Abstract
MLL-rearranged infant acute lymphoblastic leukemia (ALL) remains the most aggressive type of childhood leukemia, displaying a unique gene expression profile. Here we hypothesized that this characteristic gene expression signature may have been established by potentially reversible epigenetic modifications. To test this hypothesis, we used differential methylation hybridization to explore the DNA methylation patterns underlying MLL-rearranged ALL in infants. The obtained results were correlated with gene expression data to confirm gene silencing as a result of promoter hypermethylation. Distinct promoter CpG island methylation patterns separated different genetic subtypes of MLL-rearranged ALL in infants. MLL translocations t(4;11) and t(11;19) characterized extensively hypermethylated leukemias, whereas t(9;11)-positive infant ALL and infant ALL carrying wild-type MLL genes epigenetically resembled normal bone marrow. Furthermore, the degree of promoter hypermethylation among infant ALL patients carrying t(4;11) or t(11;19) appeared to influence relapse-free survival, with patients displaying accentuated methylation being at high relapse risk. Finally, we show that the demethylating agent zebularine reverses aberrant DNA methylation and effectively induces apoptosis in MLL-rearranged ALL cells. Collectively these data suggest that aberrant DNA methylation occurs in the majority of MLL-rearranged infant ALL cases and guides clinical outcome. Therefore, inhibition of aberrant DNA methylation may be an important novel therapeutic strategy for MLL-rearranged ALL in infants.
Introduction
Although long-term survival rates in childhood acute lymphoblastic leukemia (ALL) exceed 80%,1 the survival chances of infants (< 1 year of age) still range between 20% and 50%.2 Approximately 80% of infants with ALL carry chromosomal translocations involving the MLL (mixed lineage leukemia) gene,3 fusing the N-terminal portion of the MLL gene to the C-terminal region of one of its translocation partner genes. The most frequent MLL translocations among infant ALL patients are t(4;11), t(11;19), and t(9;11),2,4 giving rise to the fusion proteins MLL-AF4, MLL-ENL, and MLL-AF9. These chimeric MLL fusion proteins exhibit pronounced transforming capacities5 and independently contribute to an unfavorable prognosis.2,6
As a member of the trithorax gene family, MLL is involved in transcriptional regulation.7 Therefore, structural alterations of this gene may be expected to affect its function, presumably leading to transcriptional deregulation. Not surprisingly, in recent gene expression profiling studies,8,9 the authors characterized MLL-rearranged ALL as a unique type of leukemia that is genetically clearly separable from other ALL subtypes. Because epigenetic modifications affect gene expression patterns,10 we hypothesized that the specific gene expression profiles associated with MLL-rearranged infant ALL may well be driven by epigenetic changes, which recently have been established to play important roles in the development and progression of leukemia.11
The most widely studied epigenetic event in hematologic malignancies constitutes transcriptional gene silencing by promoter CpG island hypermethylation.11,12 This phenomenon either leads directly to the silencing of tumor suppressor genes, or indirectly to the up-regulation of other genes, when silencing of certain regulatory genes relaxes the suppression on their target genes. Hence, genome-wide promoter hypermethylation potentially results in abnormal gene expression profiles that favor malignant transformation. For example, we recently demonstrated that FHIT, a putative tumor suppressor gene, is characteristically silenced in MLL-rearranged infant ALL cells by CpG hypermethylation and that reexpression of this gene induced apoptosis in these cells.13
Here we applied differential methylation hybridization (DMH), an array-based technique that allows genome-wide screening of DNA methylation, by using 2 different microarray platforms to explore the DNA methylation patterns underlying MLL-rearranged infant ALL. We show that different types of MLL translocations are associated with distinct patterns of DNA methylation, and we found that the degree of DNA methylation influences clinical outcome, identifying subgroups of MLL-rearranged infant ALL patients that may particularly benefit from therapeutic strategies containing demethylating agents.
Methods
Patient samples
We studied 57 newly diagnosed infant ALL patients enrolled in the international INTERFANT-99 treatment protocol (patient characteristics are listed in supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).2 Forty-seven patients (77%) carried MLL translocations, and 13 (23%) harbored untranslocated (wild-type) MLL genes. Among the MLL translocated patients, 21 were positive for t(4;11), 17 for t(11;19), and 6 patients carried translocation t(9;11). Written informed consent and institutional review board approval from Erasmus MC–Sophia Children's Hospital were obtained for all patients in accordance with the Declaration of Helsinki. Whole normal bone marrow samples obtained from 8 nonleukemic pediatric patients were included as controls. Leukemic cell isolation and enrichment to achieve more than 90% leukemic blasts, as well as DNA and RNA extractions, were performed as described previously.14
Leukemia cell lines
RS4;11, SEMK2, and BEL-1 represent t(4;11)-positive precursor B-cell ALL cell lines. SEMK2 was originally derived from a 5-year-old girl at relapse15 and was kindly provided by Dr Scott Armstrong (Dana-Farber Cancer Institute). BEL-1 was a generous gift from Dr Ruoping Tang (University Laboratory, Paris).16 RS4;11 was established from the bone marrow of a 32-year-old woman,17 and was, like all other cell lines used in this study, purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ). KOPN-8 harbors translocation t(11;19) and was derived from a 3-month-old infant girl with B-cell precursor ALL. REH and TOM-1 represent precursor B-lineage ALL cells exhibiting a TEL-AML1 fusion and a Philadelphia chromosome, respectively. JURKAT and HSB2 both are T-lineage ALL cell lines, and Kasumi-1 and MV4;11 are AML cell lines. Kasumi-1 carries the t(8;21) AML1-ETO fusion gene, and MV4;11 harbors MLL translocation t(4;11). All cell lines were maintained as suspension cultures in RPMI 1640 with L-alanyl-L-glutamine (Invitrogen) supplemented with 10% fetal calf serum (Integro), 100 IU/mL penicillin, 100 μg/mL streptomycin, and 0.125 μg/mL fungizone (Invitrogen) at 37°C in humidified air containing 5% CO2.
Differential methylation hybridization using CpG island microarrays
DMH was performed essentially as described by Yan et al (supplemental Methods).18,19 DMH was applied on 2 different CpG island microarray platforms with limited overlap in CpG island probes. The first was the custom spotted 9K microarray chip developed by Huang et al18 containing 8640 MseI fragment probes. In addition, we also used the first commercially available genome-wide CpG island microarrays (Agilent Technologies). These high-resolution microarrays contain 243 497 60-mer oligonucleotide probes, including 67 487 CpG island probes located in or near gene promoters. For the present study, only these probes located in gene promoters were used. Because of the restricted availability of patient material, DNA methylation profiling by use of the Agilent microarrays was performed in 49 of the 57 infant ALL patients.
Gene expression profiling with the use of Affymetrix GeneChips
Gene expression profiles were generated for t(4;11)-positive (n = 15) and t(11;19)-positive (n = 14) infant ALL cases by use of the same samples for which DNA methylation profiles were already produced on Agilent microarray chips. Expression profiles were also generated for whole healthy pediatric bone marrow samples; however, these did not correspond to the samples in which the DNA methylation patterns were determined. RNA processing, microarray hybridization (HU133 Plus 2.0 Affymetrix GeneChips), and washing steps were performed according to the manufacturer's protocol (Affymetrix). The infant ALL gene expression data and DNA methylation data presented in this study have been deposited in the Gene Expression Omnibus (National Center for Biotechnology Information) and are accessible through GEO Series accession number GSE18400 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18400).
In vitro cytotoxicity assay and exposure to zebularine
In vitro sensitivity of leukemia cell lines to the demethylating agent zebularine20,21 was determined by 4-day 3-(4,5-dimethylthiazol-2-Yl)-2,5-diphenyltetrazolium bromide assays as described previously.22 Zebularine was a generous gift from Dr Victor E. Marquez (National Cancer Institute–Frederick). To study the effects of demethylation on MLL-rearranged ALL cells, the cell lines SEMK2 and RS4;11 were cultured for 10 days in the presence or absence of 100μM zebularine.
Statistical analyses
Normalization of the CpG island microarray data was performed with the use of global locally weighted scatterplot smoothing (loess) normalization,23 and differentially methylated CpG islands were identified by the use of the linear models for microarray data (limma) package in the R statistical environment (R Development Core Team, 2007; supplemental Methods).24 The resulting list of P values was corrected for multiple testing by the false-discovery rate (FDR) step-up procedure of Benjamini and Hochberg.25 An FDR-adjusted P less than .01 was regarded as significant. As a measure of internal validation for the subtype-specific methylation signatures, permutation testing (global test)26 was applied to evaluate whether genes were significantly associated with a certain type of MLL translocation. For this, the tendency of repeated reassignment of individual samples to their original cluster was assessed (supplemental Methods).
Relapse-free survival was computed with the Kaplan-Meier estimator. The duration of relapse-free survival was defined as the time from diagnosis until the date of leukemia relapse or the last follow-up. The probability of relapse in complete remission was estimated by applying the cumulative incidence estimator. The log-rank test was used to compare outcomes between different patient groups, and a 1-step Cox model was applied to estimate the hazard of relapse for these patients, adjusting for already established risk stratification according to the international INTERFANT-06 treatment protocol (supplemental Methods).
Results
Unsupervised analysis based on DNA methylation patterns separates different infant ALL subtypes
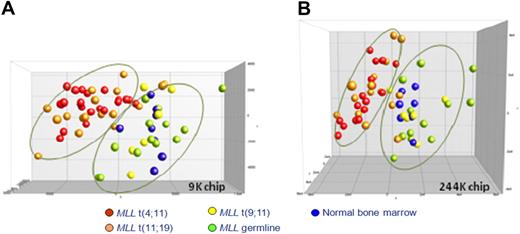
By using DMH on 2 different microarray platforms, we generated genome-wide promoter DNA methylation profiles for infant ALL patients carrying MLL translocations t(4;11), t(11;19), or t(9;11) and infant ALL patients bearing wild-type MLL genes. To explore whether these samples showed leukemia-specific increases in promoter CpG island methylation, these profiles were compared with DNA methylation patterns obtained from bone marrow samples derived from healthy children. Initially, we performed a principal component analysis (PCA), using all CpG island probes present on each array without any selection. On the basis of the first 3 components of the PCA, which explain 41.8% (9K chip) and 32.2% (244K chip) of the total variance, the patient samples were visualized (Figure 1). Interestingly, for both microarray platforms, this unsupervised analysis separated 2 major groups. Infant ALL samples that carry t(9;11) or wild-type MLL genes clustered together with normal bone marrow samples, whereas infant ALL samples carrying t(4;11) or t(11;19) clustered tightly together separately from the other samples. Although the cluster comprising t(9;11)-positive, untranslocated infant ALL samples and normal bone marrow samples appeared more heterogeneous, it has to be taken into account that this cluster consists of 3 different types of samples. Moreover, epigenetic heterogeneity is already present among the normal bone marrow samples. Finally, we emphasize that this analysis is completely unguided.
Unsupervised clustering analysis of DNA methylation in infant ALL. PCA of the CpG island methylation data from infant ALL patients and normal bone marrows using all probes present on each microarray platform. Each case is color-coded, indicating the specific infant ALL subgroups. (A) Data from the custom-spotted 9K CpG island microarray. t(4;11) (n = 21; red), t(11;19) (n = 17; orange), t(9;11) (n = 6; yellow), MLL wild-type ALL (n = 13, green), and normal bone marrow (n = 8; blue). (B) Data from the commercially available 244K CpG island microarray (Agilent). t(4;11) (n = 16; red), t(11;19) (n = 15; orange), t(9;11) (n = 6; yellow), MLL wild-type ALL (n = 12; green), and normal bone marrow (n = 7; blue). Because of restricted availability of patient material, Agilent DNA methylation profiles were generated for 49 infant ALL patients and 7 normal bone marrow samples.
Unsupervised clustering analysis of DNA methylation in infant ALL. PCA of the CpG island methylation data from infant ALL patients and normal bone marrows using all probes present on each microarray platform. Each case is color-coded, indicating the specific infant ALL subgroups. (A) Data from the custom-spotted 9K CpG island microarray. t(4;11) (n = 21; red), t(11;19) (n = 17; orange), t(9;11) (n = 6; yellow), MLL wild-type ALL (n = 13, green), and normal bone marrow (n = 8; blue). (B) Data from the commercially available 244K CpG island microarray (Agilent). t(4;11) (n = 16; red), t(11;19) (n = 15; orange), t(9;11) (n = 6; yellow), MLL wild-type ALL (n = 12; green), and normal bone marrow (n = 7; blue). Because of restricted availability of patient material, Agilent DNA methylation profiles were generated for 49 infant ALL patients and 7 normal bone marrow samples.
Specific DNA methylation patterns further separate the different infant ALL subtypes
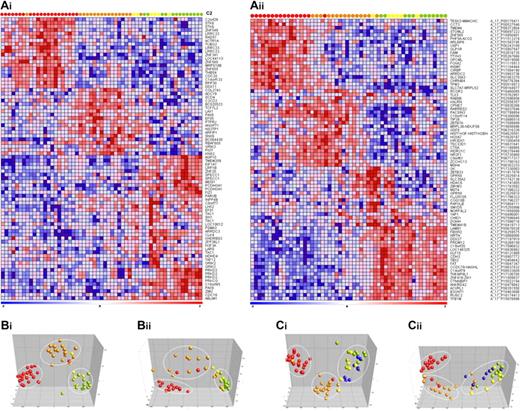
Subsequently, to explore whether specific DNA methylation profiles could define the genetic subgroups of infant ALL more accurately, the 20 most discriminative hypermethylated genes for each group (compared with all other relevant subgroups combined) were selected. For both microarray platforms the gene names, log-fold changes, and P values are listed in supplemental Tables 2 and 3. Permutation testing validated the robustness of the subtype-specific methylation signatures. By using the selected genes, we generated heatmaps in which both the genes and samples were clustered hierarchically (Euclidean distance and complete linkage; Figure 2A). This semisupervised analysis revealed that MLL t(4;11)- and t(11;19)-positive patients could clearly be separated from one another and from the other samples. In contrast, hypermethylated genes that unambiguously separate t(9;11)-positive samples from MLL wild-type (untranslocated) infant ALL samples could not be identified. Moreover, the most significantly hypermethylated genes shared by t(9;11)-positive and wild-type MLL samples were also methylated in healthy bone marrow samples (supplemental Figure 1) and therefore likely reflect normal methylation in healthy hematopoietic cells. Importantly, these genes are hypomethylated in infant ALL harboring translocation t(4;11) or t(11;19).
Infant ALL subtype-specific CpG island hypermethylation. (Ai-ii) Heatmaps showing the 20 most significantly hypermethylated probes for each infant ALL subtype. Columns represent patient samples and rows represent genes. Relative DNA methylation levels are shown in red (high) and blue (low). Genes and samples were ordered by the use of hierarchical cluster analysis (Euclidean distance, complete linkage), and gene identifiers are listed at the right. (Bi-ii) PCAs separating t(4;11) (red), t(11;19) (orange), t(9;11) (yellow), and MLL wild-type infant ALL (green). (Ci-ii) shows the PCA when normal pediatric bone marrow samples (blue) are included in the analysis. (Ai, Bi, and Ci) Data from the custom 9K CpG island microarray. (Aii, Bii, and Cii) Data from the commercially available 244K CpG island microarray (Agilent).
Infant ALL subtype-specific CpG island hypermethylation. (Ai-ii) Heatmaps showing the 20 most significantly hypermethylated probes for each infant ALL subtype. Columns represent patient samples and rows represent genes. Relative DNA methylation levels are shown in red (high) and blue (low). Genes and samples were ordered by the use of hierarchical cluster analysis (Euclidean distance, complete linkage), and gene identifiers are listed at the right. (Bi-ii) PCAs separating t(4;11) (red), t(11;19) (orange), t(9;11) (yellow), and MLL wild-type infant ALL (green). (Ci-ii) shows the PCA when normal pediatric bone marrow samples (blue) are included in the analysis. (Ai, Bi, and Ci) Data from the custom 9K CpG island microarray. (Aii, Bii, and Cii) Data from the commercially available 244K CpG island microarray (Agilent).
Next, PCA was used to better visualize these different clusters, emphasizing the separation of the samples into the 3 expected groups characterized by t(4;11), t(11;19), or t(9;11) together with translocation-negative infant ALL (Figure 2B). When included, the normal bone marrow samples remained within the cluster comprising samples carrying t(9;11) or wild-type MLL genes (Figure 2C). In concordance with this, no significant aberrant DNA methylation could be detected in t(9;11)-positive or untranslocated (wild-type MLL) infant ALL when separately compared with normal bone marrow.
Correlation between promoter methylation and gene expression
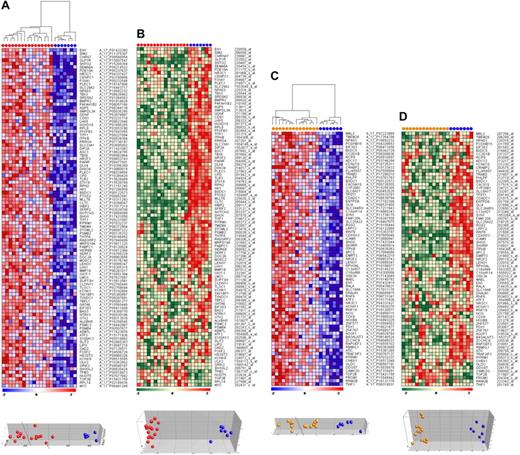
Given the aberrant methylation patterns in t(4;11)- and t(11;19)-positive infant ALL samples, we investigated the effects of promoter hypermethylation (Agilent platform) on gene expression (Affymetrix platform) of corresponding genes. Compared with normal bone marrow samples, infant ALL cells carrying t(4;11) displayed a total of 794 hypermethylated CpG island probes (FDR < 0.01), and 75 probes were significantly hypermethylated in t(11;19)-positive infant ALL (FDR < 0.01). From these analyses, the most significantly hypermethylated genes were selected. Gene names, log-fold changes in methylation, and P values for these genes are listed in supplemental Tables 4 and 5. Next, DNA methylation array data were compared with gene expression profiles from the same samples and visualized as heatmaps and PCA plots (Figure 3). Promoter hypermethylation and down-regulated gene expression correlated for approximately 90% to 95% of the genes in both t(4;11)-positive and t(11;19)-positive infant ALL. However, for the remaining genes we observed the opposite; despite extensive hypermethylation, these genes were greater expressed in leukemic samples than in normal bone marrow.
Correlation between CpG island methylation and gene expression. (A) Heatmap and PCA showing the most significantly hypermethylated genes in t(4;11)-positive infant ALL (red dots) compared with normal bone marrows (blue dots). Relative DNA methylation levels are shown in red (high) and blue (low). (B) Heatmap and PCA showing the corresponding gene expression levels from the same genes and samples as presented in panel A. Relative gene expression values are shown in red (high) and green (low). Similarly, the (C) CpG methylation data and (D) gene expression data are presented for t(11;19)-positive infant ALL samples (orange dots) compared with normal bone marrow (blue dots).
Correlation between CpG island methylation and gene expression. (A) Heatmap and PCA showing the most significantly hypermethylated genes in t(4;11)-positive infant ALL (red dots) compared with normal bone marrows (blue dots). Relative DNA methylation levels are shown in red (high) and blue (low). (B) Heatmap and PCA showing the corresponding gene expression levels from the same genes and samples as presented in panel A. Relative gene expression values are shown in red (high) and green (low). Similarly, the (C) CpG methylation data and (D) gene expression data are presented for t(11;19)-positive infant ALL samples (orange dots) compared with normal bone marrow (blue dots).
The degree of methylation influences clinical outcome in MLL-rearranged infant ALL
In both the heatmaps and PCA plots that represent the most significantly hypermethylated genes among t(4;11)-positive and t(11;19)-positive infant ALL, a clustering of patient samples into 2 subgroups appeared. Ostensibly, one cluster represents patient samples that, at least for the selected genes, seem to be more densely hypermethylated than the samples in the other cluster (Figure 3). To better visualize this difference in degree of methylation, we plotted the normalized methylation log-ratios of the genes. This semiquantitative representation of the data indeed confirmed differences in the degree of methylation between both clusters (supplemental Figure 2). To explore the clinical relevance of these subgroups, we computed risk of relapse statistics for these patient groups (supplemental Figure 3). Four patients received bone marrow transplantation in complete remission. For these patients data were censored at bone marrow transplantation. One patient died before the start of treatment (referred to as early death) and was excluded from further analyses. Twelve of 16 patients (75%) from the “heavily” methylated subgroup had a relapse after achieving complete remission, whereas among the “lightly” methylated patients, relapses occurred in 5 of 12 of the cases (42%). The cumulative incidence of relapse at 1 year after diagnosis was significantly (P < .05) different for the “heavily” and “lightly” methylated subgroups with incidences of 52.5 (SE, 13.7) and 35.7 (SE, 15.5), respectively.
The number of patients that could be included in these analyses is not sufficient to evaluate the impact of the degree of DNA methylation adjusted for known prognostic factors (ie, age, white blood cell count, and the in vivo response to prednisone) separately. Therefore, we used the INTERFANT-06 risk stratification, which represents a combination of these factors (supplemental Methods).2 Although these results must be interpreted with caution, the Cox regression model indicates that heavy methylation confers an increased risk of relapse (hazard ratio 5.77; 95% confidence interval, 1.57-21.2; P = .01; supplemental Table 6). The separate clustering of these 2 patient groups, however, did not appear in the gene expression profiles (Figure 3). This finding implies that the grouping of these patients and the observed variance in relapse-free survival rather reflects progressive accumulation of genome-wide methylation, than direct differences in gene expression. In line with this hypothesis we show that the division into “heavy” and “light” methylation remains present when all significantly hypermethylated probes are used in a semiquantitative representation (supplemental Figure 2).
MLL t(4;11)-positive cell lines as models for demethylation
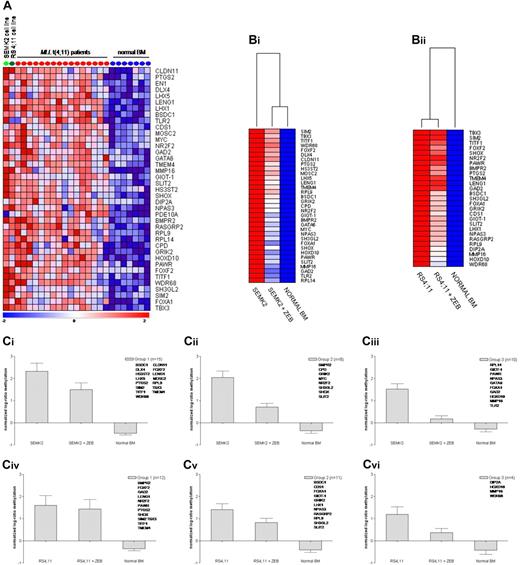
DNA methylation patterns of 2 t(4;11)-positive precursor B-cell ALL cell lines (ie, RS4;11 and SEMK2) were compared with the profiles from the t(4;11)-positive infant ALL samples. Nearly 50% of the 100 most significantly hypermethylated genes in t(4;11)-positive infant ALL were also hypermethylated in these cell lines (Figure 4A). Representing reasonable models for t(4;11)-positive infant ALL samples, we next studied the effects of demethylation on these genes by comparing DNA methylation profiles of these cell lines before and after a 10-day exposure to 100μM of the demethylating agent zebularine. In the SEMK2 and RS4;11 cell lines, respectively, 72% (33/46) and 59% (27/46) of the hypermethylated genes showed notable decreases in methylation upon exposure to zebularine. The genes display varying degrees of drug-induced demethylation (Figure 4B-C). For some of the genes the methylation status could be restored to nearly normal levels as observed in healthy hematopoietic cells.
ALL cell lines as models for (de)methylation. (A) Heatmap showing methylation levels in the t(4;11)-positive B-ALL cell lines SEMK2 (light green dot) and RS4;11 (dark green dot) of genes most significantly methylated in t(4;11)-positive infant ALL patients (red dots) compared with normal bone marrow (blue dots). (B) Heatmaps showing methylation levels of these genes after exposure to zebularine. These methylation levels were compared with the average methylation levels as determined from normal bone marrow samples (n = 7). (Bi) SEMK2 cell line and (Bii) RS4;11 cell line. (Ci-vi) Graphs displaying the mean and the standard error of the mean of changes in methylation levels after zebularine exposure. Genes were divided into 3 groups for each cell line according to the degree of responsiveness to zebularine. (Ci-iii) SEMK2 cell line; (Civ-vi) RS4;11 cell line.
ALL cell lines as models for (de)methylation. (A) Heatmap showing methylation levels in the t(4;11)-positive B-ALL cell lines SEMK2 (light green dot) and RS4;11 (dark green dot) of genes most significantly methylated in t(4;11)-positive infant ALL patients (red dots) compared with normal bone marrow (blue dots). (B) Heatmaps showing methylation levels of these genes after exposure to zebularine. These methylation levels were compared with the average methylation levels as determined from normal bone marrow samples (n = 7). (Bi) SEMK2 cell line and (Bii) RS4;11 cell line. (Ci-vi) Graphs displaying the mean and the standard error of the mean of changes in methylation levels after zebularine exposure. Genes were divided into 3 groups for each cell line according to the degree of responsiveness to zebularine. (Ci-iii) SEMK2 cell line; (Civ-vi) RS4;11 cell line.
Specific zebularine sensitivity in MLL-rearranged ALL cells
To further investigate the sensitivity of ALL cells to in vitro demethylation, cytotoxicity assays were performed by the use of escalating dosages of zebularine. Also 2 AML cell lines were added to the dataset. AML cells (with or without an MLL translocation) seem to be less sensitive to the demethylating agent zebularine than MLL-rearranged ALL cells, but the MLL-rearranged AML cell line MV4-11 does appear more sensitive than the t(8;21)-positive AML cell line Kasumi-1. Clearly, MLL-rearranged ALL cells were significantly more sensitive to zebularine than the other cell lines (P < .01; Figure 5A-B). As shown in Figure 5B, on average the IC50 value (ie, the concentration inhibitory to 50% of the cells) in MLL-rearranged ALL cells was approximately 50μM, whereas zebularine failed to reach an IC50 value in other types of ALL cell lines.
In vitro cytotoxicity to zebularine. (A) Dose-response curves showing the in vitro cytotoxic response to zebularine in individual leukemia cell lines with or without MLL rearrangements or (B) the mean cytotoxic response for MLL-rearranged ALL cell lines (n = 4) and for the other ALL cell lines (n = 4). Error bars represent SEM. The differences between the means of the groups were statistically analyzed by the use of the 2-tailed Student t test (P < .01 for each concentration used).
In vitro cytotoxicity to zebularine. (A) Dose-response curves showing the in vitro cytotoxic response to zebularine in individual leukemia cell lines with or without MLL rearrangements or (B) the mean cytotoxic response for MLL-rearranged ALL cell lines (n = 4) and for the other ALL cell lines (n = 4). Error bars represent SEM. The differences between the means of the groups were statistically analyzed by the use of the 2-tailed Student t test (P < .01 for each concentration used).
Discussion
We here present the first global view of the DNA methylome in infant MLL-rearranged ALL. MLL-rearranged infant ALL represents an aggressive and difficult-to-treat type of leukemia characterized by a unique gene expression profile that clearly separates this malignancy from other ALL subtypes.8,9 Because epigenetic modifications directly influence gene expression patterns,10 we hypothesized that specific DNA methylation patterns may underlie the characteristic gene signature as observed for MLL-rearranged infant ALL. Our data largely support this hypothesis because the majority of MLL-rearranged infant ALL cases [ie, those characterized by t(4;11) or t(11;19)] represent hypermethylated leukemias, whereas t(9;11)-positive and MLL translocation-negative (wild-type MLL) infant ALL display DNA methylation patterns that closely resemble that of normal bone marrow. Moreover, distinct leukemia-specific DNA methylation patterns could be identified for the different MLL-rearranged infant ALL subtypes as defined by the type of MLL translocation or absence of such translocations. Interference of nonleukemia-related epigenetic differences in DNA methylation (such as age, sex-specific differences in methylation, and differences related to B-cell maturation stages of leukemic cells) with our results could be excluded (supplemental Results and supplemental Tables 7-8).
Thus, the presence as well as the patterns of aberrant DNA methylation in infant ALL appear, at least to some extent, dependent on the presence and type of MLL fusion, which may reflect a mechanism proposed recently.27 Apart from DNA methylation, a second component of the epigenetic code involves histone modifications,28 shaping the chromatin in an open (transcriptionally active) or closed (inactive) conformation. An inactive chromatin state usually is associated with hypermethylated CpG promoter regions, whereas active chromatin marks, such as H3K4 trimethylation and H3K79 dimethylation, denote unmethylated promoters, allowing transcription. Interestingly, the MLL gene itself has specific histone methyltransferase activity,29,30 which is lost during fusion of the MLL gene to one of its translocation partners. Therefore, MLL fusions can be expected to result in altered chromatin structures because of aberrant histone modifications.
Recently, Krivtsov and Armstrong27 proposed that the recruitment of different histone methyltransferases by different MLL fusion proteins may indeed result in inappropriate histone modifications directed by the MLL fusion partner. Given the sound interplay between histone modification and CpG island methylation, this proposed influence of different MLL fusion genes on histone modifications, and the apparent influence of the MLL fusion partner on DNA methylation as shown in the present study, are presumably linked. In addition, this work by Krivtsov and Armstrong,27 as well as a study by Mueller et al,31 demonstrated the recruitment of a transcriptional elongation complex to MLL target genes, resulting in gene activation (ie, expression). These studies suggest that MLL fusion proteins trigger or maintain the leukemia by the activation of specific target genes.
In contrast, our present study shows that, apart from specific gene activation, MLL-rearranged ALL is also characterized by severe gene inactivation, which may well be driven by the same MLL fusion. MLL-rearranged ALL cells typically mirror highly immature B cells. Possibly, the MLL fusion ignores the activation of many genes that should have been activated (by wild-type MLL) at this stage of B-cell development and are necessary for proper differentiation toward mature and functional B cells. This would suggest that our observed patterns of gross genome-wide DNA methylation are in favor of blocking B-cell differentiation while simultaneously the MLL fusion activates several (proto-onco)genes in favor of uncontrolled cell proliferation and survival. Alternatively (or additionally), inappropriate activation of certain genes by the MLL fusion may in turn induce abnormal inactivation (silencing) of several other genes. However, these proposed mechanisms are highly speculative and remain to be confirmed.
Nonetheless, we can conclude from our data that MLL-AF4 and MLL-ENL represent MLL fusion proteins that both alter histone modifications that result in strongly altered DNA methylation patterns. The differences found in DNA methylation patterns between MLL-AF9 and MLL-ENL may then seem surprising given the apparent common mechanism of transformation involving the recruitment of DOT1L as put forward by others.27,31 Surprisingly, the MLL-AF9 fusion did not lead to significant aberrant DNA methylation in infant ALL. This finding suggests that oncogenic transformation in t(4;11)- and t(11;19)-positive infant ALL patients may be facilitated or largely driven by gross epigenetic changes, whereas t(9;11)-positive infant ALL cells presumably transform via alternative mechanisms. In concordance with this finding is that t(9;11)-positive ALL patients characteristically seem to be different from other MLL-rearranged infant ALL patients. For example, t(9;11)-positive infant ALL is typically diagnosed at a later stage during infancy and usually is characterized by a more mature immunoglobulin gene rearrangement pattern (immunophenotype) than t(4;11)- and t(11;19)-positive infant ALL.2,4 However, no significant differences in survival exist between infant ALL patients carrying either t(4;11) or t(11;19) and patients with t(9;11).2
By studying the genes most significantly hypermethylated in t(4;11)- and t(11;19)- positive infant ALL samples, we found that the expression of the majority of these genes (∼ 90%-95%) was indeed down-regulated. Among the hypermethylated genes we found genes that were previously described to be silenced as the result of DNA hypermethylation in MLL-rearranged ALL, such as the tumor suppressor gene FHIT13 and the DLX3 gene,32 demonstrating the integrity of our data. Moreover, most of these genes responded well to exposure to the demethylating agent zebularine in t(4;11)-positive cell line models. Among the most significantly hypermethylated genes for either t(4;11)-positive or t(11;19)-positive infant ALL, a limited overlap was observed. Nevertheless, global gene ontology analysis showed that most of the down-regulated genes in both subgroups are involved in transcriptional regulation (supplemental Table 9). This pronounced epigenetic deregulation of the transcriptional machinery may indeed have contributed to the unique gene expression profile characteristic for MLL-rearranged ALL.8,9 Yet, this would not be true for t(9;11)-positive infant ALL because no aberrant promoter hypermethylation was observed in these samples. This apparent contradiction, however, is easily explained by the fact that most of the published MLL-specific gene expression signatures, including the signatures reported by Armstrong et al,8 are predominantly based on t(4;11)- and t(11;19)-positive samples. Therefore, gene expression profiling studies, including t(9;11)-positive infant ALL samples, may well come to demonstrate that profiles associated with t(9;11) are different from those obtained in t(4;11)- and t(11;19)-positive samples.
Remarkably, approximately 5% of the most significantly hypermethylated genes in t(4;11)- and t(11;19)-positive infant ALL remained highly expressed. This observation controverts the dogma that promoter methylation per definition induces suppression of gene expression. However, Weber et al33 recently nuanced this dogma by demonstrating the influence of promoter CpG density on the ability to induce transcriptional repression. Therefore, these methylated but highly expressed genes may well exhibit promoters containing weak CpG islands (ie, a low or intermediate CpG density), unable to repress transcription even when methylated. Another possible explanation for this would again be the involvement of the MLL fusion protein, which may have induced activating histone modifications on otherwise inactive regions in the chromatin associated with promoter methylation. In turn, this newly acquired open chromatin state may have overruled the relatively weak DNA methylation, allowing transcription despite earlier established epigenetic silencing. If so, this group of genes may well represent potential therapeutic targets directly influenced by the MLL fusion itself.
Most of the genes that were methylated in t(9;11)-positive infant ALL and infant ALL carrying wild-type MLL genes were also methylated in normal bone marrow. Presumably, these represent genes that were already silenced in normal hematopoietic cells but became hypomethylated in t(4;11)- and t(11;19)-positive infant ALL cells. Interestingly, among these were several genes with oncogenic potential, such as CDH3, TBX2, ERCC1, and NPR2 (Figure 2A-B), that have been reported to be involved in proliferation, tumor aggressiveness, and prognosis in a wide range of human cancers.34,35 Interestingly, among these hypomethylated genes also appeared the HOXA9 gene, which was previously described to be protected from methylation by the MLL fusion itself.36 Thus, the present study not only characterizes epigenetically down-regulated genes but also identifies protooncogenes that may be inappropriately expressed in t(4;11)- and t(11;19)-positive MLL-rearranged ALL in infants. Obviously, such genes represent yet another set of candidate target genes for future therapeutic intervention.
Ofmain therapeutic interest is our finding that the degree of DNA methylation among t(4;11)- and t(11;19)-positive infant ALL patients is related to relapse-free survival, with patients presumably carrying heavily methylated genomes being at an increased risk of relapse. Therefore, these children in particular should be considered candidates for therapies, including inhibitors of DNA methylation, especially because we show here that MLL-rearranged ALL cells are highly sensitive to zebularine in vitro. We believe that this increased sensitivity to demethylation is rather based on the presence of a general methylator phenotype (ie, globally deregulated DNA methylation) than on the actual reexpresssion of a fixed number of hypermethylated genes. Apparently, genome-wide demethylation is sufficient to cause MLL-rearranged ALL cells to undergo apoptosis. This finding is in concordance with the identification of a heavily and a lightly methylated subgroup of MLL-rearranged infant ALL, which is also based on a widespread phenotype with more or less pronounced levels of DNA methylation that are in fact not visible at the gene expression level. In conclusion, the findings presented here urgently require gene per gene validation studies and mandate additional studies using demethylating agents in the currently only available genuine mouse model for MLL-rearranged ALL, recently described by Krivtsov et al.37
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the members and participating hospitals of the INTERFANT-99 study for supporting our research by providing leukemic samples. Members of INTERFANT-99 include M. Campbell (Programa Infantil Nacional de Drogas Atineoplasicas), M. Felice (Argentina), A. Ferster (Children's Leukemia Group), I. Hann, and A. Vora (UK Children's Cancer Study Group); L. Hovi (Nordic Society of Paediatric Haematology and Oncology), G. Janka-Schaub (Cooperative Study Group for Treatment of ALL), C. K. Li (Hong Kong), G. Mann (Berlin-Frankfurt-Münster Group-Austria), T. LeBlanc (French ALL Group), R. Pieters (Dutch Childhood Oncology Group), G. de Rossi, and A. Biondi (Associazione Italiana Ematologia Oncologia Pediatrica); and J. Rubnitz (St Jude Children's Research Hospital), M. Schrappe (Berlin-Frankfurt-Münster Group-Germany), L. Silverman (Dana-Farber Cancer Institute), J. Stary (Czech Paediatric Haematology), R. Suppiah (Australian and New Zealand Children's Haematology/Oncology Group), T. Szczepanski (Polish Paediatric Leukemia and Lymphoma Study Group), M. Valsecchi, and P. de Lorenzo (Trial Operating Center). Furthermore, T. H. Huang and P. Yan are gratefully acknowledged for contributing the 9K CpG island clone library and protocols for differential methylation hybridization.
This study was financially supported by a grant from the Sophia Foundation for Medical Research (SSWO grant 495). RXM has been partially funded by the Center of Medical Systems Biology (CMSB) established by the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NGI/NWO). The institutions financially supporting this study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Authorship
Contribution: D.J.P.M.S. designed and performed research and wrote the paper; P.S. performed research; E.H.J.v.R. provided technical assistance; R.X.M. designed the statistical analyses; P.L. and M.G.V. gathered patient information and computed survival statistics; R.P. and J.M.B. reviewed the paper; and R.W.S. designed and guided research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ronald W. Stam, PhD, Erasmus Medical Center–Sophia Children's Hospital, Department of Pediatric Oncology/Hematology, Rm Ee15-14a, Dr Molewaterplein 50, 3015 GE Rotterdam, The Netherlands; e-mail: r.stam@erasmusmc.nl.
References
Author notes
*P.S. and E.H.J.v.R. contributed equally to this work.