Abstract
Leukocyte integrins of the β2 family are essential for immune cell-cell adhesion. In activated cells, β2 integrins are phosphorylated on the cytoplasmic Thr758, leading to 14-3-3 protein recruitment to the β2 integrin. The mutation of this phosphorylation site impairs cell adhesion, actin reorganization, and cell spreading. Thr758 is contained in a Thr triplet of β2 that also mediates binding to filamin. Here, we investigated the binding of filamin, talin, and 14-3-3 proteins to phosphorylated and unphosphorylated β2 integrins by biochemical methods and x-ray crystallography. 14-3-3 proteins bound only to the phosphorylated integrin cytoplasmic peptide, with a high affinity (Kd, 261 nM), whereas filamin bound only the unphosphorylated integrin cytoplasmic peptide (Kd, 0.5 mM). Phosphorylation did not regulate talin binding to β2 directly, but 14-3-3 was able to outcompete talin for the binding to phosphorylated β2 integrin. X-ray crystallographic data clearly explained how phosphorylation eliminated filamin binding and induced 14-3-3 protein binding. Filamin knockdown in T cells led to an increase in stimulated cell adhesion to ICAM-1–coated surfaces. Our results suggest that the phosphorylation of β2 integrins on Thr758 acts as a molecular switch to inhibit filamin binding and allow 14-3-3 protein binding to the integrin cytoplasmic domain, thereby modulating T-cell adhesion.
Introduction
Integrins are heterodimeric plasma membrane receptors that mediate binding to the extracellular matrix and to ligands present on the surface of other cells. Their function is tightly regulated; they bind ligands only after activation. Modulation of integrin activity occurs through tightly regulated interactions between cytoplasmic molecules and integrin intracellular tails. Factors binding to integrin cytoplasmic domains regulating integrin adhesiveness include the cytoskeletal proteins talin1,2 and filamin,3 and the 14-3-3 proteins, which are molecular adaptors that bind to phosphorylated serine or threonine (pSer/pThr) containing polypeptide sequences.4
The β2 integrins are expressed exclusively on leukocytes and bind ICAM molecules on other leukocytes and endothelial cells after cell activation.5,6 Talin binds to β2 integrins in vitro and in cells and is involved in activating the β2 integrins, resulting in binding to ICAMs.1,4,7-9 The β2 integrin polypeptide chain is phosphorylated on the intracellular domain on several residues after cell stimulation with various agents.10 Thr758 is a physiologically important amino acid residue in the β2 cytoplasmic tail, and becomes phosphorylated after T-cell stimulation with T-cell receptor (TCR) antibodies or with phorbol esters.11-13 After its phosphorylation, β2 binds to 14-3-3 proteins both in vitro and in cells.4 Blocking of this interaction with a β2 Thr758 to Ala mutation, or by expression of constructs that bind to 14-3-3 proteins and block their interactions with target proteins, leads to abrogation of actin cytoskeleton rearrangements, cell spreading, and adhesion to ICAM ligands.4 β2-Thr758 phosphorylation leads to the activation of the actin cytoskeleton modulators, Rac1/Cdc42, in cells.13
The region in the β2 cytoplasmic tail that binds 14-3-3 proteins has been reported to interact with filamin in other integrins,14 and for the strong filamin-binder β7 integrin, phosphorylation mimicking substitutions of 3 threonine residues (TTT) reduces filamin affinity.3 Filamin has been reported to associate with β2 integrins in vivo15,16 and it binds a β2 integrin peptide containing the TTT sequence in vitro.7 The filamin-integrin cytoplasmic tail interaction negatively regulates talin binding and talin-dependent integrin activation3 ; it also regulates cell migration.14
In this study, we investigated the binding of the β2 integrin cytoplasmic domain to filamin, talin, and 14-3-3. We show that the β2 Thr758 phosphorylation abrogates filamin binding but is required for 14-3-3 binding. Phosphorylation does not directly regulate talin binding to β2 integrin, but 14-3-3 can outcompete talin for binding to phosphorylated β2 integrin. Filamin knockdown by siRNA increases cell adhesion to ICAM-1, indicating that filamin plays a negative role in regulating adhesion. We solved the crystal structures of Thr758-phosphorylated β2 peptide/14-3-3 complex and unphosphorylated β2 peptide/filamin complex. These structures show that the interaction sites are mostly overlapping and explain how β2 phosphorylation switches the binding specificity between filamin and 14-3-3.
Methods
Materials
The peptides β2-35, β2-35A, β2-35pT, β2-12, ARAApSAPA, and pαL (Table 1) were synthesized by Fmoc chemistry as in Valmu et al.7 The α-actinin antibody (MAB1682) and the filamin antibody (MAB1678) were from Chemicon (Temecula, CA), the 14-3-3 blotting antibody (K-19) was from Santa Cruz Biotechnology (Santa Cruz, CA), and the actin antibody (A5060) was from Sigma-Aldrich (St Louis, MO). The pThr758-β2 antibody and the R2E7B antibody to β2 have been described previously.13,17 Predesigned siRNAs for filamin A (siGenome smart pool, M-012579-00) and controls cyclophilin B (siGLO, D-001610-01-05) and nontargeting siRNA (D-001210-02-05) were from Dharmacon (Lafayette, CO).
Peptide affinity chromatography
Peptides (β2-35, β2-35A, β2-35pT, and pαL) were coupled to vinyl sulphon–activated agarose. T cells were isolated from buffy coats as described16 and lysed in 1% Tx-100, 10 mM sodium phosphate, pH 7.4, 50 mM NaCl, 10 mM EDTA, 50 mM NaF, and protease inhibitor cocktail. In some experiments, the T-cell lysates were pretreated with the peptide ARAApSAPA (10 μM). Peptide affinity chromatography with the T-cell lysates was performed as in Valmu et al.16 The competition assay between purified 14-3-3ζ and talin was performed in 1% Tx-100, 150 mM NaCl, 50 mM NaF, 10 mM EDTA, 50 mM Tris, pH 7.4. Proteins were incubated for 30 minutes with 10 μL peptide-coupled agarose. The samples were then washed 5 times with buffer, and eluted with NuPAGE LDS Sample Buffer (Invitrogen, Frederick, MD) at 70°C for 10 minutes.
Transfection with siRNA
The Jurkat cell clone E6.1 (ATCC, Manassas, VA) was maintained in RPMI 1640 medium supplemented with 10% FBS, l-glutamine, and antibiotics. Jurkat E6.1 cells (1.0 × 106 per transfection) were washed with PBS, suspended in 0.1 mL siPORT siRNA electroporation buffer (Ambion, Austin, TX), and mixed with 200 nM siCONTROL Non-Targeting no. 2, siGLO Cyclophilin B, and siGENOME SMART pool human filamin A (FLNa) siRNAs (Dharmacon). Electroporation was performed twice at 325 V, 175 Ω, and 1 μF with pulse width of 0.2 ms and pulse interval of 2 seconds. Cells were grown for 4 days.
Cell stimulation, lysis, SDS–polyacrylamide gel electrophoresis, and Western blot
For pThr758-detection, T cells were isolated from buffy coats as described.16 They were left untreated or preincubated with 1.5 μM okadaic acid and stimulated for 30 minutes with either 200 mM protein kinase C stimulating phorbol ester, phorbol 12,13-dibuturate (PDBu; Sigma-Aldrich), or 10 μg/mL of the T-cell receptor stimulating antibody OKT3 (clone CRL 8001; ATCC). After cell lysis, extracts were run on SDS–polyacrylamide gel electrophoresis (PAGE) and detected with the pThr758 polyclonal antibody as described.13 Membranes were stripped and reprobed with a β2 integrin blotting antibody (R2E7B) to confirm equal loading.
For detection of filamin knockdown, 106 siRNA-treated cells were centrifuged and the pellet was washed once with PBS. The cells were lysed in NET-buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) including protease inhibitors (Complete; Roche, Indianapolis, IN). Cell extracts were analyzed by Western blotting and detected with Amersham Biosciences (Arlington Heights, IL) HRP-labeled secondary antibodies (na931v and na934v) and Pierce enhanced chemiluminescence (ECL) Western Blotting Substrate (32106; Rockford, IL). The FLNa knockdown levels were detected with mouse antibody (mab) against FLNa, and the loading was controlled with polyclonal antibody against actin. The expression levels were quantified from scanned films using TINA 2.0 software.
For detection of proteins bound to peptide columns in the peptide affinity chromatography experiments, eluates were subjected to Western blotting with anti-FLNa (Raytest, Straubenhardt, Germany) or with α-actinin or 14-3-3 antibodies and detected by ECL or were subjected to staining with Simply Blue Safe Stain (Invitrogen).
Adhesion assay
Recombinant soluble human ICAM-1 (0.3 μg/well; R&D Systems, Minneapolis, MN) was coated on flat-bottom 96-well microtiter plates (Nunc, Rochester, NY) by overnight incubation at 4°C. The wells were blocked with 1% dry milk for 1.3 hours at 37°C. Cells were suspended in binding medium (RPMI with 40 mM HEPES, 0.1% BSA, 2 mM MgCl2) and stimulated with 200 nM PDBu or left untreated, added to each well, and allowed to adhere for 30 minutes at 37°C. After incubation, unbound cells were removed by gentle washing. The bound cells (or the total amount of added cells without washing) were lysed in 100 μL per well 0.3 mg/mL p-nitrophenylphosphate (Calbiochem, San Diego, CA) containing lysis buffer (1% Triton X-100, 50 mM sodium acetate, pH 5.0) and incubated for 45 minutes at 37°C. The reaction was terminated by adding 50 μL per well of 1 M NaOH and the absorbance at 405 nm was measured.
Protein production and purification
The human filamin A immunoglobulin-like domain 21 (IgFLNa21) was expressed and purified as previously described.3 The bovine 14-3-3ζ cDNA, which encodes a protein 100% identical with the human protein, in pET-15b plasmid (Novagen, Madison, WI) was a generous gift from Dr R. C. Liddington (Burnham Institute for Medical Research, La Jolla, CA). The recombinant protein contained a thrombin digestion site between 14-3-3ζ sequence and an N-terminal His6 tag.18 Protein expression in Escherichia coli BL21 (DE3) was induced for 3 hours at 37°C in the presence of 0.4 mM IPTG. The cells were disrupted by sonication and soluble proteins were then separated by centrifugation (36 900g for 30 minutes at 4°C). The 14-3-3 fusion protein was purified by a Ni-NTA agarose column (Qiagen, Valencia, CA),19,20 and after overnight thrombin (GE Healthcare, Little Chalfont, United Kingdom) digestion at 23°C the 14-3-3ζ part was eluted. The sample was further purified by HiTrap Q ion exchange chromatography (Amersham Biosciences) in 20 mM Tris-HCl, pH 8.0, with NaCl gradient elution. Final purification was achieved by gel filtration in a Superdex G75 24 mm × 600 mm column (Amersham Biosciences) in 20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM DTT. The pGEX2 plasmid encoding mouse talin F2/F3 head domain (residues 206-405)21 was a generous gift from Dr D. A. Calderwood (Yale University, New Haven, CT). The fusion protein was expressed in E coli BL21 Gold in the presence of 4 mM IPTG. The cells were disrupted with a French press and the soluble proteins were separated by centrifugation (36 900g for 30 minutes at 4°C). The fusion protein was purified by glutathione Sepharose column (GE Healthcare), and after overnight digestion with 500 U thrombin (GE Healthcare) at 23°C the talin part was eluted. For surface plasmon resonance experiments, the remaining thrombin traces were removed using a p-aminobenzamidine column (Sigma-Aldrich). Protein was further purified by gel filtration in a Superdex G75 24 mm × 600 mm column (Amersham Biosciences) in 150 mM NaCl, 20 mM Tris-HCl, pH 7.5.
Quantitative interaction analysis
The real-time binding of 14-3-3ζ, talin, and IgFLNa21 to β2 integrin peptides was analyzed by surface plasmon resonance (SPR) using the Biacore system (Biacore, Uppsala, Sweden). In this system, the binding of soluble analytes to immobilized ligands was measured in arbitrary units (resonance units [RU]). The phosphorylated (β2-35p) and unphosphorylated (β2-35) β2 integrin peptides were coupled to the matrix of a CM5 sensor chip by amine coupling chemistry. Serial dilutions of 14-3-3, talin F2/F3, or IgFLNa21 in running buffer (10 mM HEPES, pH 7.5, 150 mM NaCl, 3 mM EDTA, 0.005% p20 in the case of 14-3-3; 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM DTT in the case of IgFLNa21; and 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT in the case of talin) were injected over the peptide surfaces at a flow rate of 20 μL/min. Binding to an empty negative control surface was subtracted from the obtained sensorgrams. The equilibrium rate constants (Kd values) were calculated using a Langmuir model with the Biacore evaluation software 3.1 provided by the manufacturer for the 14-3-3–β2 and talin–β2 integrin interaction. For the filamin–β2 integrin interactions, which did not fit the Langmuir model, we used an equilibrium binding analysis with Sigmaplot (Systat Software, San Jose, CA). In both interactions, a simple 1:1 interaction was assumed.
Protein crystallography
An equimolar mixture of 1 mM human IgFLNa21 and the β2 integrin peptide β2-21 (Table 1) was crystallized in 1.6 M (NH4)2SO4, 0.2 M NaCH3COO, pH 4.6. The crystals were flash frozen in liquid nitrogen in 30% ethylene glycol containing crystallization solution. The diffraction data were collected at 100 K at the European Synchrotron Radiation Facility (Grenoble, France) beamline ID23-1.
The bovine 14-3-3ζ protein (8-10 mg/mL) was crystallized by the hanging drop vapor diffusion method with similar conditions as described in Liu et al22 (18%-19% PEG3350, 100 mM Tris-HCl, pH 8.5, 10 mM CaCl2, 1 mM NiCl2 at 4°C). The grown 14-3-3ζ crystals were soaked with surplus β2 cytoplasmic integrin phosphopeptide β2-10pT (Table 1). Crystals were transferred in 3 steps into cryoprotectant solution (5%-15% ethylene glycol, 19%-23% PEG3350, 100 mM Tris-HCl, pH 8.5, 10 mM CaCl2, 1 mM NiCl2) and flash frozen in liquid nitrogen.23 The diffraction data were collected at 100 K at the European Synchrotron Radiation Facility beamline ID14-3.
The data were processed and scaled with the XDS program package (Max Planck Institute for Medical Research, Heidelberg, Germany).24 Both of the structures were solved by molecular replacement with Phaser (University of Cambridge, Cambridge, United Kingdom).25 Protein Data Bank (PDB) entry 1A4O21,22,26 was used as the search model for 14-3-3ζ and PDB entry 2BRQ3 for IgFLNa21. Peptide backbone of the IgFLNa/β2 structure was built with the program ARP/wARP version 6.1 (European Molecular Biology Laboratory, Hamburg, Germany).27 The structures were refined with Refmac528 (CCP4, Daresbury Laboratory, Warrington, United Kingdom) and the molecular graphics program O (DatOno AB, Uppsala, Sweden).29 Water molecules were added to the IgFLNa/β2 structure with the program ARP/wARP. Because of hemihedral twinning (twinning fraction 0.306) 14-3-3ζ refinement was further continued with CNS 1.2 program package30 (Yale University, New Haven, CT) using strict NCS restraints. Structure figures were generated using PyMOL (DeLano Scientific, San Carlos, CA).
Results
Characterization of the effect of phosphorylation of Thr758 on 14-3-3, filamin, and talin binding to the β2-cytoplasmic peptide
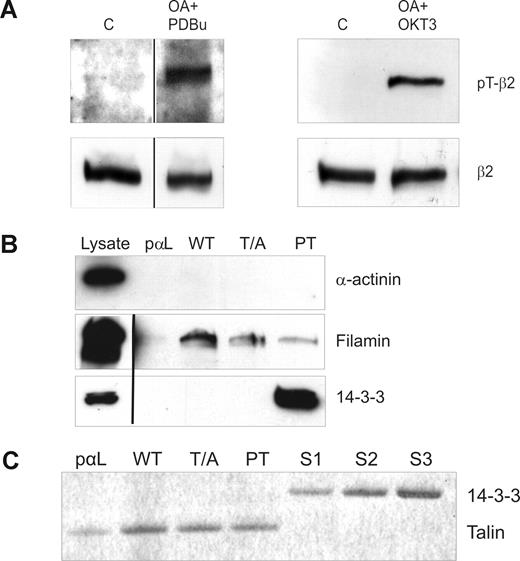
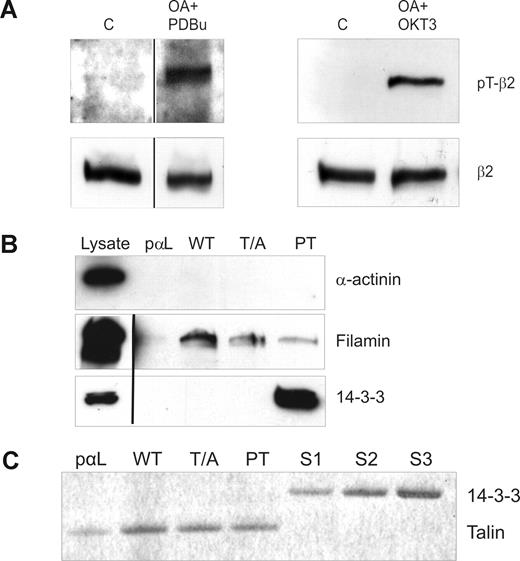
The TTT motif in integrin β chain cytoplasmic domains is important for integrin regulation, and both β7 and β2 integrins become phosphorylated on this motif in leukocytes.11-13 Using a recently developed anti-pThr758 rabbit polyclonal antibody,13 we investigated the phosphorylation status of β2 integrins in T cells. As previously reported, β2 integrins indeed become phosphorylated on the cytoplasmic Thr758 after stimulation of cells with TCR antibodies or with phorbol ester (Figure 1A). These are stimuli that are well known to lead to increased binding of T cells to the β2 integrin ligand ICAM-1.31
Analysis of β2-phosphorylation status in stimulated T cells and peptide affinity chromatography with phosphorylated and nonphosphorylated β2 cytoplasmic peptides. (A) β2 integrins are phosphorylated on Thr758 in T cells after phorbol ester and TCR ligation. Isolated human T cells were either nonstimulated (C) or preincubated with okadaic acid (OA) and stimulated with PDBu or OKT3. The cells were lysed and extracts subjected to Western blotting with the pThr758-specific antibody (pT-β2) or with the R2E7B antibody (β2). Vertical lines have been inserted to indicate a repositioned gel lane. (B) Binding of filamin and 14-3-3 to a phosphorylated control peptide (pαL), β2-35 (WT), β2-35A (T/A), and β2-35pT (PT) peptide affinity columns. The lysates and the bound materials were analyzed by Western blotting with α-actinin, 14-3-3, and filamin antibodies. Vertical lines have been inserted to indicate a repositioned gel lane. (C) Binding of purified talin head domain and 14-3-3ζ to peptide affinity columns. Talin head fragment (44 nM) binds to all β2 integrin peptides (WT, T/A, PT) in a similar way, thus indicating that talin binding is independent of β2 integrin Thr758. The control peptide (pαL) binding is much weaker, which implies specific binding. The binding of talin to PT is completely outcompeted when a similar molar amount of 14-3-3ζ was added in the incubation volume. The concentrations of the 14-3-3 added were 36 nM (S1), 71 nM (S2), and 143 nM (S3), and 44 nM talin was present in each column.
Analysis of β2-phosphorylation status in stimulated T cells and peptide affinity chromatography with phosphorylated and nonphosphorylated β2 cytoplasmic peptides. (A) β2 integrins are phosphorylated on Thr758 in T cells after phorbol ester and TCR ligation. Isolated human T cells were either nonstimulated (C) or preincubated with okadaic acid (OA) and stimulated with PDBu or OKT3. The cells were lysed and extracts subjected to Western blotting with the pThr758-specific antibody (pT-β2) or with the R2E7B antibody (β2). Vertical lines have been inserted to indicate a repositioned gel lane. (B) Binding of filamin and 14-3-3 to a phosphorylated control peptide (pαL), β2-35 (WT), β2-35A (T/A), and β2-35pT (PT) peptide affinity columns. The lysates and the bound materials were analyzed by Western blotting with α-actinin, 14-3-3, and filamin antibodies. Vertical lines have been inserted to indicate a repositioned gel lane. (C) Binding of purified talin head domain and 14-3-3ζ to peptide affinity columns. Talin head fragment (44 nM) binds to all β2 integrin peptides (WT, T/A, PT) in a similar way, thus indicating that talin binding is independent of β2 integrin Thr758. The control peptide (pαL) binding is much weaker, which implies specific binding. The binding of talin to PT is completely outcompeted when a similar molar amount of 14-3-3ζ was added in the incubation volume. The concentrations of the 14-3-3 added were 36 nM (S1), 71 nM (S2), and 143 nM (S3), and 44 nM talin was present in each column.
The TTT-containing region of the integrin β-chains is important for the binding of cytoplasmic molecules filamin3 and 14-3-3 proteins to the β2 integrin intracellular domain.4,32 To investigate the effect of phosphorylation of Thr758 on filamin and 14-3-3 protein interaction with the integrin cytoplasmic domain, we synthesized unphosphorylated and Thr758-phosphorylated integrin cytoplasmic domain peptides, as well as peptides with Thr758 mutated to alanine (Table 1). As expected, 14-3-3 proteins from T-cell lysates interacted only with pThr758 peptides and not at all with the unphosphorylated or Thr758Ala-mutated β2 peptide (Figure 1B). In contrast, filamin from cell lysates interacted well with the unphosphorylated peptide, but binding was significantly weaker to the phosphorylated peptide (Figure 1B). We could not detect binding of α-actinin, another protein that has been reported to bind to β2 integrin peptides and interact with β2 integrin in vivo,33 to any of the peptides (Figure 1B).
Talin, a major player in integrin activation, binds β2 integrins in vitro and in cells.1,7 The talin head domain can activate both the wt-β2 and Thr758Ala-β2 integrins in cells.4 Next, we assessed the role of Thr758 phosphorylation in talin binding to the integrin. We could not detect any full-length talin from T-cell lysates binding to the integrin peptides (data not shown). Thus, we instead examined binding of the main integrin-binding domain of talin, purified talin F2/F3 domains, to the β2 integrin peptides. As expected, because the talin-binding site in integrins does not overlap with Thr758,21,34 phosphorylation of Thr758 did not influence talin binding to the integrin (Figure 1C). However, the phosphorylation indirectly influences talin binding, as addition of 14-3-3 led to detachment of talin from the phosphorylated integrin peptide (Figure 1C). This result suggests that 14-3-3– and talin-binding sites in the integrin are partially overlapping.
Affinity measurement of 14-3-3–β2, talin-β2, and filamin-β2 interactions
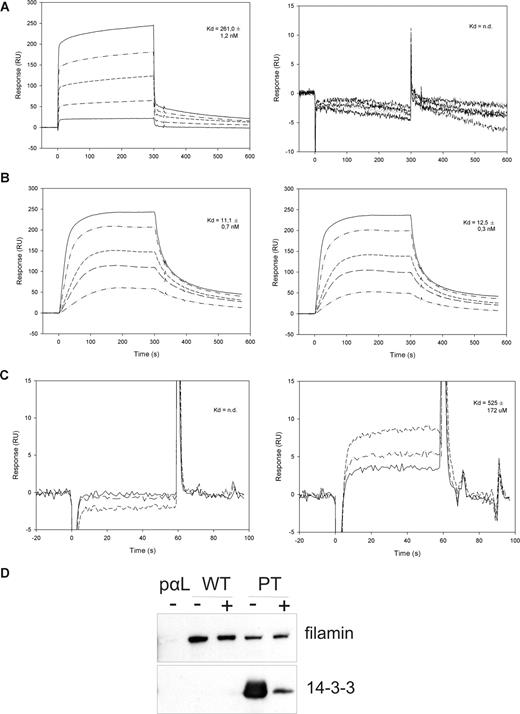
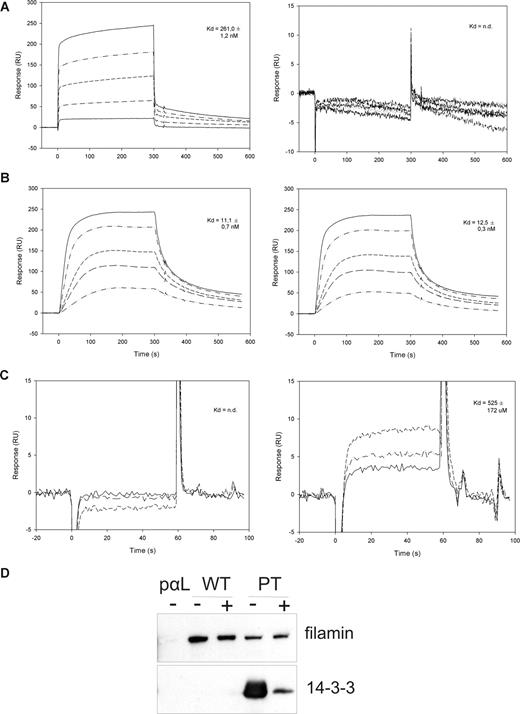
To measure the affinity of the studied protein-protein interactions, we performed surface plasmon resonance (Biacore) measurements. The peptides β2-35 and β2-35pT were coupled to individual flow cells of the Biacore sensor chip CM5, and different concentrations of the 14-3-3 protein, IgFLNa21, or talinF2/F3 were added in the solution. The 14-3-3 protein interacted with the phosphorylated but not the unphosphorylated β2 peptide, as expected. The interaction was very strong: the Kd was of the order of 260 nM (Figure 2A), which is comparable to other 14-3-3 interactions with their phosphorylated target sequences. Talin F2/F3 interacted with similar affinity with both the unphosphorylated and phosphorylated β2 peptide (Kd, 11 and 12.5 nM, respectively) (Figure 2B). The main integrin-binding site of filamin A, immunoglobulin domain 21 (IgFLNa21), interacted only with the nonphosphorylated β2 peptide, and the interaction was considerably weaker than for the 14-3-3 and talin interactions, in the range of 0.5 mM (Figure 2C). Nevertheless, the interaction of full-length filamin from T-cell lysates with the unphosphorylated β2 peptide was readily detected in our assays even if abundant amounts of 14-3-3 was present (Figure 1B), indicating that 14-3-3 could not outcompete filamin binding to the unphosphorylated integrin.
Surface plasmon resonance and competition studies of 14-3-3, talin, and filamin interactions with β2 cytoplasmic peptides. (A) Surface plasmon resonance (Biacore) sensorgrams of 14-3-3 binding to the phosphorylated (pT) and unphosphorylated (T) β2 peptide. The 14-3-3 concentrations used were 40, 160, 320, 640, and 1000 nM. (B) Surface plasmon resonance (Biacore) sensorgrams of talinF2/F3 binding to the phosphorylated (pT) and unphosphorylated (T) β2 peptide. The concentrations of talin used were 4, 10, 20, 40, and 60 nM. (C) Surface plasmon resonance (Biacore) sensorgrams of IgFLNa21 binding to the phosphorylated (pT) and unphosphorylated (T) β2 peptide. The concentrations of FLN used were 100, 200, and 400 μM. (D) Binding of filamin and 14-3-3 from ARAApSAPA-treated lysates to integrin peptide affinity columns. ARAApSAPA significantly reduced the interaction between 14-3-3 and β2 integrin peptide. The binding of filamin to wt integrin peptide or to the pT integrin peptide is not significantly increased in ARAApSAPA-treated lysates. Equal loading was confirmed by Western blotting of 14-3-3 from lysates (not shown). − indicates no ARAApSAPA treatment; +, 10 μM ARAApSAPA treatment.
Surface plasmon resonance and competition studies of 14-3-3, talin, and filamin interactions with β2 cytoplasmic peptides. (A) Surface plasmon resonance (Biacore) sensorgrams of 14-3-3 binding to the phosphorylated (pT) and unphosphorylated (T) β2 peptide. The 14-3-3 concentrations used were 40, 160, 320, 640, and 1000 nM. (B) Surface plasmon resonance (Biacore) sensorgrams of talinF2/F3 binding to the phosphorylated (pT) and unphosphorylated (T) β2 peptide. The concentrations of talin used were 4, 10, 20, 40, and 60 nM. (C) Surface plasmon resonance (Biacore) sensorgrams of IgFLNa21 binding to the phosphorylated (pT) and unphosphorylated (T) β2 peptide. The concentrations of FLN used were 100, 200, and 400 μM. (D) Binding of filamin and 14-3-3 from ARAApSAPA-treated lysates to integrin peptide affinity columns. ARAApSAPA significantly reduced the interaction between 14-3-3 and β2 integrin peptide. The binding of filamin to wt integrin peptide or to the pT integrin peptide is not significantly increased in ARAApSAPA-treated lysates. Equal loading was confirmed by Western blotting of 14-3-3 from lysates (not shown). − indicates no ARAApSAPA treatment; +, 10 μM ARAApSAPA treatment.
To further investigate whether the stronger affinity of the 14-3-3–β2 integrin interaction would mean that 14-3-3 would outcompete the filamin binding to the β2 integrin, we performed further experiments. The binding ability of 14-3-3 proteins in T-cell lysates was blocked by preincubation with the 14-3-3–binding peptide ARAApSAPA. This peptide binds to the 14-3-3 phosphate–binding groove and prevents the protein from interacting with phosphorylated targets, including β2 integrin.4 As expected, treatment of T-cell lysate with this peptide indeed significantly inhibited 14-3-3 binding to the phosphorylated β2 integrin peptide (Figure 2D). However, filamin binding was not significantly increased either to the nonphosphorylated or the phosphorylated β2 peptide (Figure 2D). This result implicates that it is not 14-3-3 binding but phosphorylation of β2 integrin that regulates the filamin interaction with the integrin. Taken together, both our peptide affinity chromatography results and Biacore results suggest that phosphorylation of Thr758 inhibits β2 integrin binding to filamin and enables its interaction with 14-3-3 proteins. We also showed that 14-3-3 competes with talin binding to the Thr758-phosphorylated β2 integrin tail.
Structures of filamin/β2 and 14-3-3/phospho-β2 complexes
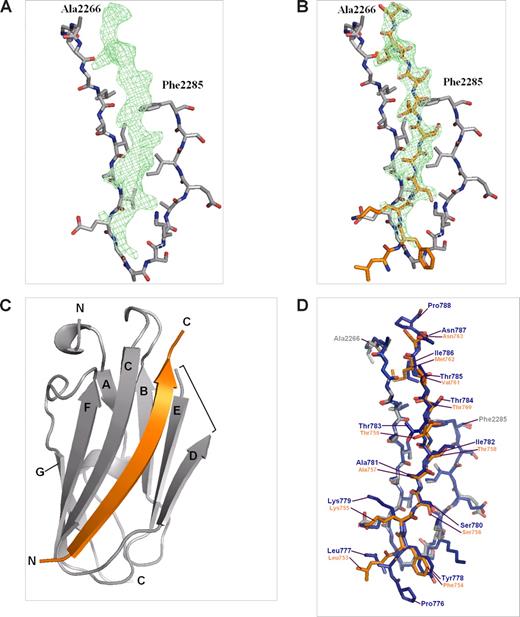
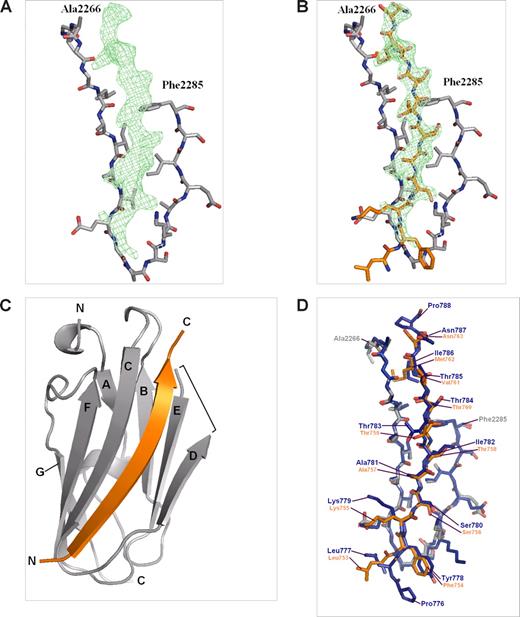
To understand the structural basis of the β2 integrin cytoplasmic domain binding to filamin and 14-3-3 we cocrystallized the β2 peptide with IgFLNa21 and soaked the β2 phosphopeptide in crystals of 14-3-3ζ. The IgFLNa-β2 peptide crystals belong to space group P213 and diffracted to 2.2-Å resolution. The structure was solved by molecular replacement using the previously solved IgFLNa21 (chain A from Protein Data Bank id 2BRQ) as the search model. Data collection and refinement statistics are shown in Table 2. In addition to the search model, electron density of the β2 peptide was observed next to the β-strand C of IgFLNa21, and 11 residues of the peptide could be built in the final model (Figure 3A-C). In the current crystal, the IgFLNa21 structure was otherwise identical (root-mean-square deviation for 86 Cα atoms 0.48 Å) with the previously published structure except that the residues of the DE loop were missing in the electron density map and could not be included in the final model (Figure 3C,D). The final refinement R-values were slightly higher (R = 24.3% and Rfree = 27.8%) than in an average 2.2-Å structure in the PDB database (Rave = 20.3% and Rfreeave = 25.3%), which is apparently caused by the disorder in the DE loop. The β2 peptide bound to IgFLNa21 in a very similar way as the β7 peptide,3 forming a hydrogen-bonded β-strand next to the strand C of IgFLNa21 and interacting hydrophobically with the side chains of strand D (Figure 3C,D). The main hydrophobic contacts are mediated by the side chain methyl groups of Thr758 and Thr760 in β2. As Thr758 resides in a hydrophobic site of IgFLNa21, it is most probable that addition of a negatively charged phosphate group to this residue strongly disfavors its interaction. Thus, the structure of IgFLNa21/β2 peptide complex explains why Thr758-phosphorylated β2 integrin is not able to interact with filamin.
Crystal structure of IgFLNa21/β2 complex. (A) Electron density map (Fo − Fc) of the β2 peptide calculated from the final model without the peptide, shown at σ = 2. Only β-strands C and D of IgFLNa21 are shown for clarity. (B) The final model of the peptide (orange) built in the electron density map. (C) Overall structure of the IgFLNa21 (gray) and the peptide shown as ribbon diagram. N and C termini are indicated and the β-strands are named. The loop between β-strands D and E that is missing from the final model is indicated with a bracket. (D) Comparison of IgFLNa21/β7 (blue) (PDB entry 2BRQ) and IgFLNa21/β2 (gray and orange) complexes. The numbering of integrin residues is shown.
Crystal structure of IgFLNa21/β2 complex. (A) Electron density map (Fo − Fc) of the β2 peptide calculated from the final model without the peptide, shown at σ = 2. Only β-strands C and D of IgFLNa21 are shown for clarity. (B) The final model of the peptide (orange) built in the electron density map. (C) Overall structure of the IgFLNa21 (gray) and the peptide shown as ribbon diagram. N and C termini are indicated and the β-strands are named. The loop between β-strands D and E that is missing from the final model is indicated with a bracket. (D) Comparison of IgFLNa21/β7 (blue) (PDB entry 2BRQ) and IgFLNa21/β2 (gray and orange) complexes. The numbering of integrin residues is shown.
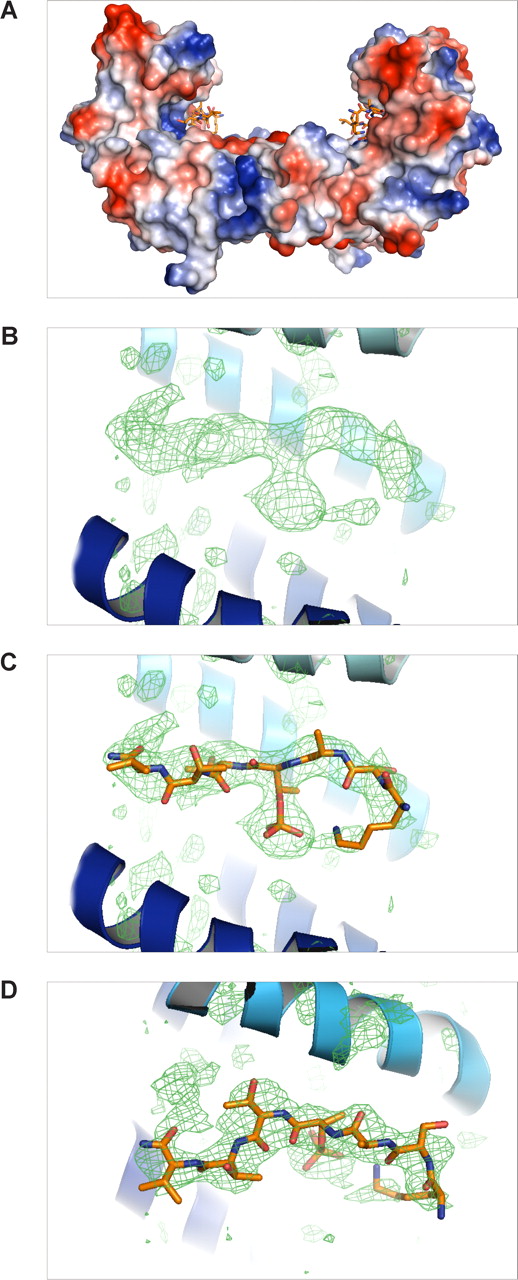
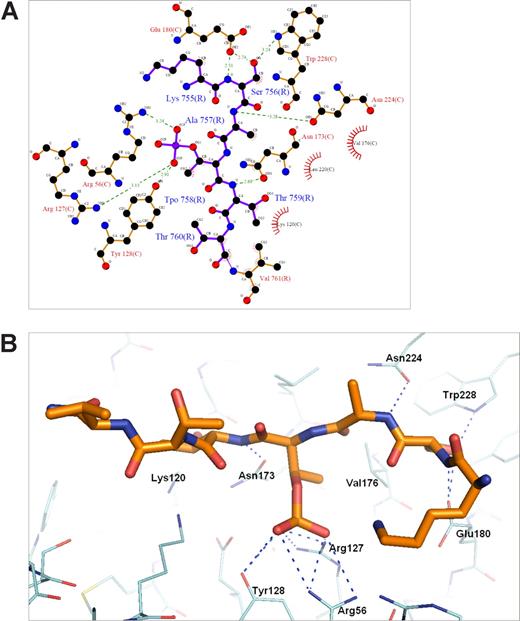
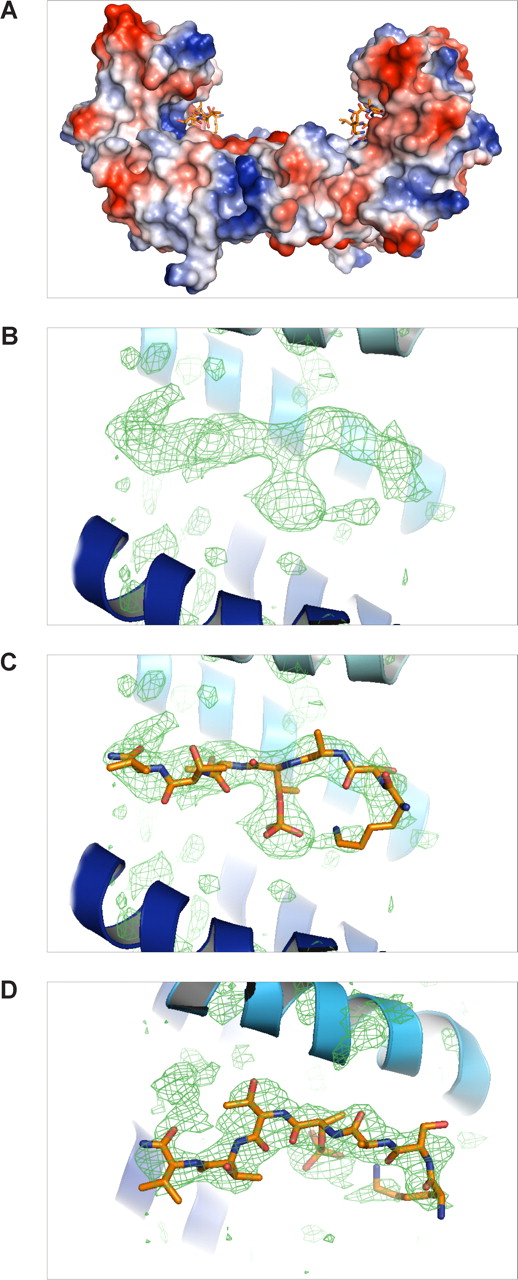
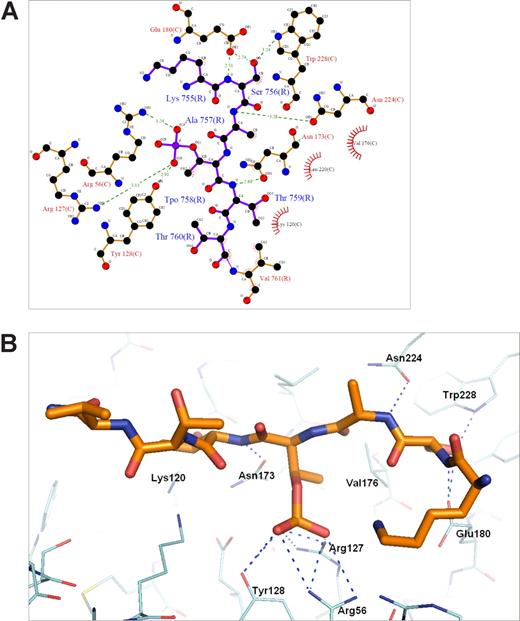
The 14-3-3ζ crystallized in the space group P65 and data up to 2.5-Å resolution were used for structure calculations (Table 2). The cumulative intensity distribution of the diffraction data indicated that the best crystals were merohedral twins with a twinning fraction of 0.306. Nontwinned crystals were not found. Despite this, the structure could be solved and the electron density maps could be clearly interpreted to yield a good quality model (R = 22.7% and Rfree = 27.3%; Table 2). The soaked β2 phosphopeptide was bound in all 4 14-3-3ζ monomers of the asymmetric unit, and noncrystallographic restrains were used in the final refinements (Figure 4). The phosphopeptide was bound to the well-characterized binding pocket between α-helices E and F of each 14-3-3 molecules.35 The main electrostatic interactions were between Thr758 phosphate group of the β2 peptide and 14-3-3ζ Arg residues 56 and 127, which form a typical basic batch in the binding site (Figures 4A,5). A hydrogen bond was observed between the phosphate group and Tyr128 of 14-3-3 (Figure 5A,B). The side chain of Ser756 formed hydrogen bonds with Trp228 and Glu180 (Figure 5A,B). The interaction features between β2 phosphopeptide and 14-3-3ζ were very similar to other published phosphopeptide/14-3-3 structures where the phosphate group has been shown to be the key determinant for binding.35
Structure of the 14-3-3ζ/phospho-β2 complex. (A) Overall structure of the 14-3-3ζ dimer and 2 peptides. The protein is shown as a surface representation colored according to surface charge as implemented in PyMOL. (B) The difference electron density map (Fo − Fc) of the β2 phosphopeptide calculated from the final model without the peptide, shown at σ = 2. The α-helices E and H of 14-3-3 are located below and above the peptide, respectively, and shown as a ribbon diagram. (C) Same as panel B, but the final model of the peptide is also shown. (D) The same as panel C, but shown from above.
Structure of the 14-3-3ζ/phospho-β2 complex. (A) Overall structure of the 14-3-3ζ dimer and 2 peptides. The protein is shown as a surface representation colored according to surface charge as implemented in PyMOL. (B) The difference electron density map (Fo − Fc) of the β2 phosphopeptide calculated from the final model without the peptide, shown at σ = 2. The α-helices E and H of 14-3-3 are located below and above the peptide, respectively, and shown as a ribbon diagram. (C) Same as panel B, but the final model of the peptide is also shown. (D) The same as panel C, but shown from above.
Details of the 14-3-3ζ–phospho-β2 interaction. (A) An illustration of the atomic interactions between the protein and peptide. Hydrogen bonds are shown as dashed lines with distances and atoms participating in hydrophobic interactions marked. The figure was generated by the program Ligplot (University College London, London, United Kingdom).36 (B) The hydrogen bonds indicated in panel A are shown in the actual model. Note that only the pThr758 (marked Tpo in A) and Ser756 have side chain hydrogen bonds with the protein.
Details of the 14-3-3ζ–phospho-β2 interaction. (A) An illustration of the atomic interactions between the protein and peptide. Hydrogen bonds are shown as dashed lines with distances and atoms participating in hydrophobic interactions marked. The figure was generated by the program Ligplot (University College London, London, United Kingdom).36 (B) The hydrogen bonds indicated in panel A are shown in the actual model. Note that only the pThr758 (marked Tpo in A) and Ser756 have side chain hydrogen bonds with the protein.
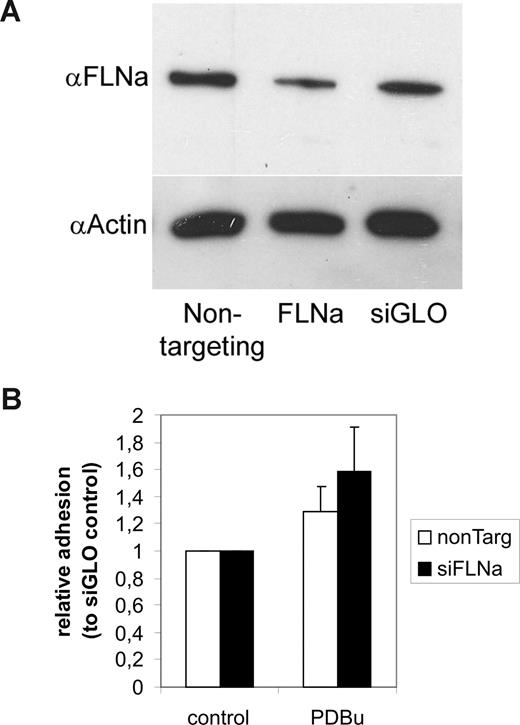
Filamin knockdown increases activated Jurkat cell adhesion to ICAM-1
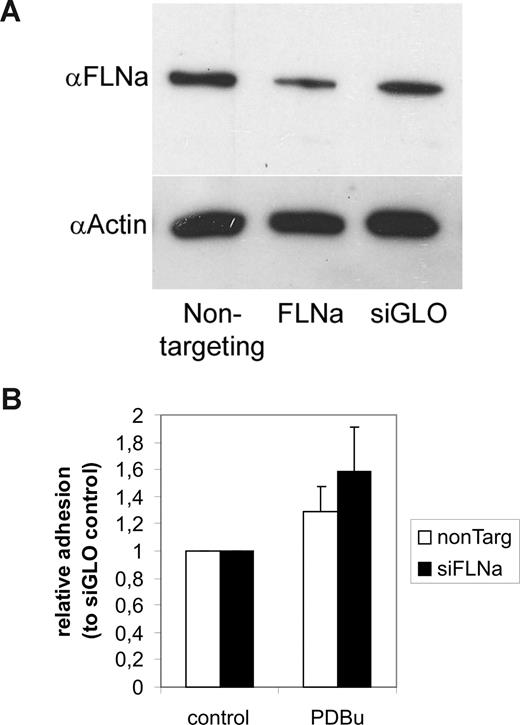
Blocking 14-3-3 binding to β2 by a Thr758Ala mutation or by 14-3-3 blocking (R18) constructs inhibits β2 integrin–dependent binding to ICAM-1.4 To investigate the role of filamin binding in regulating β2 integrin–mediated adhesion, we performed knockdown experiments (siRNA) of filamin in Jurkat cells (Figure 6A). Filamin knockdown did not influence basal adhesion to ICAM-1 (not shown). However, it increased binding of PDBu-stimulated Jurkat cells to ICAM-1 (Figure 6B). Thus, filamin plays an inhibitory role in regulating T-cell adhesion to ICAM-1.
Effect of filamin knockdown on β2 integrin–dependent T-cell adhesion to coated ICAM-1. (A) Jurkat T cells were transfected with siGENOME SMART pool human filamin A (FLNa) siRNAs or control siRNAs. Filamin knockdown with filamin-specific siRNAs, but not control siRNAs, resulted in knockdown of filamin protein levels, as shown by Western blotting of corresponding cell extracts with antifilamin antibodies. The blot was stripped and reprobed with actin antibodies to show equal loading. (B) siRNA-transfected cells were left untreated or stimulated with 200 nM PDBu and cell binding to coated ICAM-1 was assayed for as described in “Adhesion assay.” Treatment of cells with FLNa siRNAs, but not control siRNAs, increased the stimulated cell binding to ICAM-1 (P < .05 for PDBu-stimulated samples). The experiment was repeated 9 times with similar results. Error bars represent SD.
Effect of filamin knockdown on β2 integrin–dependent T-cell adhesion to coated ICAM-1. (A) Jurkat T cells were transfected with siGENOME SMART pool human filamin A (FLNa) siRNAs or control siRNAs. Filamin knockdown with filamin-specific siRNAs, but not control siRNAs, resulted in knockdown of filamin protein levels, as shown by Western blotting of corresponding cell extracts with antifilamin antibodies. The blot was stripped and reprobed with actin antibodies to show equal loading. (B) siRNA-transfected cells were left untreated or stimulated with 200 nM PDBu and cell binding to coated ICAM-1 was assayed for as described in “Adhesion assay.” Treatment of cells with FLNa siRNAs, but not control siRNAs, increased the stimulated cell binding to ICAM-1 (P < .05 for PDBu-stimulated samples). The experiment was repeated 9 times with similar results. Error bars represent SD.
Discussion
In this article, we have investigated the binding of 3 cytoplasmic proteins, the pSer/pThr-binding adaptor protein 14-3-3 and the large cytoskeletal proteins talin and filamin, to the unphosphorylated and phosphorylated β2 integrin cytoplasmic tails. The main findings are that the phosphorylation of β2 on the physiologically relevant Thr758 leads to impairment of filamin interaction and binding of 14-3-3, thus acting as a “molecular switch” for these protein-protein interactions, and that 14-3-3 can outcompete talin for binding to the β2 integrin in its phosphorylated state.
IgFLNa21 has been shown to be the main interaction site in filamin A for β1 and β7 integrins.3 Here we report the structure of IgFLNa21 with a peptide from the β2 cytoplasmic tail. The interaction is very similar to the previously reported IgFLNa21/β7 integrin complex.3 The main difference of the 2 structures derives from the fact that the TTT motif is one residue earlier in β2 than in β7. Because of this, the main hydrophobic interaction with the Phe2285 of FLNa is mediated by Thr758 in β2 instead of Ile782 in β7. The methyl group of Thr758 points toward the interface, whereas the hydroxyl group is partially exposed. Based on the structure, it is apparent that the addition of a phosphate group to Thr758 would be very unfavorable for the β2-IgFLNA21 interaction. We could also show this in phosphopeptide affinity chromatography and in surface plasmon resonance assays. To the best of our knowledge, this is the first report where the inhibitory effect of integrin phosphorylation on filamin binding has been directly demonstrated. In an earlier study, phosphate mimicking Thr to Glu mutations were used in the β7 integrin.3
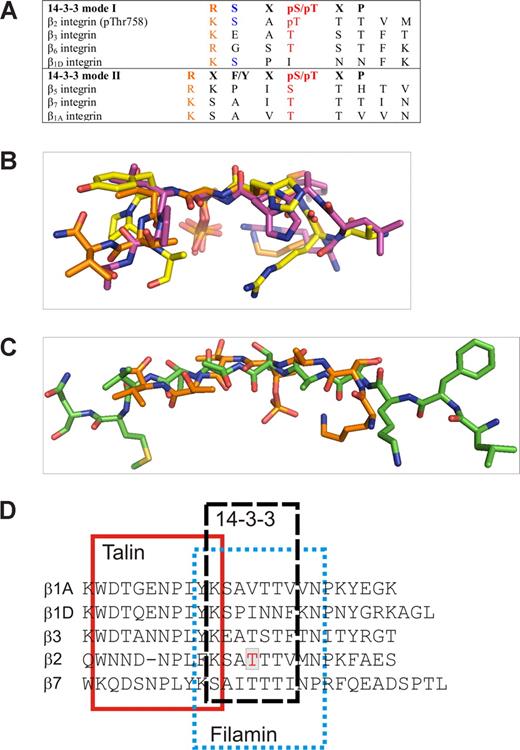
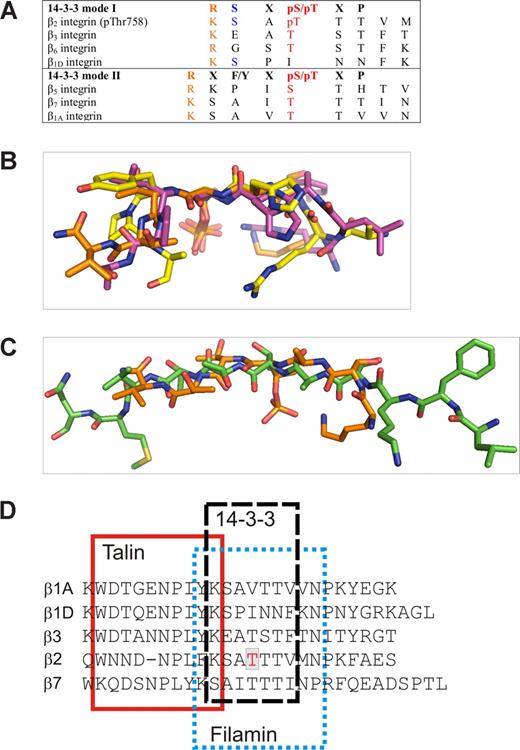
We showed here that 14-3-3 bound only to Thr758-phosphorylated β2. This is in line with the general principle that most 14-3-3 interactions are dependent on phosphorylation. The β2 sequence is quite close to the mode 1 14-3-3–binding consensus motif RSXpS/TXP37 (Figure 7A). The structure of the pThr758 β2/14-3-3 complex revealed that the phosphate group is the major determinant of the interaction. There are only few other side chain interactions between β2 and IgFLNa21 in the complex. Again, this is very similar to other 14-3-3 complexes (Figure 7B).
Comparison of 14-3-3–binding peptides and integrins. (A) Alignment of mode 1 and mode 2 14-3-3–binding consensus sequences with integrin sequences. (B) Superimposition of our 14-3-3 bound β2 (orange) with mode 1 (PDB entry 1QJB, purple) and mode 2 (PDB entry 1QJA, yellow) peptide structures. The only common features are the Arg/Lys and the pThr/pSer indicated in red in panel A. (C) Superimposition of β2 peptides in 14-3-3–binding conformation (orange) and IgFLNa21-binding conformation (green) shows that in both complexes the peptides are extended, but the side chain orientations differ markedly. (D) Sequence alignment of talin-, filamin-, and 14-3-3–binding sites in different integrin β-chains. The binding sites are based on the β3-talin complex structure 1MIZ21 and our current structures (β2/14-3-3 and β2/filamin).
Comparison of 14-3-3–binding peptides and integrins. (A) Alignment of mode 1 and mode 2 14-3-3–binding consensus sequences with integrin sequences. (B) Superimposition of our 14-3-3 bound β2 (orange) with mode 1 (PDB entry 1QJB, purple) and mode 2 (PDB entry 1QJA, yellow) peptide structures. The only common features are the Arg/Lys and the pThr/pSer indicated in red in panel A. (C) Superimposition of β2 peptides in 14-3-3–binding conformation (orange) and IgFLNa21-binding conformation (green) shows that in both complexes the peptides are extended, but the side chain orientations differ markedly. (D) Sequence alignment of talin-, filamin-, and 14-3-3–binding sites in different integrin β-chains. The binding sites are based on the β3-talin complex structure 1MIZ21 and our current structures (β2/14-3-3 and β2/filamin).
When we compare the conformation of the β2 peptide bound to 14-3-3 and to filamin, we see that they are very different (Figure 7C). Both of them are extended, but the peptide does not form a β-strand in the 14-3-3 complex as seen in the filamin complex. This indicates that the integrin cytoplasmic tail can adopt different conformations depending of its binding partner. In the crystal structures, we see only those parts of the cytoplasmic tail peptides that are in direct contacts, whereas the other regions apparently are disordered. It is possible that integrin β tails are disordered also in vivo, unless they are in contact with other molecules.
We have now shown that inhibition of filamin binding to β2 integrins by filamin “knockdown” leads to an increase in stimulated T-cell adhesion to ICAM-1. In combination with previously reported results showing that inhibition of 14-3-3 binding to β2 dramatically reduces adhesion,4 this result shows that the phosphorylation of Thr758 is an important molecular switch that regulates adhesion. We have previously shown that a Thr758-phosphorylated β2 peptide, which was introduced into cells with the aid of a hydrophobic penetratin sequence, activated cell adhesion and further signaling to the actin regulating small G-proteins Rac1 and Cdc42.13 Thus, phosphorylation of β2 on Thr758, which occurs after cell stimulation, leads to a detachment of filamin and a binding of 14-3-3 to the β2 tail, and the combination of further downstream molecular events leads to increased cell adhesion.
Talin is an important regulator of αLβ2 integrins in T cells stimulated through the T-cell receptor.8 We found that phosphorylation of β2 integrin does not directly affect binding of talin to the integrin. However, it appears that 14-3-3 binding to the phosphorylated integrin may lead to detachment of talin from the integrin tail. The talin-binding site in the β3 integrin21 and the 14-3-3–binding site in β2 (this study) show only one amino acid overlap, but that might be enough to prevent the simultaneous binding of these 2 molecules to phosphorylated integrin tails (Figure 7D). One could envision a sequential model by which these proteins influence αLβ2 integrin–mediated adhesion. Filamin is bound to the integrin in resting cells (where β2 is unphosphorylated) and plays an inhibitory role in integrin activation. Talin is directly involved in β2 integrin activation by regulating both integrin affinity and clustering,8 and talin and filamin may compete for the β2 integrin, as has been described for the β7 integrin.3 The importance of talin in β2 integrin–mediated cell-cell binding is especially pronounced at early stages (30 seconds to 5 minutes) after T-cell receptor stimulation, although whether there is a constitutive or stimulated association between talin and αLβ2 is not known.8 14-3-3 does not bind to the integrin until the β2-chain becomes phosphorylated at Thr758, at later time points after TCR stimulation.11 At these later time points, the contact between T-cell and antigen-presenting cell matures and talin is no longer crucial for adhesion.8 Phosphorylation of Thr758 directly inhibits filamin binding, and indirectly inhibits talin binding to the β2 integrin. The phosphorylated integrin/14-3-3 complex then mediates actin reorganization downstream of the integrin, strengthening the cell-cell contact.
The 14-3-3 proteins are involved in many cellular processes but do not have any enzymatic activity. There are at least 2 possible mechanisms by which 14-3-3s can function in signaling. As they are dimers, either they can work as scaffolding proteins that bind to 2 phosphorylated ligands simultaneously, or they can function as allosteric modulators that stabilize further interactions of the ligand. In β2 integrin signaling, both of these mechanisms are possible. Because the 2 phosphopeptide-binding grooves in the 14-3-3 dimer are quite close to each other (the distance between the 2 phosphate groups is about 30 Å) and the binding pockets are partly covered, 2 folded domains cannot fit in the binding pocket simultaneously. However, 2 extended peptides such as integrin cytoplasmic tails could bind the 14-3-3 dimer simultaneously. Thus, 14-3-3 could act by inducing integrin clustering, in a similar way that has been observed for 14-3-3s to induce hexamer formation of a P-type H+-ATPase.38 On the other hand, 14-3-3 could induce some further interactions of the integrin cytoplasmic domain, for instance by favoring certain conformations of the tail. It is apparent that Rac1/Cdc42 activation is downstream of 14-3-3–integrin interaction.13 However, the intermediate steps between 14-3-3 and these small G-proteins are currently not known.
Do other integrins bind to 14-3-3 or is the binding mechanism described here specific for β2 integrins? When we compared other integrin cytoplasmic tail sequences to the region surrounding Thr758 of β2, we found that Thr777 of β3 (or Thr751 without signal peptide) and Thr766 of β6 locate in a very similar consensus position as Thr758 of β2, resembling the mode 1 14-3-3–binding phosphopeptide sequence. The β3 integrin has been reported to be phosphorylated on Thr777 in cells.39-41 β1, β7, and β5 sequences have some resemblance to the mode 2 14-3-3–binding motif (Figure 8B). Of these, both β1 and β7 integrins are known to be phosphorylated in cells.12,42 There is conflicting evidence about the involvement of 14-3-3 proteins in β1 integrin–mediated adhesion.4,43,44 To our knowledge, however, the direct effects of β1 phosphorylation on 14-3-3 interactions have not been studied. The sequence comparisons suggest that many phosphorylated β integrin tails may interact with 14-3-3 in a similar way to the Thr758-phosphorylated β2.
Only limited structural information is available on integrin β tails. Nuclear magnetic resonance (NMR) methods have given an overall picture of the conformational flexibility of integrin tails.45,46 Crystal structures of the integrin/talin21 and integrin/filamin complexes3 are available as well as detailed NMR data of the integrin-talin interaction.34 In this paper, we have described the structure of 14-3-3 with the integrin cytoplasmic tail phosphopeptide and a second integrin/filamin complex. It reveals that the phosphorylation of Thr758 in β2 switches the binding between filamin and 14-3-3. We believe that this is an important new concept of integrin function.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The coordinates have been deposited to PDB under codes 2V7D (14-3-3ζ/β2 complex) and 2JF1 (IgFLNa21/β2 complex). We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities, and we thank Dr Didier Nurizzo and Dr Joanna Timmins for assistance in using beamlines ID23-1 and ID14-3.
This study has been supported by Academy of Finland grants 105211, 114717 (J.Y.), and 210390 (C.G.G.); the Sigrid Jusélius Foundation (C.G.G.); the Finnish Cancer Society (C.G.G.); the Finnish Medical Association (C.G.G.); Magnus Ehrnrooth Foundation (S.C.F.); the Liv och Halsa foundation (S.C.F., C.G.G.); and the Ruth and Nils-Erik Stenback foundation grants (S.C.F.; all Helsinki, Finland).
Authorship
Contribution: H.T. performed research, analyzed data, and contributed to writing the paper; E.N., S.M.N., M.A., T.S., T.K., and M.T. performed research and analyzed data; C.G.G. contributed to design of research and writing the paper; and J.Y. and S.C.F. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Susanna Fagerholm, Section of Immunology, Division of Pathology and Neuroscience, Ninewells Hospital and Medical School, Dundee University, DD1 9SY, Dundee, United Kingdom; e-mail: s.c.fagerholm@dundee.ac.uk; or Jari Ylänne, Department of Environmental and Biological Science and Nanoscience Center, University of Jyäskylä, PO Box 35, 40014 Jyväskylä, Finland; e-mail: jylanne@bytl.jyu.fi.
References
Author notes
*H.T., E.N., and S.M.N. contributed equally to this work.