Abstract
CD4+CD56+ hematodermic neoplasm (CD4+CD56+HN) is an aggressive hematopoietic malignancy with distinct clinicopathologic and immunophenotypic features that commonly involve the skin, bone marrow, and blood. Differentiation from cutaneous myelomonocytic leukemia (c-AML) may be exceedingly difficult and requires extensive phenotyping. The molecular mechanisms involved in the development of CD4+CD56+HN are largely unresolved. Moreover, recurrent chromosomal alterations have not yet been precisely defined in CD4+CD56+HN and c-AML. In the present study an integrated genomic analysis using expression profiling and array-based comparative genomic hybridization (CGH) was performed on lesional skin biopsy samples of patients with CD4+CD56+HN and c-AML. Our results demonstrate that CD4+CD56+HN and c-AML show distinct gene-expression profiles and distinct patterns of chromosomal aberrations. CD4+CD56+HN is characterized by recurrent deletion of regions on chromosome 4 (4q34), chromosome 9 (9p13-p11 and 9q12-q34), and chromosome 13 (13q12-q31) that contain several tumor suppressor genes with diminished expression (Rb1, LATS2). Elevated expression of the oncogenes HES6, RUNX2, and FLT3 was found but was not associated with genomic amplification. We noted high expression of various plasmacytoid dendritic-cell (pDC)–related genes, pointing to the cell of origin of this malignancy.
Introduction
CD4+CD56+ hematodermic neoplasm (CD4+CD56+HN), previously termed blastic natural killer (NK)-cell lymphoma, is a recently recognized disease entity with distinct clinicopathologic and immunophenotypic features. Clinically, this malignancy generally presents in the skin, often followed by bone marrow and blood involvement. CD4+CD56+HN is highly resistant to conventional chemotherapy and has a poor prognosis.1-3 Histopathologic examination of CD4+CD56+HN skin lesions shows diffuse monotonous infiltrates of medium-sized cells with fine chromatin resembling lymphoblastic or myeloblastic sarcoma. The neoplastic cells characteristically express CD4 and CD56, may be positive for TdT, and show a dotlike staining for CD68 in some cases, whereas they are negative for T-cell, B-cell, NK-cell, and myelomonocytic markers.3
In a recent study in which 63 published cases of CD4+CD56+HN and 10 cases of cutaneous myeolomonocytic leukemia (c-AML) were compared, striking similarities in clinical presentation, histology, immunophenotype, and prognosis were noted.2 c-AML cells also frequently coexpress CD4 and CD56 and, as CD4+CD56+HN, may precede the leukemic phase of the disease. Until recently, differentiation between the two conditions depended on the presence or absence of classic myelomonocytic markers, such as CD33, myeloperoxidase, or lysozyme. More recent studies showed that most CD4+CD56+HN but only few c-AML patients express CD123, TCL1a, and BDCA-2, markers characteristically expressed by plasmacytoid dendritic cells (pDCs).3-5 Expression of these pDC markers is not only helpful in differential diagnosis but has also led to the current view that CD4+CD56+HN is derived from precursor pDCs and that the term blastic NK-cell lymphoma is no longer justified.3,6
Few studies have addressed the molecular genetic features of these CD4+CD56+HNs. Using low-resolution comparative genomic hybridization (CGH), recurrent chromosomal abnormalities have been detected in 70% of cases, while fluorescent in situ hybridization studies demonstrated translocations involving chromosome 5q and 6q.3,7-9 Similar studies on c-AML are completely lacking.
In this report we describe the results of an integrated genomic analysis using both gene-expression profiling and array-based CGH in selected groups of CD4+CD56+HN and c-AML. The aims of this study were to find consistent gene-expression profiles and genetic abnormalities that may aid in differential diagnosis, support the pDC origin of the group of CD4+CD56+HNs, and possibly provide new targets for therapeutic intervention.
Patients, materials, and methods
Approval was obtained from the Leiden University Medical Center institutional review board for these studies. Informed consent was provided in accordance with the Declaration of Helsinki.
Patient selection and material
Pretreatment skin biopsies from 5 patients with a CD4+CD56+HN (cases 1 to 5) and 6 patients with a c-AML (cases 6 to 11) were included in this study. The clinical details of these 11 patients are presented in Table 1. The neoplastic cells of the CD4+CD56+HN cases expressed CD4, CD56, CD123, TCL1A, and TdT (4 of 5 cases) but were consistently negative for B-cell, T-cell, and myelomonocytic markers (Table 2). In all 6 c-AML cases expression of myelomonocytic markers CD33, MPO, CD68, and/or lysozyme was observed (Table 2). Selected tumor samples contained at least 90% tumor cells.
DNA from all patient biopsies (cases 1 to 11) was used for array-based CGH analysis. In addition, from 8 of these 11 patients (cases 1 to 6, 8, and 11) gene-expression profiling on mRNA also was performed.
Extraction of RNA and DNA
RNA and DNA were isolated from biopsies of CD4+CD56+HN and c-AML patients. RNA was extracted from 50 × 20 μM frozen sections using the RNeasy kit (Qiagen, Hilden, Germany), yielding 25 to 60 μg total RNA. DNA was isolated from 25 × 20 μM frozen sections using the Genomic-tips 20/G kit (Qiagen), yielding 10 to 60 μg genomic DNA.
Oligonucleotide microarrays
Samples and microarrays (Human Genome U133plus2.0 array [Affymetrix Santa Clara, CA] interrogating more than 47 000 human transcripts and variants) were processed according to the manufacturer's protocol (available from Affymetrix) as described previously.10
The array images were quantified using the Genechip operating system (GCOS) v1.2 software (Affymetrix). The average fluorescence intensity was determined for each microarray, and then the output of each experiment was globally scaled to a target value of 200. Normalization of the data and analysis of the gene-expression patterns were done using BRB ArrayTools v3.3.011 and the DNA Chip Analyzer,12,13 and results were visualized using GeneMaths XT (Applied Maths, Austin, TX). Expression results were compared with publicly available data.14
Array-based CGH
Genomewide analysis of DNA copy number changes of patient samples was performed using high-resolution array-based CGH containing about 3500 bacterial artificial chromosomes (BACs). Fabrication and validation of the array, hybridization methods, and analytic procedures have been described elsewhere in detail,15 while the clone content is available in the “Cytoview” window of the Sanger Centre mapping database site, Ensembl.16 Data were analyzed as described previously.17
Quantitative real-time PCR
Complementary DNA synthesis was performed on 1 μg total RNA after treatment with RQ1 DNase I (Promega, Madison, WI) using IScript reverse transcriptase (RT) (Bio-Rad, Veenendaal, the Netherlands), oligo(dT)12-18, and random hexamer priming (Bio-Rad) in a final volume of 20 μL. Real-time polymerase chain reaction (PCR) was performed with the MyIQ instrument and the SYBR Green Supermix (Bio-Rad). The cycle parameters for transcripts of interest and for the reference genes U1A and RPS11 used for normalization were as follows: denaturing for 15 seconds at 95°C and annealing and extension for 20 seconds at 60°C for 40 cycles. For primer sequences (oligos synthesized by Invitrogen [Breda, The Netherlands]) of selected transcripts, see Table S1 (available on the Blood website; see the Supplemental Table link at the top of the online article). Data were evaluated using MyIQ software (Bio-Rad) and the second derivative maximum algorithm, while confirmation of the specificity of the PCR product and standard curves were performed as previously described.10
Results
Gene-expression profiling
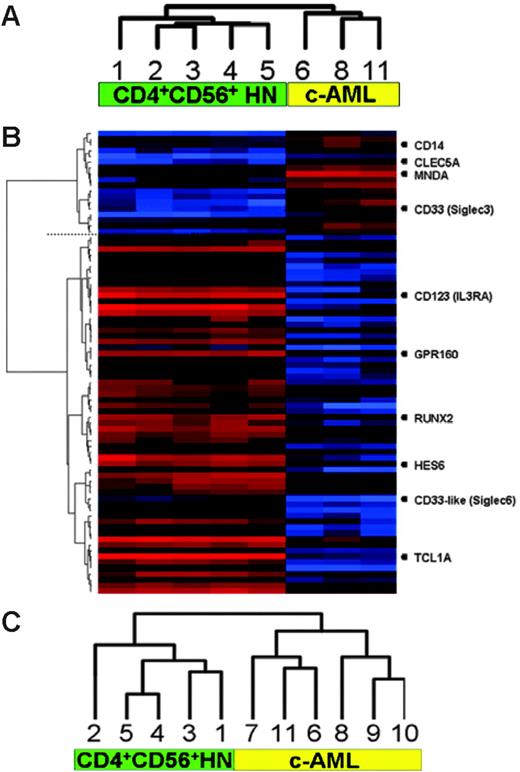
Analysis of hybridization signals of 54 675 probes represented on the U133plus2.0 genechip revealed that in CD4+CD56+HN an average of 42% and in c-AML an average of 40.8% of the genes were expressed. Using unsupervised hierarchic clustering based on the gene-expression values of all transcripts present at the U133plus2.0 genechip, CD4+CD56+HN and c-AML split into 2 distinct groups consisting of a group of 5 CD4+CD56+HNs and a group of 3 c-AMLs (Figure 1A). Next we excluded all genes encoding proteins used for diagnosis and repeated the unsupervised cluster analysis. Exclusion did not change the result: The 2 disease entities classified separately in an identical manner.
Clustering of gene-expression and high-resolution array-based CGH data. (A) Unsupervised hierarchic clustering of gene-expression profiles demonstrating 2 distinct groups. (B) Supervised analysis revealing differentially expressed genes between the 2 groups (genes discussed in this paper are listed on the right). (C) Unsupervised hierarchic clustering of array-based CGH data. Numbers correspond to cases as described in Tables 1 and 2.
Clustering of gene-expression and high-resolution array-based CGH data. (A) Unsupervised hierarchic clustering of gene-expression profiles demonstrating 2 distinct groups. (B) Supervised analysis revealing differentially expressed genes between the 2 groups (genes discussed in this paper are listed on the right). (C) Unsupervised hierarchic clustering of array-based CGH data. Numbers correspond to cases as described in Tables 1 and 2.
Next, supervised clustering was used to obtain a more detailed comparison between CD4+CD56+HN and c-AML (Figure 1B), revealing that more than 795 genes were significantly differentially expressed (P ≤ .01) with fold changes ranging from 0.021 to 450.
The most highly differentially expressed genes, using a fold change difference of at least 10, are given in Table 3. Within the c-AML group increased expression of numerous characteristic myelomonocytic transcripts such as SIGLEC3 (CD33), CD14, CLEC5A, and MNDA could be distinguished.18,19 In the CD4+CD56+HN group, this analysis demonstrated selective expression of markers supporting derivation from pDCs and revealed putative candidate oncogenes as well as novel membranebound markers.
Cell of origin
The genes significant in separating CD4+CD56+HN from c-AML included a large number of genes known to be expressed by plasmacytoid dendritic cells (indicated in Table 3 with an “X”). Expression of TCL1a and IL3RA (CD123) was consistent, with the results of immunostaining presented in Table 2. Furthermore, we detected selective expression of Toll-like receptors (TLRs) TLR9 and TLR10, TLRs typically expressed by pDCs, and absence of TLR2, TLR4, and TLR8 transcripts, which are considered to be prototypic for dendritic cells of myelomonocytic origin.20
Putative oncogenes
High expression of genes physiologically involved in Notch signaling (HES6, RUNX2) but also recognized as putative oncogenes was encountered in the CD4+CD56+HN biopsies when compared with c-AML. Expression of HES6 and RUNX2 was not only elevated relative to c-AML but also when comparing with normal pDCs, suggesting functional dysregulated expression of these oncogenes in CD4+CD56+HN.
Novel membranebound markers for CD4+CD56+HN
Selective expression of SIGLEC6 encoding the CD33-like protein and elevated expression of the orphan G protein–coupled receptor GPR160 was identified. Whereas SIGLEC6 was exclusively expressed in CD4+CD56+HN, SIGLEC3, encoding CD33, was only expressed in c-AML.
Finally, we noticed overexpression of a set of genes thus far not described for CD4+CD56+HN or other hematopoietic cells but all highly expressed in neuronal cells and thought to play a crucial role in the development of the central nervous system (Table 3).
Global genomic aberrations
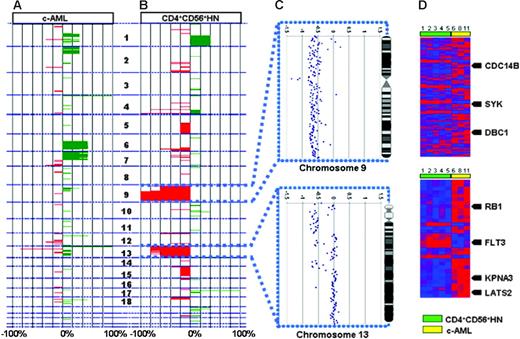
The array-based CGH analysis using an array with a resolution of approximately 1 Mb enables detection of both small and whole-chromosome areas of low copy number gain (DNA copy number 3 or 4) as well as deletions (DNA copy number 0 or 1) in both groups. When comparing the patterns of chromosomal aberrations, clearly 2 distinct patterns were revealed, reinforcing the notion that CD4+CD56+HN is also pathogenetically distinct from c-AML (Figure 1C). An overview of chromosomal imbalances as a karyogram is depicted in Figure 2, while a cumulative overview of array-based CGH results is shown in Figure 3A-B.
Overview of chromosomal imbalances. Imbalances are shown in (A) 5 CD4+CD56+HN cases and (B) 6 cAML cases. Red bars on the left of each ideogram represent losses, and green bars of the right represent gains of the corresponding chromosomal regions.
Overview of chromosomal imbalances. Imbalances are shown in (A) 5 CD4+CD56+HN cases and (B) 6 cAML cases. Red bars on the left of each ideogram represent losses, and green bars of the right represent gains of the corresponding chromosomal regions.
Integrated genomic analysis. The analysis shows chromosomal imbalances in 6 c-AML cases (A) and 5 CD4+CD56+HN cases (B). Green represents gains; red, losses of the corresponding chromosomal regions. (C) High-resolution analysis of chromosomes 9 and 13 for a representative individual case of CD4+CD56+HN. (D) Heat map depicting all significantly differentially expressed genes in CD4+CD56+HN versus c-AML within regions on chromosomes 9 and 13 that show recurrent chromosomal abnormalities. Arrows indicate known tumor suppressor genes and oncogenes; numbers correspond to cases described in Tables 1 and 2.
Integrated genomic analysis. The analysis shows chromosomal imbalances in 6 c-AML cases (A) and 5 CD4+CD56+HN cases (B). Green represents gains; red, losses of the corresponding chromosomal regions. (C) High-resolution analysis of chromosomes 9 and 13 for a representative individual case of CD4+CD56+HN. (D) Heat map depicting all significantly differentially expressed genes in CD4+CD56+HN versus c-AML within regions on chromosomes 9 and 13 that show recurrent chromosomal abnormalities. Arrows indicate known tumor suppressor genes and oncogenes; numbers correspond to cases described in Tables 1 and 2.
The mean number of alterations per patient in the c-AML group was 97 (range, 8 to 205) with low copy number gains (n = 86; range, 6 to 200) more frequent than losses (n = 10; range, 2 to 27). No highly recurrent alterations (present in more than 50% of cases) could be detected, with trisomy 8 detected in 2 patients (33%) as the most recurrent alteration in this group.
In the CD4+CD56+HN patient group, the mean number of chromosomal aberrations was 328 (range, 266 to 497 aberrations per case). Losses (n = 270; range, 151 to 382) were more frequent than low copy number gains (n = 58; range, 8 to 115).
The most frequent findings in CD4+CD56+HN (present in at least 80% of cases) were deletions of chromosomes 4q34.1-4q34.2 (80%), 9q (100%), 9p (80%), and 13q (80%). No recurrent low copy number gains could be detected in this group. The detected recurrent chromosomal deletions are not a result of structural rearrangement of chromosomes but more the loss of a given full chromosome.
Detailed analysis of chromosomal regions with recurrent deletion in CD4+CD56+HN
To identify altered gene-expression levels in chromosomal areas of gain or loss, we made an attempt to integrate both expression profiling and array-based CGH data. To this end we compared expression levels of genes located in recurrent regions of deletion in CD4+CD56+HN with expression of these genes in c-AML. We noted that positive correlations primarily could be made between deletion of genes and loss of expression, while increased expression as a result of gene amplification was hardly encountered. Interestingly, we also detected a considerable number of negative correlations (ie, monoallelic loss of a gene locus associated with elevated expression of a gene located herein).
Deletion of chromosome 9q12-9q34.3
All 5 CD4+CD56+HN patients showed deletion of a large part (46.0 to 134.9 Mb) of chromosome 9q (Figure 3C). This region of deletion encompasses more than 300 known genes. Analysis of the expression levels of these genes revealed that expression of 238 of these genes was unaltered and expression of 20 genes was elevated when compared with c-AML samples. Fifty-four genes located in this region showed diminished expression. Among these are the known tumor suppressor genes CDC14B, DBC1, and SYK.
Deletion of chromosome 13q12.11-13q31.1
Deletion of chromosome 13q12.11-13q31.1 was detected in 4 of 5 CD4+CD56+HN biopsies but not in any of the c-AML cases (Figure 3C). One of these samples showed only partial deletion of chromosome 13. In this latter sample distinct regions of deletion could be detected: 13q12.11 (0 to 21.0 Mb), 13q12.12-13q14.11 (24.2 to 42.4 Mb), 13q14.2-13q14.3 (46.8 to 51.5 Mb), 13q21.32-13q21.33 (63.7 to 69.4 Mb), and 13q31.1 (82.8 to 84.9 Mb). The 5 regions contain more than 100 annotated genes, of which 27 genes showed altered expression (Figure 3D). Interestingly, expression of FLT3, which functions as an oncogene in hematopoietic malignancies, showed a negative correlation: Its expression was elevated compared with c-AML despite monoallelic deletion. Twenty-one genes displayed decreased expression, among them the tumor suppressor genes KPNA3, LATS2, and RB1.
Deletion of chromosomes 4q34.1-4q34.2 and 9p13.2-9p11.2
Deletion of a discrete region of chromosome 4 was observed in 4 of 5 CD4+CD56+HN samples, while loss of this region was not observed in any of the c-AML cases. This region of loss was centered at 175.7 to 177.9 Mb, a region containing 8 annotated genes. However, no decreased expression of any of these genes could be detected.
Finally, 4 of 5 CD4+CD56+HN samples showed loss of chromosome 9p13.2-9p11.2. This region of loss was centered at 38.0 to 44.5 Mb, a region containing 5 known genes. However, no significant reduction in expression in any of these genes could be detected.
Confirmation of microarray expression data by real-time PCR
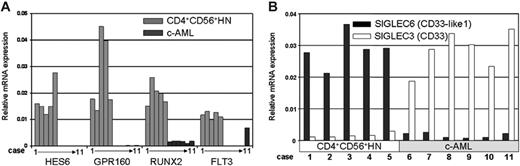
To validate microarray results, we selected 14 genes from different “classes” and biologic functions and quantified their expression by real-time PCR in all 11 tumor biopsies. In addition, we determined FLT3 mRNA levels, because this gene was up-regulated in all CD4+CD56+HN biopsies despite the loss of 1 allele. For all samples we could validate the microarray results (an example for a selection of genes is depicted in Figure 4A). Finally, we confirmed the mutually exclusive expression of SIGLEC3 (CD33) and SIGLEC6 (CD33-like1) genes in c-AML and CD4+CD56+HN, respectively (Figure 4B).
Messenger RNA expression as measured by real-time PCR. (A) Real-time RT-PCR confirmation of genes of interest: HES6, GPR160, RUNX2, and FLT3. (B) Real-time RT-PCR reveals differential expression of SIGLEC3 (CD33) and SIGLEC6 (CD33-like1) when comparing CD4+CD56+HN and c-AML. Expression values are presented as relative to RPS11 expression.
Messenger RNA expression as measured by real-time PCR. (A) Real-time RT-PCR confirmation of genes of interest: HES6, GPR160, RUNX2, and FLT3. (B) Real-time RT-PCR reveals differential expression of SIGLEC3 (CD33) and SIGLEC6 (CD33-like1) when comparing CD4+CD56+HN and c-AML. Expression values are presented as relative to RPS11 expression.
Discussion
In the present study an integrated genomic analysis using expression profiling and array-based CGH was performed on selected groups of CD4+CD56+HN and c-AML.
In this study a first attempt was made to unravel expression profiles and consistent genetic abnormalities that may aid in differential diagnosis between these 2 groups, give insight in the molecular pathogenesis and cellular differentiation of CD4+CD56+HN, and provide targets that can possibly be used for diagnosis and therapeutic intervention.
Gene-expression profiling showed a clear distinction between the 2 groups and identified putative oncogenes and novel diagnostic/therapeutic markers within the CD4+CD56+HN group of patients. Also, array-based CGH analysis showed distinct patterns of chromosomal alteration: In c-AML hardly any recurrent alterations were detected, while in CD4+CD56+HN returning patterns of chromosomal alterations occurred in all patients. To date, cytogenetic analysis has been performed in only 23 patients, all using conventional CGH, and resulted in a heterogeneous pattern of abnormalities. Our data demonstrate that recurrent chromosomal alterations mainly involve chromosomes 4, 9, and 13. However, we could not confirm the previously published high percentage of 5q and 12p aberrations, which may relate to differences in patient selection and/or techniques used.3 The candidate loci on chromosomes 4, 9, and 13 include a number of known or suspected tumor suppressor genes that show diminished expression (CDC14B, DBC1, SYK, KPNA3, LATS2, and RB1). By combining expression and array-based CGH data we also identified a number of genes paradoxically overexpressed in CD4+CD56+HN despite monoallelic chromosomal loss, suggesting deregulation of control of transcription.
The most highly differentially expressed genes between CD4+CD56+HN and c-AML can in general be categorized as either pDC-related genes, highly expressed by the CD4+CD56+HN (eg, SpiB),21 or myelomonocytic genes (eg, CD14, CD33), which are exclusively expressed in c-AML. This observation not only underscores that both disease entities are distinct but also supports the proposition that the pDC precursor is the cell of origin of CD4+CD56+HN.1,22 This is further confirmed by selective expression of the recently defined CD4+CD56+HN markers TCL1a, TdT, CD123, and BDCA-2 (the latter 2 with fold changes of 9 and 6, respectively, when compared with c-AML) in CD4+CD56+HN.4
Gene-expression profiling additionally revealed high expression of 2 putative oncogenes, HES6 and RUNX2, not associated with chromosomal imbalances. High expression of FLT3 in CD4+CD56+HN was paradoxically associated with monoallelic deletion of the locus containing this protooncogene.
HES6 is a basic helix-loop-helix transcription factor from the family of mammalian homologs of Drosophila hairy and enhancer of split. HES family members are involved in the Notch signaling pathway, long thought to play a role in the proliferation of a broad variety of cell types, both normal and transformed.23 HES6 expression has thus far only been detected in embryonic tissues.24,25 Although limited data are available concerning the physiologic role of HES6, it has been shown that HES6 is able to interfere with the E-box binding capability of HES1 and therefore is able to inhibit HES1 activity.26 Aggressive forms of cancer often show loss of HES1 expression,27 which suggests HES1 to be a candidate tumor suppressor gene. RUNX2 overexpression has been examined using RUNX2 transgenic mice, where overexpression leads to a block of T-cell differentiation and an accumulation of cells in the preleukemic phase.28 Nevertheless, the cells lack the capacity to proliferate, indicating that additional oncogenic events are necessary to rescue cells from the growth-suppressive effects of RUNX2.29
When combining array-based CGH data and gene-expression profiles, a remarkable observation was the association of very high expression of the oncogene FLT3 with chromosomal loss of a minimal region in 13q12 in CD4+CD56+HN. Clinical and experimental evidence both indicate that FLT3 is a protooncogene with the capacity to enhance survival and proliferation of leukemia blast cells.30 Wild-type FLT3 is expressed in a wide range of hematopoietic malignancies, including acute lymphoid leukemia and mixed lineage leukemia, and it is expressed in 70% to 100% of AML (first described by Birg et al31 ). FLT3 is targeted by specific inhibitors in the experimental treatment of acute myeloid leukemia that have activating mutations in the FLT3 gene. Suppression of FLT3 signaling by these inhibitors could potentially also be of benefit to patients with CD4+CD56+HN.32 Finally, we detected aberrant high expression of 2 membranebound receptors with largely unknown function, GPR160 and Siglec6. GPR160 is an “orphan” G protein–coupled receptor and has recently been described as having relevance in the pathology of prostate cancer.33 Siglec6 is a family member of the sialoadhesins.34 Human siglecs are expressed in a cell type–specific fashion, suggesting involvement in discrete functions. Interestingly, expression of Siglec6 was hitherto only observed in human placenta, and its specific expression in CD4+CD56+HN might provide a starting point for future research to validate its potential as a novel diagnostic tool and/or therapeutic target.
In sum, the used integrated genomic analysis revealed clear distinction between CD4+CD56+HN and c-AML, which may aid in the differential diagnosis, and supports the notion that the group of CD4+CD56+HNs is a malignancy arising from (precursor) pDCs.
Authorship
Contribution: R.D., R.v.D., R.W., M.H.V., and C.P.T. designed the study and wrote the paper; R.D., R.v.D., K.S., and C.P.T. performed the research and analyzed the data; and all authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: C. P. Tensen, Department of Dermatology, B1-Q, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: c.p.tensen@lumc.nl.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Aat Mulder, Gemma van Venrooij, and Jeroen Knijnenburg for their excellent technical assistance. Analyses were performed using BRB ArrayTools developed by Dr Richard Simon and Amy Peng Lam.