Abstract
Current evidence supports a model in which the low-affinity state of the platelet integrin αIIbβ3 results from αIIbβ3 adopting a bent conformation. To assess αIIbβ3 biogenesis and how αIIbβ3 initially adopts the bent conformation, we mapped the conformational states occupied by αIIb and β3 during biogenesis using conformation-specific monoclonal antibodies (mAbs). We found that αIIbβ3 complex formation was not limited by the availability of either free pro-αIIb or free β3, suggesting that other molecules, perhaps chaperones, control complex formation. Five β3-specific, ligand-induced binding site (LIBS) mAbs reacted with much or all free β3 but not with β3 when in complex with mature αIIb, suggesting that β3 adopts its mature conformation only after complex formation. Conversely, 2 αIIb-specific LIBS mAbs directed against the αIIb Calf-2 region adjacent to the membrane reacted with only minor fractions of free pro-αIIb, raising the possibility that pro-αIIb adopts a bent conformation early in biogenesis. Our data suggest a working model in which pro-αIIb adopts a bent conformation soon after synthesis, and then β3 assumes its bent conformation by virtue of its interaction with the bent pro-αIIb.
Introduction
Integrin receptors are composed of heterodimers of α and β subunits. The β3 family of integrin receptors is composed of αIIbβ3, which is specific for platelets and their precursor megakaryocytes, and αvβ3, which is more widely distributed on many cells, including osteoclasts and endothelial cells.1 αIIbβ3 plays an important role in platelet aggregation, and inhibitors of the receptor have demonstrated efficacy in preventing thrombotic complications of percutaneous coronary interventions.2 Previous studies of αIIbβ3 integrin biogenesis have provided important information on the synthesis of the αIIb and β3 subunits, αIIbβ3 complex formation, carbohydrate processing, transport of the assembled complexes from the endoplasmic reticulum (ER) to the Golgi, cleavage of αIIb into heavy and light chains in the Golgi, and subsequent transport of the mature receptor to granule and platelet surface membranes.3–11 In particular, they suggested that the availability of either free pro-αIIb or free β3 limits αIIbβ3 complex formation.7,9
The crystal structure of the extracellular domain of αvβ3 unexpectedly revealed that the receptor had a bent conformation,12 and electron microscopy suggested that activation and ligand binding are associated with the adoption of an extended conformation.13 It is presumed that αIIbβ3 also undergoes a transformation from a bent to an extended conformation on activation,14 which occurs when platelets are stimulated with one or more agonists, resulting in binding of ligands, including fibrinogen. The crystal structure of the headpiece of αIIbβ3 in complex with ligand-mimetics also identified a swing-out motion of the β3 hybrid domain at its juncture with the βA (I-like) domain that is thought to contribute to ligand binding and/or the initiation of outside-in signaling.15 The bent, inactive conformations of these integrins thus play an important role in the function of these receptors. In the case of αIIbβ3, it permits platelets to circulate in plasma containing high concentrations of one of its ligands, fibrinogen, without spontaneous aggregation. Thus, it is important to understand how the bent, compact conformation is achieved during biogenesis (Figure 1).
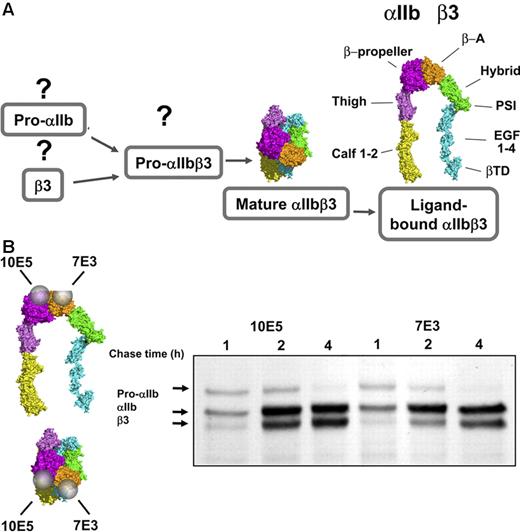
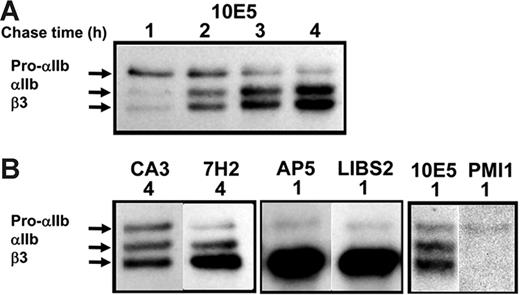
Progression of αIIbβ3 conformations during biogenesis. (A) The conformations of free pro-αIIb, free β3, and pro-αIIbβ3 are unknown. The conformation of mature, bent, unactivated αIIbβ3 and the proposed extended structure of activated, ligand-bound αIIbβ3 are modeled after the crystal structures of αVβ3 and the αIIbβ3 headpiece, respectively. The domains of αIIb and β3 are indicated.12,15 (B) Immunoprecipitation with αIIbβ3 complex-specific mAbs 10E5 and 7E3 following pulse-chase labeling demonstrates that both antibodies recognize the pro-αIIbβ3 complex in addition to the mature αIIbβ3 complex. The figure on the left indicates the locations of the 10E5 and 7E3 epitopes on models of the extended and bent forms of αIIbβ3. This figure has been adapted from its original published form with the permission of the journal Clinical Pharmacology & Therapeutics.
Progression of αIIbβ3 conformations during biogenesis. (A) The conformations of free pro-αIIb, free β3, and pro-αIIbβ3 are unknown. The conformation of mature, bent, unactivated αIIbβ3 and the proposed extended structure of activated, ligand-bound αIIbβ3 are modeled after the crystal structures of αVβ3 and the αIIbβ3 headpiece, respectively. The domains of αIIb and β3 are indicated.12,15 (B) Immunoprecipitation with αIIbβ3 complex-specific mAbs 10E5 and 7E3 following pulse-chase labeling demonstrates that both antibodies recognize the pro-αIIbβ3 complex in addition to the mature αIIbβ3 complex. The figure on the left indicates the locations of the 10E5 and 7E3 epitopes on models of the extended and bent forms of αIIbβ3. This figure has been adapted from its original published form with the permission of the journal Clinical Pharmacology & Therapeutics.
We recently reported that the αIIb N-linked glycan at amino acid 15, which is located on a solvent-exposed loop in the β-propeller domain, is important for αIIbβ3 complex formation, presumably by mediating attachment to the ER membrane-bound lectin calnexin.16 In the present study we have used pulse-chase analysis of HEK293 cells synthesizing αIIb and β3, as well as megakaryocyte lineage cells expressing αIIbβ3 derived from umbilical cord blood and a series of monoclonal antibodies (mAbs) specific for different regions and conformations of αIIb and β3 to chart the progress of αIIb and β3 folding during biogenesis. Our data demonstrate that neither free pro-αIIb nor free β3 limits pro-αIIbβ3 complex formation, indicating that control of this vital step in biogenesis is achieved by some other mechanism, perhaps the availability of one or more chaperone molecules. In addition, the mAb immunoprecipitation patterns support a model in which pro-αIIb becomes bent during or immediately after synthesis, perhaps as a result of the engagement of the head region, which is synthesized first, with the membrane-bound lectin/chaperone calnexin. The β3 subunit achieves its final bent conformation only after complexing with pro-αIIb.
Materials and methods
Antibodies
The antibodies used in these studies and their specificities are detailed in Table 1 based on previous reports17–25 and the current studies. Several of the mAbs (AP5, AP6, LIBS1, LIBS2, PMI-1, B1B5) are categorized as ligand-induced binding site (LIBS) antibodies because they preferentially or exclusively bind to the activated and/or ligand-bound conformation(s) of αIIbβ3.18,21
Human umbilical cord blood culture
Leukocytes were separated from 3 to 6 units of human umbilical cord blood judged to be inadequate for clinical purposes (generously provided by the New York Blood Center) by Dextran 70 sedimentation (Amersham Biosciences, Piscataway, NJ), and then enriched for CD34+ progenitor cells by negative selection using a combination of antibodies against maturation/lineage-specific markers (RosetteSep; StemCell Technologies, Vancouver, BC) concomitant with density sedimentation (Ficoll-Paque Plus; Amersham Biosciences). These cells were then cultured in serum-free medium (StemCell Technologies) with 50 ng/mL thrombopoietin (TPO) plus 10 ng/mL IL-11 for 3 days (both StemCell Technologies), followed by culture in the same medium with 50 ng/mL TPO alone for another 6 to 8 days.
HEK293-cell culture
HEK293 cell lines that stably expressed normal human αIIbβ3 receptors were established as previously described.30 Transfections were performed using Lipofectamine 2000 (Gibco-BRL, Carlsbad, CA) according to the manufacturer's instructions, followed by selection in media containing 800 μg/mL G418 for 2 to 4 weeks. To obtain a population of cells uniformly expressing high levels of αIIbβ3, cells were labeled with the mAb 10E5 (anti-αIIbβ3) and sorted using a FACSVantage SE cell sorter (Becton-Dickinson, Rutherford, NJ).
Biosynthetic labeling and immunoprecipitation
Samples were prepared as previously described.34 Briefly, cells were incubated for 30 minutes at 37°C in methionine/cysteine-free medium, followed by pulse-labeling for 15 minutes at 37°C in medium containing 35S-methionine/cysteine (300 μCi [11.1 MBq]/10 cm plate). The pulse was terminated by incubation in medium containing unlabeled methionine/cysteine (1 mg/mL each), and the cells were incubated at 37°C until lysis in 1% Triton-X 100 lysis buffer. Following cell lysis, supernatants were precleared with protein-G Sepharose beads (Amersham Biosciences), and samples containing equivalent amounts of trichloroacetic acid-precipitable radioactivity (∼ 5-6 × 106 counts/sample) were incubated 16 hours at 4°C with one or more of the antibodies listed under “Antibodies” (4 μg/reaction). Samples were incubated with protein-G Sepharose beads for 1 hour at 4°C, washed twice, and incubated with SDS sample buffer for 10 minutes at 100°C. Samples were then subjected to SDS–polyacrylamide gel electrophoresis, and the gels were dried and exposed to film. To ensure that equal amounts of protein were loaded into each well, duplicate samples were analyzed by immunoblotting. The amount of each mAb used for immunoprecipitation was determined to be at a near-saturating concentration by titration experiments using 0 to 20 μg of each mAb (data not shown). Nonspecific binding was determined by performing immunoprecipitation with mouse IgG on whole-cell lysates of both cell types.
Computer modeling of αIIb amino acids
A complete model of the structure of the extracellular inactive conformation of αIIbβ3 was first constructed using MODELLER 8v2.35 The αIIb chain was modeled taking the coordinates of the propeller from the crystal structure of αIIbβ3 (residues 1-453; PDB ID, 1TY6)15 and using αV coordinates as a template for the remainder of the sequence (PDB ID, 1U8C).36 Coordinates of the β3 chain were extracted from the complex of αVβ3 (PDB ID, 1U8C),36 and the missing domains, EGF1 (435-475) and EGF2 (486-522), were modeled from the crystal structures of the β2 EGF1 (PDB ID, 1YUK)37 and the αVβ3 EGF3 (PDB ID, 1U8C) domains,36 respectively. The complete system (including cations from the αVβ3 crystal structure) was then energy minimized using GROMACS 3.2.138 as follows: (1) the system was immersed in a preequilibrated 16 × 16 × 16 nm cubic box of simple point-charge water molecules, and sodium (44) counter-ions were added to neutralize the system; (2) surrounding water molecules were first minimized and then subjected to 50 ps molecular dynamics simulations at 300°K and 1 bar, while harmonically restraining all protein heavy atoms to their initial positions [force constant equal to 1000 kJ/(mol · nm2)]; and (3) the resulting system was energy minimized without any restraints.
To assess the most likely conformations of the region of αV corresponding to amino acids 842 to 873 in αIIb, a set of 200 independently optimized conformations of the 842 to 873 loop in the Calf-2 domain were generated from the minimized model of αIIbβ3 using the loop modeling routine of MODELLER 8v2.35 Briefly, the optimization relies on a protocol consisting of conjugate gradient minimization and molecular dynamics simulation with simulated annealing. The final loop prediction is the optimized conformation that has the lowest pseudo-energy score, which contains terms from a molecular mechanics force field as well as restraints based on statistical distributions derived from known protein structures. Because the accuracy of the MODELLER algorithm was only established for loops that are much shorter (up to 14 residues) than the one we modeled (32 residues), we also used the Robetta server39 to obtain 5 comparative models of the structure of the Calf-2 domain (residues 745 to 960) based on the corresponding αV structure (PDB ID, 1JV2).39 Robetta assembles loop regions from fragments and optimizes them to fit the aligned template structure using the Rosetta fragment insertion method.40 This algorithm has been reported to provide reasonable models of conformations of peptide segments containing 13 to 34 residues.41 Multiple decoy models were generated and independent simulations were carried out. From this ensemble 4 models were selected (models 1-4) using different variants of the Rosetta energy function and are presented along with the default K*Sync alignment-derived model (model 5).42
Results
Experiments with HEK293 cells transfected with αIIb and β3
αIIbβ3 Complex-dependent antibodies 10E5 and 7E3 react with pro-αIIbβ3 in addition to mature αIIbβ3.
To assess complex formation between αIIb and β3 during biogenesis, we used 2 mAbs, 10E5 and 7E3, that react with the mature αIIbβ3 complex17,19 but not with either subunit alone.43 10E5 interacts exclusively with the “Cap” subdomain of the αIIb β-propeller,15 whereas the 7E3 epitope reacts with a region adjacent to the MIDAS in the βA (I-like) domain of β3.30 Their patterns of precipitation of radiolabeled αIIb and β3 were nearly identical (Figure 1). At the 1-hour time point they precipitated pro-αIIb, mature αIIb, and β3; at 2 and 4 hours the intensity of the pro-αIIb band progressively decreased, and the intensities of the mature αIIb and β3 bands increased, consistent with ongoing αIIbβ3 biogenesis during this time period.7,16 There was minimal further increase in αIIb and β3 band intensity after 4 hours (data not shown), indicating that biogenesis was essentially complete by this time point. Immunodepletion by 10E5 removed the αIIb and β3 species recognized by 7E3 and vice versa (data not shown). Thus, both of these antibodies recognize pro-αIIbβ3 complexes in addition to mature αIIbβ3 complexes.
Anti-αIIb antibodies CA3 and anti-V5 identify an excess of free pro-αIIb.
To assess the presence of a pool of pro-αIIb that is not complexed with β3, we used the anti-αIIb–specific mAbs CA3, whose epitope is unknown, and anti-V5, which is directed at a V5 tag added to the cytoplasmic domain of the recombinant αIIb used to transfect the cells. Each of these mAbs precipitated more pro-αIIb than did either 10E5 or 7E3, but similar amounts of mature αIIb and β3, at both the 2- and 4-hour time points (compare Figure 1 with Figure 2B). Thus, they react with free pro-αIIb, the pro-αIIbβ3 complex, and the mature αIIbβ3 complex. The amount of pro-αIIb precipitated at 4 hours was less than that precipitated at 2 hours, consistent with both conversion of pro-αIIb into mature αIIb and/or degradation of pro-αIIb. Thus, there is a substantial pool of free pro-αIIb at the 4-hour time point, even though production of mature αIIbβ3 appears to be essentially complete by that time. Therefore, the availability of pro-αIIb does not appear to limit αIIbβ3 biogenesis.
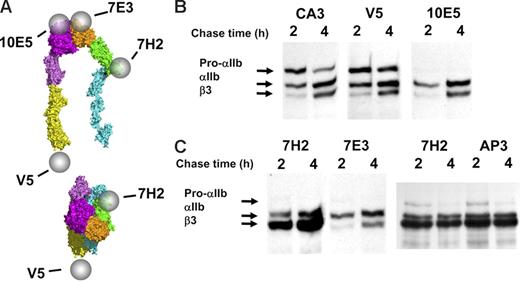
Excess free pro-αIIb and β3 exist in stably transfected HEK293 cells. (A) Diagram localizing the epitopes of antibodies 10E5, 7E3, anti-V5, and 7H2 on the bent and the extended and ligand-bound conformations of αIIbβ3; the epitope of antibody CA3 is on αIIb, but it has not been localized to a specific region. (B) Two anti-αIIb antibodies, CA3 and anti-V5, immunoprecipitated more pro-αIIb at both 2 hours and 4 hours than the complex-specific antibody 10E5. (C) Anti-β3 antibody 7H2 bound to β3 that was in complex with either pro-αIIb or mature αIIb and also immunoprecipitated more β3 at both 2 hours and 4 hours than the 2 complex-specific antibodies 10E5 (Figure 2B) and 7E3. All gel lanes in this figure are from the same immunoblot. Equivalent amounts of protein were loaded in each lane.
Excess free pro-αIIb and β3 exist in stably transfected HEK293 cells. (A) Diagram localizing the epitopes of antibodies 10E5, 7E3, anti-V5, and 7H2 on the bent and the extended and ligand-bound conformations of αIIbβ3; the epitope of antibody CA3 is on αIIb, but it has not been localized to a specific region. (B) Two anti-αIIb antibodies, CA3 and anti-V5, immunoprecipitated more pro-αIIb at both 2 hours and 4 hours than the complex-specific antibody 10E5. (C) Anti-β3 antibody 7H2 bound to β3 that was in complex with either pro-αIIb or mature αIIb and also immunoprecipitated more β3 at both 2 hours and 4 hours than the 2 complex-specific antibodies 10E5 (Figure 2B) and 7E3. All gel lanes in this figure are from the same immunoblot. Equivalent amounts of protein were loaded in each lane.
Anti-β3-specific antibodies, 7H2 and AP3, identify excess free β3.
To assess the presence of a pool of β3 not complexed with αIIb, we used the β3-specific mAbs 7H2 and AP3. 7H2 appears to react with an epitope on or near the PSI domain, because a C13A mutation in this domain results in loss of 7H2 binding (W.B.M., J.L., and B.S.C., unpublished observations, April 2005). The mAb AP3 reacts with an epitope that includes amino acids 50 and 62 from the PSI and hybrid domains, respectively20,31,44 (J. Peterson and P.J.N., unpublished observations). 7H2 precipitated more β3 than either 10E5 or 7E3 but similar amounts of pro-αIIb and mature αIIb (Figure 2C), and AP3 precipitated β3 with virtually the same pattern. Thus, these mAbs react with free β3 in addition to β3 in complex with either pro-αIIb or mature αIIb. These data indicate that there also is a substantial pool of free β3 at the 4-hour time point, even though production of mature αIIbβ3 appears to be essentially complete by this time. Thus, the availability of β3 does not appear to limit αIIbβ3 biogenesis.
Epitopes of the anti-β3 LIBS-specific antibodies are exposed on both free β3 and a subpopulation of the pro-αIIbβ3 complex.
Antibodies AP5 and LIBS2 have previously been shown to recognize epitopes on the β3 PSI and βTD domains, respectively (Figure 3A). 22,32,33,44 They bind to only a small fraction of mature αIIbβ3 complexes on unactivated platelets, and their binding is markedly enhanced by ligand binding to the receptor or by EDTA treatment. After 2 to 4 hours of chase, both of these antibodies precipitated nearly as much β3 as the non-LIBS mAb 7H2 (Figure 3B). They also precipitated some pro-αIIb [but less than that precipitated by mAbs 10E5 (data not shown) and 7H2 (Figure 3B)] but no mature αIIb. Thus, after considering the amount of β3 that is precipitated by 7H2 as part of the mature αIIbβ3 complex, it appears that AP5 and LIBS2 react with virtually the entire population of free β3 and subpopulations of the pro-αIIbβ3 complex. These data indicate that (1) the epitopes for these antibodies on the PSI and βTD domains are exposed and accessible on free β3; (2) on pro-αIIbβ3 complex formation, β3 subunits undergo conformational changes that begin to mask or alter the AP5 and LIBS2 epitopes, and/or these epitopes are blocked by association with pro-αIIb; and (3) the conformational changes leading to loss or masking of these epitopes are complete on formation of mature αIIbβ3.
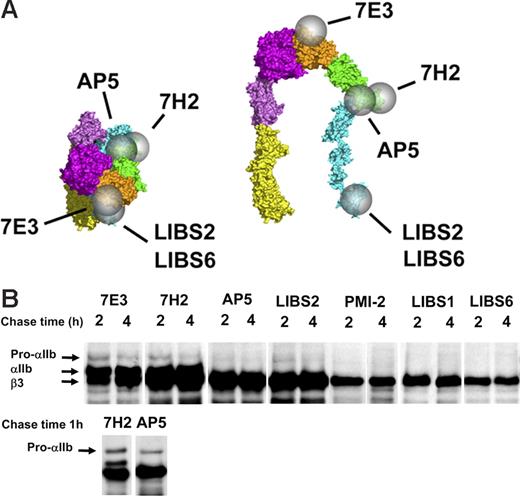
β3-Specific LIBS antibodies variably recognize free β3 and the pro-αIIββ3 complex but not the mature αIIbβ3 complex. (A) Epitopes of mAb 7E3, 7H2, and the β3-specific LIBS antibodies are indicated by spheres on the bent and extended structures of ligand-bound αIIbβ3. The epitopes of LIBS1 and PMI-2 are unknown. (B) Precipitation of αIIb and β3 by the mAbs at various times after initiating the chase. The mAbs LIBS2 and AP5 precipitated virtually the entire pool of free β3 (defined by the binding of the non-LIBS mAb 7H2), a portion of the pro-αIIb in complex with β3 but no mature αIIbβ3. The mAbs PMI-2, LIBS1, and LIBS6 precipitated subpopulations of uncomplexed β3 but no complexed β3. Data shown are from one of more than 5 experiments. The top row of blots is from a single experiment and is overexposed to show the fainter pro-αIIb bands. The bottom row is from a separate experiment and earlier time point to show the absence of mature αIIb in the AP5 precipitate. Equivalent amounts of protein were loaded in each lane.
β3-Specific LIBS antibodies variably recognize free β3 and the pro-αIIββ3 complex but not the mature αIIbβ3 complex. (A) Epitopes of mAb 7E3, 7H2, and the β3-specific LIBS antibodies are indicated by spheres on the bent and extended structures of ligand-bound αIIbβ3. The epitopes of LIBS1 and PMI-2 are unknown. (B) Precipitation of αIIb and β3 by the mAbs at various times after initiating the chase. The mAbs LIBS2 and AP5 precipitated virtually the entire pool of free β3 (defined by the binding of the non-LIBS mAb 7H2), a portion of the pro-αIIb in complex with β3 but no mature αIIbβ3. The mAbs PMI-2, LIBS1, and LIBS6 precipitated subpopulations of uncomplexed β3 but no complexed β3. Data shown are from one of more than 5 experiments. The top row of blots is from a single experiment and is overexposed to show the fainter pro-αIIb bands. The bottom row is from a separate experiment and earlier time point to show the absence of mature αIIb in the AP5 precipitate. Equivalent amounts of protein were loaded in each lane.
Anti-β3 LIBS-specific antibodies PMI-2, LIBS1, and LIBS6 react with free β3, but not with the pro-αIIbβ3 or mature αIIbβ3 complexes.
The mAb LIBS6 reacts within the βTD domain of β3, near the transmembrane domain,22,25 whereas the epitopes of PMI-2 and LIBS1, both of which inhibit platelet aggregation, are not known.22,44,45 These mAbs bind to only a small percentage of unactivated mature αIIbβ3 receptors, and their binding is greatly enhanced by ligand binding to αIIbβ3.18,22,45 Each of these antibodies precipitated β3 at 2 and 4 hours (Figure 3B). Unlike mAbs 7H2, 7E3, AP5, and LIBS2, however, these antibodies did not precipitate pro-αIIb (even when longer exposures were analyzed to adjust for the lower density of the β3 bands), and unlike mAbs 7H2 and 7E3 they did not precipitate mature αIIb. Thus, the epitopes for these mAbs are exposed on both ligand-bound and free β3, but not β3 in complex with either pro-αIIb or mature αIIb. To estimate the fraction of free β3 precipitated by these antibodies, the intensity of the β3 band precipitated by each antibody was divided by the difference in intensity of the β3 bands precipitated by 7H2 (representing free β3 plus β3 in complex with αIIb) and 7E3 (representing β3 in complex with αIIb). These values were 65% for PMI-2, 75% for LIBS1, and 40% for LIBS6.
Epitopes of 3 anti-αIIb LIBS-specific antibodies are masked on free pro-αIIb, pro-αIIbβ3 and mature αIIbβ3.
Antibody PMI-1 has previously been shown to react with a linear sequence in αIIb near the αIIb cleavage site (amino acids 844-859).18,46 Although the homologous region in αV was not defined in the αV crystal structure,12,47 it is contained in a loop, and the amino acids on either side of the loop map to the base of the Calf-2 domain (Figure 4A). Binding of mAb PMI-1 to αIIb is enhanced by ligand binding or EDTA treatment of αIIbβ3.18,21 B1B5 recognizes the αIIb light chain between residues 859 and 993 in the Calf-2 domain29 (Figure 4A). The binding of B1B5 to platelets is enhanced by incubation with tirofiban (2-fold) or treatment with either EDTA (1.5-fold) or DTT (2-fold), thus identifying B1B5 as a partial LIBS mAb (W.B.M., J.L., and B.S.C., unpublished data, March 2006). Both PMI-1 and B1B5 give strong signals on immunoblots of αIIb: PMI-1 reacts with the heavy chain and B1B5 reacts with the light chain (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article).
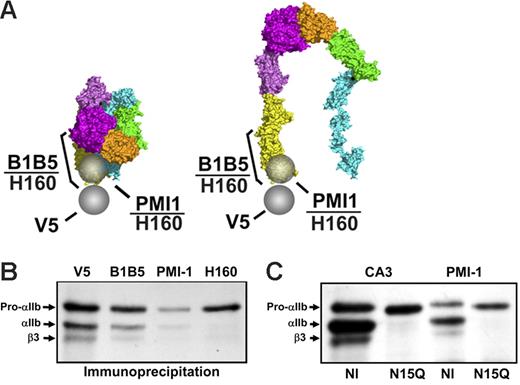
αIIb-Specific LIBS mAbs recognize only small amounts of free pro-αIIb. (A) Epitopes of the mAb anti-V5 and the αIIb-specific LIBS mAbs are indicated by spheres or bars on the bent, unactivated, and the proposed extended structure of ligand-bound αIIbβ3. (B) αIIb-Specific LIBS mAbs B1B5 and PMI-1 reacted with subpopulations of free pro-αIIb, pro-αIIbβ3, and mature αIIbβ3. Polyclonal antibody anti-αIIb (H-160), whose epitope encompasses both the B1B5 and PMI-1 epitopes, precipitated less pro-αIIb than V5. The 1.5-hour time point is depicted from 1 of more than 5 experiments. Equivalent amounts of protein were loaded in each lane. (C) PMI-1 precipitated less pro-αIIb than CA3 from cells with an αIIbN15Q mutation that eliminates the N15 glycan.
αIIb-Specific LIBS mAbs recognize only small amounts of free pro-αIIb. (A) Epitopes of the mAb anti-V5 and the αIIb-specific LIBS mAbs are indicated by spheres or bars on the bent, unactivated, and the proposed extended structure of ligand-bound αIIbβ3. (B) αIIb-Specific LIBS mAbs B1B5 and PMI-1 reacted with subpopulations of free pro-αIIb, pro-αIIbβ3, and mature αIIbβ3. Polyclonal antibody anti-αIIb (H-160), whose epitope encompasses both the B1B5 and PMI-1 epitopes, precipitated less pro-αIIb than V5. The 1.5-hour time point is depicted from 1 of more than 5 experiments. Equivalent amounts of protein were loaded in each lane. (C) PMI-1 precipitated less pro-αIIb than CA3 from cells with an αIIbN15Q mutation that eliminates the N15 glycan.
Both PMI-1 and B1B5 precipitated less pro-αIIb than did mAbs anti-V5 or CA3 at all time points (Figure 4B and data not shown), with PMI-1 precipitating less pro-αIIb than B1B5 in 6 of 6 experiments. PMI-1 also precipitated much less mature αIIb and β3 than did either anti-V5 or CA3, or the complex-dependent mAbs 10E5 and 7E3 (Figures 2B-C and 4B and data not shown). mAb B1B5 precipitated more pro-αIIb and mature αIIb than PMI-1, but less than the amount precipitated by anti-V5, consistent with its partial LIBS characteristics. The rabbit polyclonal antibody, anti-αIIb (H-160), prepared against αIIb amino acids 847 to 1006, reacted strongly with the αIIb heavy chain (Mr 120 kDa) on immunoblot, indicating that it recognizes an epitope close to that of PMI-1, and only minimally with the αIIb light chain (Mr 22 kDa) (data not shown). Anti-αIIb (H-160) immunoprecipitated only a subpopulation of free pro-αIIb and virtually none of the αIIb in complex with β3. When compared with the density of the pro-αIIb band precipitated by anti-V5, the density of the bands precipitated by PMI-1, B1B5, and H-160 were 21% ± 18%, 45% ± 6%, and 54% ± 34% of the density of the V-5 precipitated band (all n = 3). Thus, the PMI-1 and B1B5 epitopes are only expressed or available on subpopulations of free pro-αIIb. Because all of these antibodies react with denatured αIIb on immunoblots (Figure S1), the reduced immunoprecipitation is presumably due to decreased epitope expression or availability. To assess whether the PMI-1 epitope of the recombinant αIIb could be exposed by EDTA, as it is on platelets, PMI-1 binding was determined by flow cytometry before and after treatment with EDTA. Only a small amount of PMI-1 bound to untreated cells, but EDTA treatment increased PMI-1 binding more than 5-fold (Figure S1). To assess the effect of eliminating the N15 glycan on recognition of pro-αIIb by the LIBS mAb PMI-1, we performed immunoprecipitation studies on HEK293 cells expressing the αIIb N15Q mutation in combination with normal β3 (Figure 4C). At 45 minutes after pulse-chase, PMI-1 precipitated less pro-αIIb than CA3 from cells expressing N15QαIIbβ3, but a slightly greater proportion of total pro-αIIb (as judged by CA3 binding) than in cells expressing normal αIIbβ3.
Experiments with megakaryocyte-lineage cells derived from umbilical cord blood
Kinetics of αIIbβ3 biogenesis are similar in HEK293 and megakaryocyte-lineage cells derived from umbilical cord blood.
To verify that observations made in transfected HEK293 cells reflect αIIbβ3 biogenesis in a more physiologically relevant cell, we studied biogenesis of αIIbβ3 in megakaryocyte-lineage cells derived from human umbilical cord blood (CB) cells. After 10 days of culture in the presence of TPO, 95% ± 2% of CB cells expressed αIIbβ3, 83% ± 5% expressed GPIb, and 54% ± 10% expressed α2β1 (mean ± SD, all n = 4). On incubation with 10 μM thrombin receptor-activating peptide, the percentage of CB cells recognized by PAC1, an activation-dependant, ligand-mimetic anti-αIIbβ3 mAb, increased from 3% ± 2% to 16% ± 1% (mean ± SD, n = 3). The kinetics of β3 production and pro-αIIb conversion into mature αIIb were similar in CB and HEK293 cells (compare Figures 1B and 5A). Pro-αIIb disappearance, which reflects both the conversion of pro-αIIb into mature αIIb plus pro-αIIb degradation, was slightly more rapid in CB cells [1.4 ± 0.2 hours (n = 4) versus 2.3 ± 1.0 hour (n = 3) for CB cells and stably transfected HEK293 cells, respectively, P = .1]. The immunoprecipitation patterns obtained with the mAb panel were similar between the CB and HEK293 cells.
Kinetics of αIIbβ3 biogenesis in cells derived from umbilical cord blood (CB) cells. (A) Pulse-chase analysis of CB cells with the αIIbβ3 complex-specific mAb 10E5 showed a pattern of pro-αIIb, mature αIIb, and β3 kinetics similar to those in stably transfected HEK293 cells (compare with Figure 2). Similar to the results in stably transfected HEK293 cells, the anti-αIIb mAb CA3 and the anti-β3 mAb 7H2 revealed the presence of pools of free pro-αIIb and β3, respectively, in the CB cells at a time when αIIbβ3 production was essentially complete. (B) The anti–β3 LIBS mAbs AP5 and LIBS2 recognized pro-αIIbβ3 but not mature αIIbβ3, and the anti–αIIb LIBS mAb PMI-1 precipitated only a small subpopulation of pro-αIIb. Each box represents a separate experiment.
Kinetics of αIIbβ3 biogenesis in cells derived from umbilical cord blood (CB) cells. (A) Pulse-chase analysis of CB cells with the αIIbβ3 complex-specific mAb 10E5 showed a pattern of pro-αIIb, mature αIIb, and β3 kinetics similar to those in stably transfected HEK293 cells (compare with Figure 2). Similar to the results in stably transfected HEK293 cells, the anti-αIIb mAb CA3 and the anti-β3 mAb 7H2 revealed the presence of pools of free pro-αIIb and β3, respectively, in the CB cells at a time when αIIbβ3 production was essentially complete. (B) The anti–β3 LIBS mAbs AP5 and LIBS2 recognized pro-αIIbβ3 but not mature αIIbβ3, and the anti–αIIb LIBS mAb PMI-1 precipitated only a small subpopulation of pro-αIIb. Each box represents a separate experiment.
Discussion
In this study, we used conformation-specific mAbs to assess the conformational changes that the pro-αIIb and β3 subunits undergo prior to and during formation of the pro-αIIbβ3 heterodimer and the mature αIIbβ3 receptor in a transfected cell line and in megakaryocyte-lineage cells derived from human umbilical cord blood. Previous studies of αIIbβ3 biogenesis4,7,9,10 established that (1) the 2 subunits are translated from separate mRNAs; (2) αIIb has N-linked glycans that undergo processing in the Golgi, where αIIb is cleaved into heavy and light chains; (3) β3 is N-glycosylated but does not undergo processing of its N-linked glycans during receptor maturation; and (4) both αIIb and β3 are required for surface expression. Although the results of these studies were generally concordant, Duperray et al7 suggested that the rate of processing of αIIb in chronic myelogenous leukemia–derived megakaryocytes may limit the production of αIIbβ3, whereas Rosa and McEver9 reported that in HEL cells the β3 pool size may be limiting. We recently reported evidence that the pro-αIIb N15 glycan on the αIIb headpiece β-propeller domain is important in αIIbβ3 biogenesis and proposed that interaction of the β-propeller region with the membrane-associated chaperone calnexin may account for αIIb adopting the bent, inactive conformation during biogenesis.
In the current study, we identified sizable pools of both free pro-αIIb and β3 in 2 different cell types. Thus, pro-αIIbβ3 heterodimer formation does not appear to be limited by the availability of either subunit. Our data indicate that pro-αIIbβ3 complex formation is not controlled or limited by αIIb subunit availability. Thus, factors other than subunit availability must control pro-αIIbβ3 complex formation; the availability of other chaperone molecules that mediate the binding of the βA (I-like) domain of β3 to the β-propeller domain of αIIb may be one of those factors.
Our findings that mAbs 10E5 and 7E3 react with the pro-αIIbβ3 complex in addition to mature αIIbβ3,17,19 but not with free pro-αIIb or free β3, are similar to those reported for αLβ2 by Huang et al, who used mAbs specific for the αLβ2 complex with epitopes that are analogous to those of 10E5 and 78E3.48,49 They interpreted their data as indicating that the βA (I-like) domain of β2 and the β-propeller domain of αL do not become fully folded until complex formation occurs. This could also be the explanation for our findings. However, an alternative explanation for both their and our data is that the epitopes on the β-propeller and βA (I-like) domains are inaccessible to the mAbs because a chaperone(s) bind to the subunits in these regions. If this hypothesis is correct, then formation of the pro-αIIbβ3 complex most likely results from, or initiates, a conformational change that releases the complex from the chaperones, and this event may be a signal for transport to the Golgi for further processing.
Data from a variety of techniques, including electron microscopy and x-ray crystallography, have led to a working model in which αIIbβ3 undergoes dramatic conformational changes with activation and ligand binding that involve leg separation, headpiece extension, and a β3 swing-out motion at the junction of the β3 βA (I-like) domain and the hybrid domain.14,15 We have found that β3 LIBS mAbs also bind most or all free β3, and some also bind a portion of the pro-αIIbβ3 complex. Thus, it seems most likely that these LIBS epitopes become inaccessible after complex formation as a result of head-head, leg-leg, and/or head-leg interactions that stabilize the bent conformation of αIIbβ3 during the latter stages of biogenesis. Because mAbs AP5 and LIBS2 recognized virtually all of the free β3 pool, their epitopes appear to be altered or masked exclusively by their association with αIIβ. In contrast, because LIBS1, LIBS6, and PMI-2 did not recognize all of the free β3 pool, it is possible that either some β3 leg-head interactions occur relatively early, or that chaperones or other adjacent molecules partially block access to these epitopes. Moreover, because the β3 LIBS mAbs demonstrated variable reactivity with the pro-αIIbβ3 complex, it is possible that the β3 regions recognized by the mAbs that are able to precipitate at least a fraction of the pro-αIIbβ3 complexes (PSI for AP5, and βTD for LIBS2) are the last regions to adopt their mature conformations. Alternatively, separate subpopulations of pro-αIIbβ3 complexes may adopt different conformations after complex formation, either exposing or masking these epitopes. Of note, studies by Huang et al49 on αLβ2 (LFA-1) biogenesis demonstrated that an activating mAb whose epitope localized to the hybrid/PSI/EGF region of β2, recognized free β2, but not β2 in complex with αL. These data are consistent with our results with mAb AP5, which is also an activating mAb and binds to the analogous region of β3, and suggest that our observations on the step-wise nature of β3 subunit association with αIIb during biogenesis may be applicable to other integrin receptors.
In contrast to anti-β3 LIBS, the anti-αIIb LIBS mAb PMI-1, whose epitope is near the αIIb cleavage site within the Calf-2 domain,46 recognized approximately 20% of pro-αIIb subunits. This suggests that the majority of pro-αIIb subunits adopt a conformation that conceals the PMI-1 epitope immediately after subunit synthesis; that a chaperone, another molecule, and/or the platelet membrane conceals the epitope; or that the PMI-1 epitope is not formed until ligand binds to αIIbβ3. Because PMI-1 readily identifies both pro-αIIb and mature αIIb after denaturation in SDS by immunoblotting, and PMI-1 binds to a linear peptide,46 it seems more likely that its epitope is present but inaccessible. If the pro-αIIb headpiece does bind to calnexin soon after synthesis, then the proximity between the membrane-associated headpiece and the tail region of αIIb, which contains the PMI-1 epitope, may be responsible for the inaccessibility of the PMI-1 epitope. We also studied the binding of the antibody B1B5, whose epitope is on the extracellular portion of the αIIb light chain, and the polyclonal anti-αIIb antibody H-160, prepared against a peptide overlapping both the PMI-1 and B1B5 epitopes (Table 1). Like PMI-1, both of these antibodies precipitated less free pro-αIIb than anti-V5. Although care must be used in the interpretation of decreased antibody binding, the finding that all 3 antibodies have limited reactivity is highly suggestive that the tail region of pro-αIIb, which contains these epitopes, is not completely accessible during biogenesis.
Because the αVβ3 region corresponding to the αIIb loop containing the PMI-1 epitope (844-859) was not identified in the αVβ3 crystal structure,12 we do not have αVβ3 structural information to help understand the conformation of the analogous region in αIIb. To address this deficiency, we have applied 2 loop-determination programs, MODELLER 8v250 and Robetta,39 to predict the structure of the αIIb loop between amino acids 842 to 873 in the Calf-2 domain, which encompasses the PMI-1 epitope, the dominant H-160 epitope(s), and perhaps some or all of the B1B5 epitope. It should be noted that reliable prediction of the conformation of a 32–amino acid loop is not possible at the present time because the accessible conformational space increases exponentially with increasing chain length. These data are presented only to suggest the likely conformational space available for this loop.
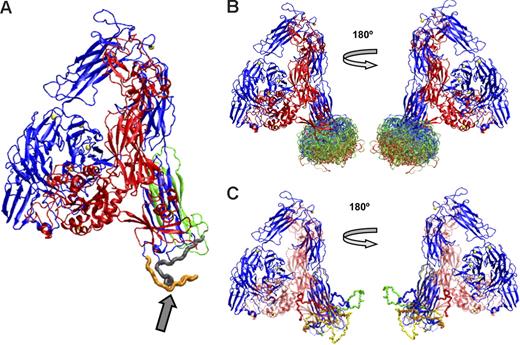
Figure 6A shows the αIIb subunit containing the conformation of the loop between amino acids 842 and 873 initially predicted by MODELLER, and Figure 6B shows the superimposition of 200 conformations for the loop, with the most favored shown in red, the least favored in blue, and those in between in green. Figure 5C shows 5 energetically favorable loop conformations derived using Robetta, a program that has been shown to provide reasonable structures for loops of 13 to 34 residues.41 The conformational space sampled by the Robetta loop modeling routine is quite broad and similar to that sampled by MODELLER. Some of the conformations from both programs that orient the loop below the Calf-2 region may not be possible in αIIb on the platelet surface because of the location of the plasma membrane. Conformations that orient the loop between the β-propeller and the Calf-2 domain, such as the red loop in Figure 6C, may not be accessible when the receptor is bent but may become accessible if the receptor adopts an extended position. In fact, Calzada et al51 reported that mAbs directed at epitopes on the β-propeller domain could interfere with the binding of mAbs directed at epitopes on the Calf-2 domain, indicating the proximity of these regions in mature αIIbβ3 on the platelet surface.
Computer models constructed using MODELLER or Robetta of the energetically most-favored conformations of the αIIb loop between amino acids 842 and 873. (A) αIIb with the loop (arrow) in an energetically favored conformation derived from MODELLER. The loop colors indicate an αIIb cleavage site (R859) that separates the heavy (orange) and light (gray) chains. The PMI-1 epitope is immediately proximal to the cleavage site, as is at least a portion of the anti-αIIb (H-160) epitope. The B1B5 epitope is on the light chain, indicated by the gray and green ribbons. (B) The 200 most energetically favorable conformations of the loop were derived using MODELLER. The most favorable conformations are in red, less favorable ones in green, and least favorable ones in blue. (C) Five loop conformations were derived using Robetta.
Computer models constructed using MODELLER or Robetta of the energetically most-favored conformations of the αIIb loop between amino acids 842 and 873. (A) αIIb with the loop (arrow) in an energetically favored conformation derived from MODELLER. The loop colors indicate an αIIb cleavage site (R859) that separates the heavy (orange) and light (gray) chains. The PMI-1 epitope is immediately proximal to the cleavage site, as is at least a portion of the anti-αIIb (H-160) epitope. The B1B5 epitope is on the light chain, indicated by the gray and green ribbons. (B) The 200 most energetically favorable conformations of the loop were derived using MODELLER. The most favorable conformations are in red, less favorable ones in green, and least favorable ones in blue. (C) Five loop conformations were derived using Robetta.
To obtain more information on the conformations of pro-αIIb and mature αIIb, we also assessed the ability of thrombin to cleave pro-αIIb and mature αIIb. Previous studies by Fujimura and Phillips52 and Phillips et al53 demonstrated that thrombin does not cleave mature αIIb when it is complexed with β3, but it does cleave mature αIIb when the complex is dissociated with EDTA.52,53 The predicted thrombin cleavage sites in αIIb are both within solvent-exposed regions of the β-propeller that are not likely to be affected by the bending of αIIb (Figure S2). However, they are likely to be masked by αIIbβ3 complex formation. We found that the free pro-αIIb subunit is susceptible to thrombin cleavage, confirming that the cleavage sites are accessible in the free pro-αIIb subunit (Figure S2). Thus, the inaccessibility of the PMI-1 and H-160 epitopes on the Calf-2 domain of free pro-αIIb is specific to that region.
If adoption of the bent conformation of αIIb limits access to the Calf-2 domain, then our data suggest a working model in which pro-αIIb adopts a bent conformation soon after synthesis, and then free β3 assumes its bent conformation by virtue of its interaction with the bent pro-αIIb. One possible mechanism by which pro-αIIb could adopt a bent conformation would be through interaction of the pro-αIIb headpiece with a membrane-associated chaperone, and we have previously proposed that the N15 glycan of αIIb, which is important in αIIbβ3 biogenesis, interacts with the membrane-associated calnexin. In the current study, however, we did not observe a major increase in the fraction of pro-αIIb precipitated by PMI-1 when the N15 glycan was eliminated by an N15Q substitution. Thus, it is possible that either the αIIb headpiece can interact with some other membrane-associated chaperone, that pro-αIIb interacts with calnexin via nonglycan interactions,54 or that pro-αIIb headpiece association with the membrane is not necessary for limiting access to the pro-αIIb Calf-2 domain. Additional studies are required to assess this working model.
Understanding the biogenesis of integrins should provide insights into the mechanism(s) by which mutant receptors fail to form complexes or form abnormal complexes that do not progress to the Golgi. Such understanding may also provide insights into the energetics involved in the conformational changes associated with activation and ligand binding. Moreover, because antagonists to αIIbβ3, α4, and αL have all demonstrated therapeutic efficacy,55–58 understanding integrin biogenesis may help in designing pharmacologic approaches to prevent integrin biogenesis or the conformational changes associated with integrin activation and/or ligand binding.
Authorship
Contribution: W.B.M. designed and performed experiments, analyzed and interpreted data, and drafted the manuscript; J.L. and N.V. designed and performed experiments; M.M. performed molecular modeling; P.J.N. designed experiments and analyzed and interpreted data; B.S.C. designed experiments, analyzed and interpreted data, and drafted the manuscript.
Conflict-of-interest disclosure: B.S.C. is an inventor of abciximab, a derivative of 7E3, and in accord with federal law and the policies of the Research Foundation of the State University of New York, shares in royalty payments made to the Foundation based on sales of abciximab. All other authors declare no competing financial interests.
Correspondence: W. Beau Mitchell, New York Blood Center, 310 E 67th St, New York, NY 10021; e-mail bmitchell@nybloodcenter.org.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Mortimer Poncz and Mark Ginsberg for providing antibodies, and the National Cord Blood Program of the New York Blood Center for providing umbilical cord blood.
This work was supported in part by the National Institutes of Health (grants HL19278, CTSA-UL1RR024143, and GCRC-M01RR00102) (B.S.C.), (grant HL68622 01) (W.B.M.), and (grant HL44612) (P.J.N.) and by funds from Stony Brook University.