typically situated at the leading edges of long resorption trails (Figure 6E-G). Expression of osteoclast marker proteins was confirmed by laser confocal microscopy on cells grown on a dentine substrate. For this analysis, osteoclasts generated from both H1 ESCs and MSC-iPSCs were identified with the use of a β3 integrin antibody. These β3 integrin–positive cells were actively resorbing as demonstrated by the coexpression of F-actin rings (identified by phalloidin staining) and by expression of secreted cathepsin K, a cysteine protease essential for osteoclast resorption (Figure 6H-I).
Expression of mature osteoclast markers. Mature osteoclasts derived from hESCs express F-actin rings (A-C) and VNR (E-G). H1-derived hematopoietic precursors from day 21 cultures were seeded at 105 cells/96-mm well onto dentine slices in the presence of M-CSF and RANKL and stained 14 days later with TRITC-phalloidin (A-C) or by immunohistochemistry for VNR with the 23C6 antibody (E-G). F-actin rings (white arrows, panels A-C), ruffled borders (white arrowheads, panels B-C), and strong VNR expression (black arrows, panels E-G) are present in actively resorbing, multinucleated osteoclasts. Resorption trails are clearly visible in panels E-G (*). Images were acquired with a Zeiss Axioskop2 Plus microscope equipped with epifluorescence using plan-Neofluar objectives and an Axiocam camera. Auto adjustments were performed with Adobe Photoshop software. Laser confocal microscopy demonstrates the coexpression of β3 integrin (green), F-actin rings (red), and cathepsin K (blue) in H1-dervied (H) and MSC-iPS1–derived (I) osteoclasts. F-actin ring–positive cells were never detected in cultures lacking RANKL (D). Original magnifications: ×10 (A,D); ×20 (B,C,E,F); and ×40 (G).
Expression of mature osteoclast markers. Mature osteoclasts derived from hESCs express F-actin rings (A-C) and VNR (E-G). H1-derived hematopoietic precursors from day 21 cultures were seeded at 105 cells/96-mm well onto dentine slices in the presence of M-CSF and RANKL and stained 14 days later with TRITC-phalloidin (A-C) or by immunohistochemistry for VNR with the 23C6 antibody (E-G). F-actin rings (white arrows, panels A-C), ruffled borders (white arrowheads, panels B-C), and strong VNR expression (black arrows, panels E-G) are present in actively resorbing, multinucleated osteoclasts. Resorption trails are clearly visible in panels E-G (*). Images were acquired with a Zeiss Axioskop2 Plus microscope equipped with epifluorescence using plan-Neofluar objectives and an Axiocam camera. Auto adjustments were performed with Adobe Photoshop software. Laser confocal microscopy demonstrates the coexpression of β3 integrin (green), F-actin rings (red), and cathepsin K (blue) in H1-dervied (H) and MSC-iPS1–derived (I) osteoclasts. F-actin ring–positive cells were never detected in cultures lacking RANKL (D). Original magnifications: ×10 (A,D); ×20 (B,C,E,F); and ×40 (G).
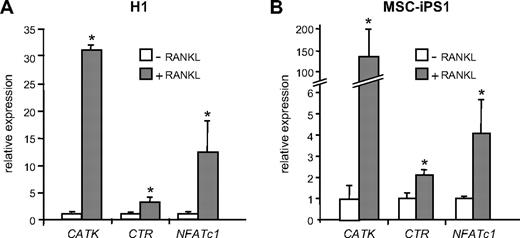
Finally, to validate the differentiation of osteoclasts using molecular markers, we performed gene expression analyses on cells cultured on a dentine substrate. Quantitative real-time PCR analysis of expression of osteoclast-specific genes and osteoclast transcription factors revealed that H1- and MSC-iPS1–derived cells expressed the osteoclast markers cathepsin K and calcitonin receptor in a RANKL-dependent manner (Figure 7A,B). H1 ESCs exhibited an approximately 30- and 4-fold increase in the expression of cathepsin K and calcitonin receptor, respectively (Figure 7A), whereas MSC-iPSCs showed an approximately 150-fold increase in cathepsin K expression and a 2-fold increase in calcitonin receptor (Figure 7B), all compared with their respective controls cultured in M-CSF only.
Quantitative PCR analysis of osteoclast marker genes. RANKL-dependent expression of osteoclast marker genes in cultures of H1-derived (A) and MSC-iPS1–derived (B) precursors. Cells from day 21 EB cultures were seeded at 105 cells/96-mm well on dentine slices in the absence or presence of RANKL as indicated and harvested 14 days later for RNA isolation and analysis of the osteoclast marker genes cathepsin K (CATK), calcitonin receptor (CTR), and NFATc1 by quantitative reverse-transcription PCR. The data represent the mean ± SD of 3 to 6 wells. *P < .05.
Quantitative PCR analysis of osteoclast marker genes. RANKL-dependent expression of osteoclast marker genes in cultures of H1-derived (A) and MSC-iPS1–derived (B) precursors. Cells from day 21 EB cultures were seeded at 105 cells/96-mm well on dentine slices in the absence or presence of RANKL as indicated and harvested 14 days later for RNA isolation and analysis of the osteoclast marker genes cathepsin K (CATK), calcitonin receptor (CTR), and NFATc1 by quantitative reverse-transcription PCR. The data represent the mean ± SD of 3 to 6 wells. *P < .05.
Expression of the M-CSF and RANKL receptors, c-FMS and RANK, respectively, generally was unchanged in RANKL-stimulated cultures in both H1- and MSC-iPS1–derived osteoclast cultures (data not shown). The expression of the c-FOS proto-oncogene, which we have shown previously to be essential for osteoclast differentiation,32 also was not altered (data not shown). However, the expression of the c-FOS target gene, NFATc1, which is also required for osteoclast differentiation,3 showed an approximately 10-fold increase in the H1-derived population and a 4-fold increase in cells generated from MSC-iPSCs after RANKL treatment compared with control cultures containing M-CSF only (Figure 7A-B). Taken together, these data demonstrate the generation of bona fide osteoclasts from myeloid precursors derived from hESCs and hiPSCs under defined conditions.
Discussion
In this study we have demonstrated the robust generation of osteoclasts from hESCs and reprogrammed hiPSCs under defined conditions involving commitment to hematopoiesis and enrichment of precursor cells in the monocyte-macrophage lineage. The osteoclasts expressed molecular markers characteristic of their differentiation and activity and were able to resorb a mineralized matrix, which represents their single most fundamental characteristic.
The recent emergence of osteoclasts as key cells that control osteoblast activity and bone formation in vivo during bone remodeling has highlighted the importance of establishing systems for generating and studying homogeneous osteoclast cultures.33-35 The current system of choice for generating human osteoclasts uses whole blood from suitable donors and isolates PBMCs by performing several physical separation techniques to yield a population of cells that is adherent and lacking in nonadherent lymphoid cells. Exposure of this mixed population to M-CSF and RANKL drives a proportion of these precursors to the monocyte-macrophage lineage and subsequently to osteoclasts. However, although this process appears to be a relatively straightforward one, it remains inefficient because questions of donor sample variation, cell purity, and dependence on different batches of serum lead to variability in precursor frequency and reproducibility of differentiation. Moreover, the derivation of osteoclasts from adult PBMCs is not suitable for studying the ontogeny of human osteoclasts during embryogenesis and from early embryonic hematopoietic precursors. We have addressed these questions in this study using pluripotent stem cells.
Several previous studies11,14,15,21 in which the authors used hESCs have demonstrated the efficient generation of hematopoietic lineages, some using defined conditions. However, comparable approaches with the use of reprogrammed hiPSCs are only recently emerging, and the authors of these studies36-38 have to date used OP9 stromal cells and/or serum to promote hematopoietic differentiation. Using this approach, Choi et al39 recently documented the development of cells with some characteristics of osteoclasts from human pluripotent stem cells. In this study, we induced hematopoietic development from both hESCs and hiPSCs with defined factors in the absence of serum and supporting feeder cell populations. This approach has the advantage in that it is possible to activate specific endogenous signaling pathways at defined times in the cultures in the absence of physical separation procedures.
Under these conditions, hematopoietic differentiation is efficient as 40% to 70% of the population expressed CD45 by 3 weeks of culture. Furthermore, these populations are enriched for macrophage progenitors and display osteoclastogenic potential. Although all 3 cell lines, H1, HES2, and MSC-iPS1, generated osteoclasts that expressed specific molecular markers and were functional, they exhibited slightly different kinetics of differentiation under identical culture conditions and indeed also in their basal commitment to hematopoiesis and hematopoietic colony formation. This finding suggests that there are intrinsic differences between each cell line. The basis for these differences is not known, but this is not unexpected in view of recent reports demonstrating that hESCs exhibit intrinsic differences not only in their basal differentiation profile but also in their capacity for hematopoiesis.40,41
On the basis of the number of osteoclasts obtained from a range of cell densities over several experiments, our preliminary estimate is that up to approximately 1:1000 hESC- or hiPSC-derived myeloid cells plated gives rise to an osteoclast under our current conditions. Although this might represent a slightly greater frequency than that commonly reported from unfractionated human PBMCs, it is difficult to directly compare precursor populations derived from ESC/iPSC and PBMCs or indeed from bone marrow hematopoietic precursors because they represent different developmental stages and all populations are grown under different conditions and have different serum requirements. In studies using murine ESCs, Okuyama et al24 have demonstrated a high variability in osteoclast differentiation potential from precursors grown in the presence of serum and on different stromal cell types. Clearly, therefore, it is important to define accurately precursor cell numbers under different conditions and in different cell lines. The stepwise protocol we are using offers the opportunity to manipulate the differentiation system at each stage, and we are currently optimizing the conditions for expanding the frequency of hESC- and hiPSC-derived precursors in a defined way, using cytokines and growth factors previously shown to be effective in enhancing osteoclastogenesis from PBMCs (eg, transforming growth factor-β1, dexamethasone, TNF-α),42,43 or at earlier stages of hematopoiesis using cytokines in concert with signaling proteins (eg, BMP, Wnt, Hedgehog) that have been implicated in expansion of the hematopoietic stem cell pool.44,45
In summary, the ability to generate large numbers of osteoclasts from hESCs and hiPSCs in a consistent, reproducible, and defined manner provides an essential tool for identifying novel antiresorptive compounds for osteoporosis as well as for the pathologic bone resorption characteristic of malignant bone disease and rheumatoid arthritis.46,47 The generation of gain- and loss-of-function mutations at different stages of hematopoietic and osteoclast differentiation in vitro will provide molecular insights into the pathways that give rise to osteoclasts during human development. Finally, the derivation of iPSCs from patients harboring cell-autonomous genetic mutations affecting osteoclast differentiation and function (eg, in autosomal-recessive osteopetrosis)48-50 provides a model for recapitulating disease pathology in vitro and establishing a basis for genetic rescue and autologous cell-based therapies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Staci Schwantz and Sunita D'Souza for expert technical assistance, Prof G. Q. Daley (Harvard Medical School) for the MSC-iPS1 cells, and Prof T. R. Arnett and Dr G. Charras (University College London) for microscopy.
This study was supported (in part) by research funding from the Nuffield Foundation-Oliver Bird Rheumatism Program to A.E.G., Arthritis Research Campaign (ARC ref 18197) to G.S., National Institutes of Health grants R01 HL080627 and P20 GM075019 to G.M.K., and Austrian Science Fund (FWF) and Anabonos EU-FP7 to E.F.W.
National Institutes of Health
Authorship
Contribution: A.E.G., M.K., E.F.W., and G.M.K. designed and performed research, analyzed data, and wrote the paper; A.B. and F.B. conducted research and analyzed data; G.S. performed research, analyzed data, and wrote the paper; and I-H.P. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Agi E. Grigoriadis, Department of Craniofacial Development, Guy's Hospital, King's College London, Tower Floor 27, London Bridge, London SE1 9RT, United Kingdom; e-mail: agi.grigoriadis@kcl.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal