In this issue of Blood, Ye and colleagues show that CD34+ cells obtained from patients with JAK2-V617F MPDs could be reprogrammed to iPSCs and be differentiated back into hematopoietic progenitors.1
Myeloproliferative disorders (MPDs) represent a group of clonal hematopoietic progenitor/stem cell disorders associated with excess production of cells of myeloid lineages, resulting in an increase in one or more mature peripheral blood elements. This group of myeloproliferative neoplasms includes polycythemia vera (PV), essential thrombocythemia (ET), primary myelofibrosis (PMF), chronic myeloid leukemia (CML), and other rarer disorders. Whereas CML was the first blood cancer known to be linked to a chromosomal translocation, the JAK2-V617F mutation associated with PV was discovered only several years ago.2 However, the mechanisms of transformation by JAK2-V617F mutation are not well understood, particularly why the same mutation causes different phenotypes including PV, ET, or PMF. It has been hypothesized that disease manifestation depends on the cell affected by the original mutation, the genetic background of the patient, or the level of JAK2-V617F activity. The work by Ye et al provides a novel approach to ask and answer important questions about MPD pathogenesis, by modeling development of myeloproliferative neoplasia in vitro using patient-specific induced pluripotent stem cells (iPSCs).1
In 2006, the Yamanaka group revealed that mouse skin fibroblasts could be reprogrammed to pluripotency via ectopic expression of 4 transcription factors.3 A year later, iPSCs were obtained from human fibroblasts.4,5 These discoveries opened opportunities to generate disease-specific iPSCs carrying a particular genetic trait at the cellular level. As proof of this concept, iPSCs have been generated from fibroblasts obtained from patients with several genetic diseases including the inherited bone marrow failure syndrome Fanconi anemia.6 However, a fibroblast-based approach would not work for acquired blood diseases such as MPDs or leukemia, because cytogenetic abnormalities defining such diseases are limited to bone marrow cells in most of the cases. Several months ago, Loh et al demonstrated that iPSCs could be generated by reprogramming mobilized peripheral blood CD34+ cells.7 The work published in this issue of Blood by Ye et al is the first description of successful reprogramming of CD34+ cells from patients with acquired blood diseases.1 Using retroviral vectors encoding Oct4, Sox2, Klf4, and c-Myc genes, Ye et al generated iPSCs from CD34+ cells obtained from healthy controls and MPD patients carrying the JAK2-V617F mutation. While MPD-derived iPSCs retained the JAK2-V617F mutation, they had a normal karyotype, embryonic stem cell–like phenotype, and pluripotent differentiation potential. When control and diseased iPSCs were differentiated back into CD34+CD45+ hematopoietic progenitors, the progenitors derived from MPD-iPSCs recapitulated the features of somatic CD34+ cells from which the iPSCs were originally derived. Similar to somatic MPD CD34+ cells, iPSC-derived CD34+CD45+ cells demonstrated enhanced erythropoiesis and up-regulation of genes known to be increased in PV.
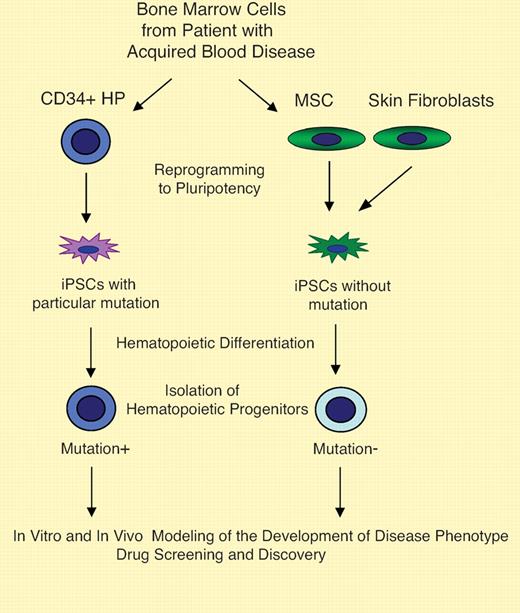
This study clearly demonstrates how iPSC technology could be used to model acquired blood diseases. This technology would be of particular value for the study of blood disorders such as myelodysplastic syndromes, paroxysmal nocturnal hemoglobinuria, and others for which animal models are not available or difficult to create. In addition, iPSCs carrying leukemia-specific cytogenetic translocation could be used to analyze how cancer stem cells develop. Importantly, the iPSC-based approach would be helpful in addressing the role of genetic background in manifestation of neoplastic blood disorders. Because iPSCs are capable of indefinite self-renewal, diseased blood cells can be generated continuously in the laboratory, eliminating the need for a constant supply of hematopoietic progenitors from the patients. In particular, a continuous supply of genetically diverse diseased blood cells for drug screening and discovery could be created. Because multiple types of cells can be generated from iPSCs, interaction of diseased blood cells with endothelial or stromal cells could be modeled in vitro. However, several important issues related to iPSC models of blood diseases remain to be addressed. It is known that the hematopoietic differentiation potential of iPSC lines generated from the same starting material varies significantly.8 If several clones were generated from iPSCs, which clones should be selected to make an appropriate conclusion regarding differences in differentiation potential? What would be an appropriate control for diseased versus nondiseased iPSCs? For studies of acquired blood diseases, iPSC lines can be generated from hematopoietic cells and fibroblasts or bone marrow mesenchymal stem cells (see figure). In this way, iPSCs with the same genetic background, but different in terms of presence or absence of acquired mutations, will be available for comparative analysis. The majority of disease-specific iPSCs have been made using retroviral vectors. Although the impact of exogenous expression is unclear, the possibility remains that retroviral integration and background expression of pluripotency genes may affect the behavior of iPSC-derived hematopoietic progenitors. Recently developed new reprogramming methods allowing for the generation of transgene-free iPSCs will be helpful to overcome this limitation.
The use of iPSCs in modeling for acquired blood disease. Bone marrow samples from patients with acquired blood diseases can be used to obtain mutation-free mesenchymal stem cells (MSCs) and CD34+ cells or other types of hematopoietic progenitors (HPs) carrying disease-associated mutation. Alternatively, diseased peripheral blood CD34+ cells and fibroblasts or other types of cells lacking mutation from the same patient can be used. By reprogramming cells with or without genetic abnormality from the same patient, iPSCs with the same genetic background but different in expression of mutation can be generated. Using an in vitro differentiation system, hematopoietic precursors at different stages of maturation and terminally differentiated cells can be obtained for studies of disease pathogenesis. Transplantation of de novo generated cells with neoplasia-specific mutation into immunocompromised mice can be used to address emergence of blood cancer stem cells. Drug screening and discovery is another obvious and immediate benefit of iPSC technology for development of new therapies for blood diseases.
The use of iPSCs in modeling for acquired blood disease. Bone marrow samples from patients with acquired blood diseases can be used to obtain mutation-free mesenchymal stem cells (MSCs) and CD34+ cells or other types of hematopoietic progenitors (HPs) carrying disease-associated mutation. Alternatively, diseased peripheral blood CD34+ cells and fibroblasts or other types of cells lacking mutation from the same patient can be used. By reprogramming cells with or without genetic abnormality from the same patient, iPSCs with the same genetic background but different in expression of mutation can be generated. Using an in vitro differentiation system, hematopoietic precursors at different stages of maturation and terminally differentiated cells can be obtained for studies of disease pathogenesis. Transplantation of de novo generated cells with neoplasia-specific mutation into immunocompromised mice can be used to address emergence of blood cancer stem cells. Drug screening and discovery is another obvious and immediate benefit of iPSC technology for development of new therapies for blood diseases.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■