Abstract
Liver and lymph node sinusoidal endothelial cell C-type lectin (LSECtin [CLEC4G]) is a C-type lectin encoded within the liver/lymph node–specific intercellular adhesion molecule-3–grabbing nonintegrin (L-SIGN)/dendritic cell–specific intercellular adhesion molecule-3–grabbing nonintegrin (DC-SIGN)/CD23 gene cluster. LSECtin expression has been previously described as restricted to sinusoidal endothelial cells of the liver and lymph node. We now report LSECtin expression in human peripheral blood and thymic dendritic cells isolated ex vivo. LSECtin is also detected in monocyte-derived macrophages and dendritic cells at the RNA and protein level. In vitro, interleukin-4 (IL-4) induces the expression of 3 LSECtin alternatively spliced isoforms, including a potentially soluble form (Δ2 isoform) and a shorter version of the prototypic molecule (Δ3/4 isoform). LSECtin functions as a pathogen receptor, because its expression confers Ebola virus–binding capacity to leukemic cells. Sugar-binding studies indicate that LSECtin specifically recognizes N-acetyl-glucosamine, whereas no LSECtin binding to Mannan- or N-acetyl-galactosamine–containing matrices are observed. Antibody or ligand-mediated engagement triggers a rapid internalization of LSECtin,which is dependent on tyrosine and diglutamic-containing motifs within the cytoplasmic tail. Therefore, LSECtin is a pathogen-associated molecular pattern receptor in human myeloid cells. In addition, our results suggest that LSECtin participates in antigen uptake and internalization, and might be a suitable target molecule in vaccination strategies.
Introduction
The identification of the lectin gene cluster at chromosome 19p13.21 has led to the realization that some C-type lectins are capable of mediating intercellular adhesion, pathogen-binding, and antigen internalization for induction of T cell responses.2 The paradigmatic example of this type of lectin is dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN), which efficiently internalizes antigens,3 mediates dendritic cell intercellular adhesions,4 and recognizes a wide range of microorganisms through binding to mannose- and Lewis-containing glycans.5 C-type lectins on dendritic cells enhance their ability for pathogen recognition6 and contribute to modulation of toll-like receptor (TLR)-initiated signals.7 Consequently, the definition of the range of dendritic cell lectins and their binding specificities might provide adequate targets for immune intervention and prevention of pathogen entrance and spreading.
The lectin gene cluster at chromosome 19p13.2 includes the genes encoding for the type II C-type lectins DC-SIGN, liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin (L-SIGN), CD23, and liver and lymph node sinusoidal endothelial cell C-type lectin (LSECtin).1,4,8,9 DC-SIGN is expressed on myeloid dendritic cells,4,10 and alternatively activated in vitro on macrophages.11 In vivo it is found on interstitial dendritic cells,12 a subset of CD14+ peripheral blood DC,13 human microvascular endothelial cells,8 and on synovial, placenta, lymph node, and alveolar macrophages.14–16 By contrast, L-SIGN is exclusively expressed on endothelial cells of the liver, lymph nodes, and placenta,17,18 but not on myeloid cells.
The LSECtin (CLEC4G) gene is located between the CD23 and DC-SIGN genes with the three genes arranged in the same orientation.9 LSECtin encodes a protein with a lectin domain followed by a 110-residue stalk region, a transmembrane domain, and a 31-residue cytoplasmic domain.9 LSECtin has been previously detected on liver and lymph node sinusoidal endothelial cells at the protein and RNA level.9 LSECtin functions as an attachment factor for Ebola virus and SARS, but it does not bind HIV or hepatitis C virus.19 We now describe the expression of LSECtin isoforms in ex vivo isolated human peripheral blood and thymic dendritic cells as well as in dendritic cells and macrophages generated in vitro. LSECtin exhibits ligand-induced internalization, and its sugar recognition specificity differs from that of DC-SIGN. The presence of LSECtin on myeloid cells should therefore contribute to expanding their antigen-capture and pathogen-recognition capabilities.
Materials and methods
The study described was approved by the Centro de Investigaciones Biologicas (CSIC) Institutional Review Board. The study did not involve any direct contact with human subjects.
Cell culture
Human peripheral blood mononuclear cells were isolated from buffy coats from normal donors over a Lymphoprep (Nycomed Pharma, Oslo, Norway) gradient according to standard procedures.20 Monocytes were purified from peripheral blood mononuclear cells by magnetic cell sorting using CD14 microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany). Monocyte-derived dendritic cells (MDDC), monocyte-derived macrophages, and alternatively (AAMØ) or classically (CAMØ) activated macrophages were generated as previously described.11,20 Isolation of peripheral blood myeloid dendritic cells (MyDC)was done with a Blood DC isolation kit (MACS; Miltenyi Biotec) with some modifications. First, peripheral blood mononuclear cells were depleted of T cells, NK cells, and monocytic cells by magnetic separation, and the remaining population was incubated with monoclonal antibodies (mAbs) to fluorescein isothiocyanate (FITC)-CD4, phycoerythrin (PE)-labelled CD11c, PE-Cy5-CD14, and PE-Cy5-CD19 and sorted in a FACSVantage cell sorter (BD Biosciences, Franklin Lakes, NJ). PE-Cy5-positive cells were discarded, and double-positive cells for CD4 and CD11c were sorted as MyDC (CD11c+ BDCA2-CD123-). The purity of the resulting population was confirmed by additional antibody staining.
Human thymic dendritic cells and macrophages were isolated from thymus fragments removed during corrective cardiac surgery of patients aged 1 month to 4 years. After Lymphoprep centrifugation, thymocyte cell suspensions were depleted of T, B, NK cells and CD34+ precursors by magnetic cell sorting (AutoMacs; Miltenyi Biotech) as described.21 Macrophages were isolated from the depleted cell fraction by using PE-labeled anti-CD14mAb and anti-PE microbeads, and exhibited a CD13+, CD11c+, CD14+ phenotype. Thymic pDC were isolated from the macrophage-depleted fraction with PE-labeled antiCD123 and antiPE microbeads, and showed a CD11c-, CD13-, BDCA2+, CD123+ phenotype. Thymic MyDC (CD13+, CD11cdim, CD14−) were isolated from the CD123-negative fraction with PE-labeled antiCD13 and anti-PE microbeads. Sorted populations proved to be over 97% pure on reanalysis. Phenotypic analysis was carried out by indirect immunofluorescence as described.20
The K562 (chronic myelogenous leukemia) and THP-1 (monocytic leukemia) cell lines were cultured as described20 and THP-1 differentiation induced with phorbol myristate acetate (PMA) at 10 ng/mL.11 HEK293T and COS-7 cells were grown in Dulbecco modified eagle medium (DMEM) with 10% fetal calf serum (FCS) and transfected with Superfect (Qiagen, Hilden, Germany). Transfection of the pCDNA3.1(-)-based constructs in K562 cells was accomplished with Superfect or by nucleofection (Amaxa GmbH, Cologne, Germany). Mutation of residues Y,6 E14E,15 W21GRW24VHW27, and both Y6 and E14E15 were done by site-directed mutagenesis on LSECtin cDNA cloned in pCDNA3.1(-), which resulted in the generation of LSECtin Y/F, LSECtin EE/AA, LSECtin 3W/3A, and LSECtin DM constructs.
Isolation and detection of LSECtin isoform mRNA in distinct cell types
LSECtin isoforms were isolated by reverse transcriptase-polymerase chain reaction (RT-PCR) on RNA from MDDC of a healthy donor. Total cellular RNA was isolated with RNeasy columns (Qiagen). Two micrograms of RNA was reverse-transcribed and amplification was carried out on 5 μL of each cDNA synthesis reaction in 50 μL of solution. After a 5 minute denaturation step, LSEctin was optimally amplified after 35 cycles of denaturation (95°C, 45 seconds), annealing (65°C, 30 seconds), and extension (72°C, 90 seconds), followed by a 10-minute extension step at 72°C. Oligonucleotides used for amplification of the coding region of the prototypical LSECtin isoform (LSECtin full-length) were LSECtin sense 5′-GGGAATTCGCCTGCATCGCCATGGACACC-3′) and LSECtin antisense 5′-CCCAAGCTTGGGCGGGGTCAGCAGTTGTGC -3′). Amplification of LSECtin isoforms was accomplished using the primer pairs LSECtin sense/LSECtin antisense, LSECtin Δ3/4sense/LSECtin antisense, and LSECtin Δ2sense/LSECtin antisense. The oligonucleotide LSECtin Δ3/4sense 5′-CCTATTGTCCAAGGGCTCGGG -′3 spans through the exon 2/exon 5 junction. Oligonucleotide LSECtin Δ2sense 5′-CCGAGGAGGTCCCCGGAGCCT-′3 spans through the exon 1/exon 3 junction. PCR fragments were resolved in 1.2% agarose gels, purified, cloned, and sequenced. For eukaryotic expression, the selected LSECtin cDNA isoforms were excised from TOPO cloning vectors (Invitrogen, Paisley, UK) with EcoRI and HindIII, gel purified, and ligated into EcoRI- and HindIII-digested pCDNA3.1(-). As a control in RT-PCR experiments, GAPDH mRNA was amplified using oligonucleotides 5′-GGCTGAGAACGGGAAGCTTGTCA-3′ and 5′-CGGCCATCACGCCACAGT TTC-3′, which together amplify a 417 bp fragment. Images were captured with GelDoc XR (BioRad, Hercules, CA) using Quantity One-4.5.2 software.
Detection and cell-surface distribution of LSECtin
The cDNA region coding for the extracellular portion of LSECtin (residues 55-293) was generated by polymerase chain reaction with the primers 5′-CACCGCCTCCACGGAGCGCGCGG-3′ and 5′-CCCAAGCTTGGGCGGGGTCAGCA GTTGTGC-3′. The resulting fragment, which contains the neck and lectin domains of LSECtin, was cloned in-frame downstream of the hexahistidine sequence of pET100/D-TOPO (Invitrogen) and sequenced. The resulting vector, as well as a positive control encoding β-galactosidase (pET100/D/lacZ), were transformed into BL21 bacteria, and HIS-LSECtin and HIS-β-gal fusion proteins were purified on Ni2+-nitrilotriacetic acid-agarose (Qiagen). The purified HIS-LSECtin fusion protein was injected intraperitoneally (three times) into Balb/c mice. After a final intraperitoneal boost, splenocytes were removed and fused to SP2 cells at a 1:1 ratio using PEG 1500 (Sigma, Barcelona, Spain), as described.10 Screening for anti-LSECtin antibodies was done by enzyme-linked immunosorbent assay using 96-well plates coated with HIS-LSECtin or HIS-β-gal (negative control) fusion proteins. Final selection of the selected hybridoma (SOTO1) was done by immunofluorescence on HEK293T transiently transfected with an LSECtin-expression vector.
Peptides based on the sequence of the LSECtin neck region (AQAKLMEQESALRELREEVTQGLA) and cytoplasmic tail (MDTTRYSKWGGSSEEVPGGP WGRWVHWSRR) were synthesized using the multiple antigen peptide system. New Zealand white rabbits were immunized by subcutaneous injection of each individual peptide or the HIS-LSECtin protein (0.5 mg of a 1-mg/mL solution in phosphate-buffered saline) in complete Freud's adjuvant 1:1 on day 0 and in incomplete Freud's adjuvant 1:1 on days 21 and 42. Rabbits were bled on day 49, and serum was assayed for LSECtin recognition by Western blot. The resulting antisera (ADS-1 against the neck domain, ADS-3 against the cytoplasmic tail and ADS-4 against the HIS-LSECtin protein) were subsequently validated by Western blot on 10 μg of cell lysates, as described.20 DC-SIGN was detected using a polyclonal antiserum against the neck region of the molecule.22 Isolation of detergent-insoluble and soluble MDDC cell membrane fractions was done by sucrose gradient ultracentrifugation as described.23
Gene-expression profiling in dendritic cells
RNA from immature and LPS-mature MDDC from two independent donors, and monocyte-derived macrophages from three independent donors generated in the presence of either granulocyte macrophage-colony stimulating factor (GM-CSF) or macrophage-colony stimulating factor (M-CSF), were labeled, processed, and independently hybridized on Codelink human whole genome DNA chip of the Codelink microarray platform (Amersham Biosciences, Uppsala, Sweden) containing 55 000 human gene targets. Scanned images were processed using the Codelink Expression Analysis Software. Raw data were normalized by the quantile method. Data corresponding to the experimental groups were analyzed by the Student t test. The raw P values obtained were adjusted using the control of the false discovery rate-based procedure and implemented in the multitest package within the Bioconductor set of routines (www.bioconductor.org).
Sugar-binding assays
For precipitation of Mannan-, N-acetyl-glucosamine-, and N-acetyl-galactosamine-binding proteins, K562 cells stably transfected with different LSECtin isoforms (3 × 106) were collected, washed with phosphate-buffered saline and 1 mM EDTA, and lysed. Then, each lysate was incubated with 50 μL of Mannan-agarose (Sigma), N-acetyl-glucosamine-agarose (Sigma), or N-acetyl-galactosamine-agarose (Sigma) for 12 hours at 4°C. After extensive washing, bound proteins were eluted either by incubation with an excess of the sugars or boiling the agarose beads in 3× Laemmli's sample buffer. SDS-eluted material was resolved by SDS-PAGE and LSECtin detection accomplished with specific polyclonal antibodies.
Ebola glycoprotein (GP)1 and virus-binding assays
Ebola virus binding was determined using lentiviral particles pseudotyped with Ebola virus GP according to a transient transfection protocol previously described.24 K562 transfectants were challenged with Ebola virus GP pseudotypes or controls: 25 × 104 cells were resuspended in 250 μL complete medium and incubated overnight with 250 μL of transfection supernatants. Cells were assayed for luciferase expression 48 hours postinfection. For inhibition experiments, cells were preincubated for 10 minutes at room temperature with the preimmune or ADS-1 polyclonal antibodies.
Ebola virus GP1 (envelope glycoprotein of the Zaire strain of Ebola virus, provided by Dr. Anthony Sanchez, CDC, Atlanta, GA) was fused with human IgG-Fc by subcloning into pEF-Fc (provided by Dr. J. M. Casasnovas, CNB, CSIC, Madrid, Spain). The pSyngp120IgG plasmid, which encodes strain JR-FL HIV gp120 fused to human IgG1Fc, was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Germantown, MD). For binding experiments, envelope constructions were transfected into 293T cells and supernatants collected after 72 hours. MDDC were incubated for 30 minutes at room temperature with 1:10 dilution of the supernatants in phosphate-buffered saline, 2% FCS, 4 mM CaCl2. Cells were then washed twice and incubated with a PE-conjugated goat antihuman IgG-Fc antibody (Immunotech, Marseille, France). Cells were analyzed in an EPICS-XL cytometer using the Expo32 software.
LSECtin internalization assays
K562 cells were transiently transfected with LSECtin by Nucleofection (Amaxa). After 24 hours, cells were washed, resuspended in complete medium, and incubated with either SOTO1 (for LSECtin), P5D2 (for CD29), or TS1/18 (for CD18) antibodies for 30 minutes at 4°C to prevent internalization. After extensive washing, cells were placed at 37°C to allow internalization to occur, and aliquots were removed after 2, 5, and 15 minutes and immediately placed at 4°C. Then, cells were incubated with a 1:100 dilution of a FITC-labeled goat anti-mouse antibody (Serotec) to detect cell surface-bound antibodies. All incubations were done in the presence of 50 μg/mL of human IgG to prevent binding through the Fc portion of the antibodies. In some cases, and to detect internalized antibodies, cells were fixed and permeabilized (CytoFIX/CytoPerm; BD Biosciences) before addition of the secondary antibody. Flow cytometry analysis was performed with an EPICS-CS (Coulter Científica, Madrid, Spain) using log amplifiers.
Results
LSECtin mRNA is found in monocyte-derived dendritic cells and is induced by IL-4
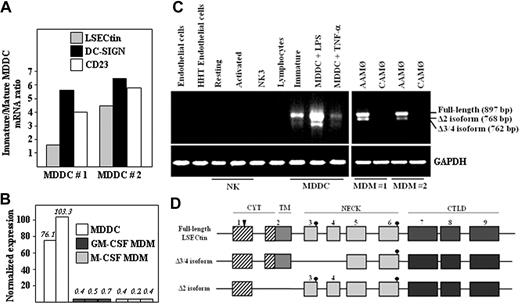
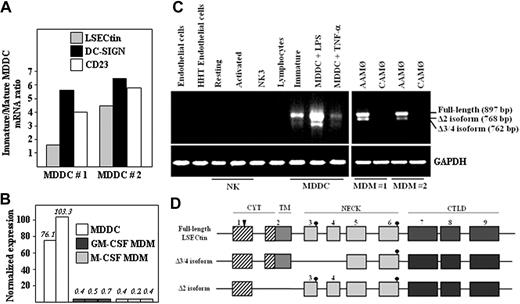
The LSECtin gene lies within the C-type lectin-encoding DC-SIGN/L-SIGN/CD23 gene cluster on chromosome 19p13.2.1 Although reported to be exclusively expressed on liver and lymph node sinusoidal endothelial cells,9 its chromosomal location prompted us to determine whether LSECtin was found in myeloid cells. Microarray gene profiling experiments showed that LSECtin mRNA is expressed in immature and mature human MDDC (Figure 1A). MDDC contained significantly higher levels of LSECtin RNA than macrophages generated either in the presence of GM-CSF or M-CSF (Figure 1B). This set of data was verified by RT-PCR, because LSECtin mRNA was readily detected in MDDC but was absent from peripheral blood lymphocytes, resting or activated NK cells, or CD34+-derived endothelial-like cells (Figure 1C).
Expression of LSECtin mRNA in human MDDC. (A) Relative levels of LSECtin, DC-SIGN, and CD23 mRNA in immature and mature MDDC as determined by gene expression profiling using Codelink Whole Genome Bioarrays. (B) Relative levels of LSECtin mRNA in immature MDDC from two independent donors and monocyte-derived macrophages generated in the presence of either GM-CSF or M-CSF from three independent donors, as determined by gene expression profiling using Codelink Whole Genome Bioarrays. Values represent the intensity of expression normalized with the median of all the intensity values in the microarray. (C) Detection of LSECtin mRNA in hematopoietic cells, cell lines, and in vitro–generated monocyte-derived macrophages and dendritic cells. Total RNA was isolated from the indicated cells and cell lines and subjected to RT-PCR for amplification of the LSECtin coding region and GAPDH (as control). Analyzed RNA was obtained from endothelial cells from normal and HHT donors, resting and activated NK cells, the NK3 NK cell clone, peripheral blood T lymphocytes, MDM (interferon-γ-activated, CAMØ; IL-4-activated, AAMØ), immature or mature (with either lipopolysaccharide or tumor necrosis factor-α) MDDC, and the THP-1 and K562 leukemic cell lines. (D) Schematic representation of the structure of LSECtin mRNA species amplified by RT-PCR from monocyte-derived dendritic cells. Boxes represent the individual exons, and the arrowhead marks the position of the initiation methionine. Dark circles indicate the potential N-glycosylation sites.
Expression of LSECtin mRNA in human MDDC. (A) Relative levels of LSECtin, DC-SIGN, and CD23 mRNA in immature and mature MDDC as determined by gene expression profiling using Codelink Whole Genome Bioarrays. (B) Relative levels of LSECtin mRNA in immature MDDC from two independent donors and monocyte-derived macrophages generated in the presence of either GM-CSF or M-CSF from three independent donors, as determined by gene expression profiling using Codelink Whole Genome Bioarrays. Values represent the intensity of expression normalized with the median of all the intensity values in the microarray. (C) Detection of LSECtin mRNA in hematopoietic cells, cell lines, and in vitro–generated monocyte-derived macrophages and dendritic cells. Total RNA was isolated from the indicated cells and cell lines and subjected to RT-PCR for amplification of the LSECtin coding region and GAPDH (as control). Analyzed RNA was obtained from endothelial cells from normal and HHT donors, resting and activated NK cells, the NK3 NK cell clone, peripheral blood T lymphocytes, MDM (interferon-γ-activated, CAMØ; IL-4-activated, AAMØ), immature or mature (with either lipopolysaccharide or tumor necrosis factor-α) MDDC, and the THP-1 and K562 leukemic cell lines. (D) Schematic representation of the structure of LSECtin mRNA species amplified by RT-PCR from monocyte-derived dendritic cells. Boxes represent the individual exons, and the arrowhead marks the position of the initiation methionine. Dark circles indicate the potential N-glycosylation sites.
LSECtin mRNA was seen in AAM∅ but absent in either naive or CAM∅ (Figure 1C). Sequencing of the two RT-PCR fragments routinely generated from MDDC and AAM∅ revealed the existence of three LSECtin mRNA species, which encode the prototypic isoform (full-length, 879 bp), an isoform lacking the transmembrane region (Δ2, 768 bp), and an isoform lacking the first two exons encoding the stalk region (Δ3/4, 762 bp) (Figure 1D). The full-length isoform was amplified from more than 20 unrelated MDDC donors, whereas the shorter isoforms were variably detected (data not shown). Therefore, LSECtin mRNA is differentially expressed in monocyte-derived macrophages and dendritic cells, with the latter containing three distinct mRNA species. Moreover, LSECtin expression can be detected in macrophages activated in the presence of IL-4 (AAM∅).
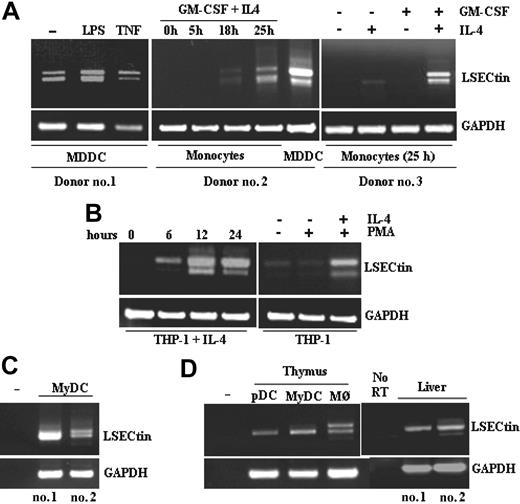
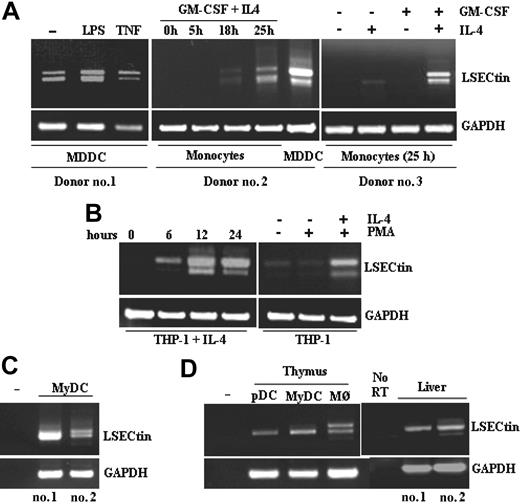
The presence of LSECtin mRNA in AAM∅, and the proximity of the LSECtin gene to that of DC-SIGN, whose expression is IL-4-dependent,4,10 led us to explore the cytokine responsiveness of the LSECtin gene. LSECtin mRNA was induced during GM-CSF + IL-4-promoted MDDC differentiation and was found to be responsive to the presence of IL-4 (Figure 2A). Besides, and in agreement with the microarray data, LSECtin mRNA was detected in MDDC matured with either lipopolysaccharide or tumor necrosis factor-α (Figure 2A). Similarly, IL-4 induced the appearance of LSECtin mRNA on proliferating or PMA-differentiated THP-1 cells, where the three LSECtin alternatively spliced isoforms were detected (Figure 2B). Because the combined addition of PMA and IL-4 promotes THP-1 cells to acquire a dendritic cell-like phenotype,11 these results are compatible with the presence of LSECtin in MDDC. More importantly, LSECtin mRNA was detected in ex vivo isolated peripheral blood myeloid dendritic cells (CD11c+ BDCA2-CD123-) (Figure 2C) as well as in myeloid and plasmacytoid thymic dendritic cells and thymic macrophages (Figure 2D). Sequencing of the amplified fragments confirmed the presence of the full-length and Δ2 isoforms in both thymic macrophages and myeloid dendritic cells, which were also seen in human liver RNA as control (Figure 2D). Altogether, these results indicate that LSECtin RNA is induced by IL-4 and is expressed by dendritic cells in vivo.
Cytokine-dependence of LSECtin mRNA levels in MDDC and detection of LSECtin mRNA in ex vivo peripheral blood and thymic dendritic cells. (A) Monocytes from three independent donors were treated for the indicated periods of time with either GM-CSF, IL-4, or both cytokines, or allowed to differentiate into MDDC and further matured with lipopolysaccharide or tumor necrosis factor-α. After RNA extraction from the distinct cell types, the coding region of the LSECtin mRNA was amplified and the resulting fragments resolved by agarose gel electrophoresis. (B) THP-1 leukemic myeloid cells were treated for the indicated periods of time with IL-4 alone (left panel) or for 96 hours in the presence of IL-4 and the differentiation-inducing agent PMA (right panel). After RNA extraction from the distinct cell types, the coding region of the LSECtin mRNA was amplified and the resulting fragments resolved by agarose gel electrophoresis. (C) RNA was extracted from human myeloid peripheral blood dendritic cells from three independent donors and LSECtin mRNA detected by RT-PCR. (D) RNA was extracted from human myeloid and plasmacytoid thymic dendritic cells, thymic macrophages, or liver biopsies from three independent donors and LSECtin mRNA detected by RT-PCR. In all cases, GAPDH mRNA was amplified as a control.
Cytokine-dependence of LSECtin mRNA levels in MDDC and detection of LSECtin mRNA in ex vivo peripheral blood and thymic dendritic cells. (A) Monocytes from three independent donors were treated for the indicated periods of time with either GM-CSF, IL-4, or both cytokines, or allowed to differentiate into MDDC and further matured with lipopolysaccharide or tumor necrosis factor-α. After RNA extraction from the distinct cell types, the coding region of the LSECtin mRNA was amplified and the resulting fragments resolved by agarose gel electrophoresis. (B) THP-1 leukemic myeloid cells were treated for the indicated periods of time with IL-4 alone (left panel) or for 96 hours in the presence of IL-4 and the differentiation-inducing agent PMA (right panel). After RNA extraction from the distinct cell types, the coding region of the LSECtin mRNA was amplified and the resulting fragments resolved by agarose gel electrophoresis. (C) RNA was extracted from human myeloid peripheral blood dendritic cells from three independent donors and LSECtin mRNA detected by RT-PCR. (D) RNA was extracted from human myeloid and plasmacytoid thymic dendritic cells, thymic macrophages, or liver biopsies from three independent donors and LSECtin mRNA detected by RT-PCR. In all cases, GAPDH mRNA was amplified as a control.
LSECtin protein expression in dendritic cells and macrophages
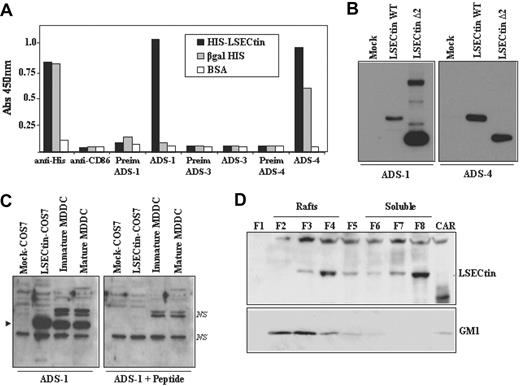
To confirm these results at the protein level, LSECtin-specific polyclonal antisera were raised and their specificity tested by enzyme-linked immunosorbent assay on purified recombinant HIS-LSECtin. Whereas an anti-HIS serum detected HIS-LSECtin and HIS-β-gal to a similar extent, ADS-1 (stalk region-specific) and ADS-4 antiserum (raised against the extracellular region of the molecule) exclusively reacted with HIS-LSECtin (Figure 3A). By contrast, and as expected, ADS-3 antiserum (specific for the LSECtin cytoplasmic tail) showed no reactivity against the HIS-LSECtin recombinant protein (Figure 3A). The LSECtin specificity of the ADS-1 and ADS-4 antisera was further demonstrated by Western blot after transient transfection of the full-length and the Δ2 isoforms in COS7 cells (Figure 3B).
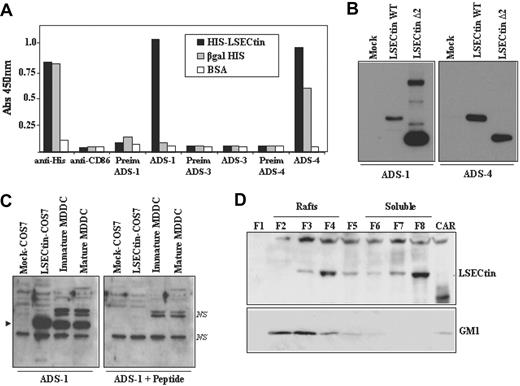
Expression of LSECtin in human MDDC. (A) Specificity of polyclonal antisera against LSECtin by enzyme-linked immunosorbent assay. High protein-binding 96-well plates were coated with either purified HIS-LSECtin, purified βgal-HIS, or BSA. After washing, wells were treated with either a monoclonal antibody against the HIS epitope (anti-His), a control monoclonal antibody (antiCD86), polyclonal antisera against LSECtin (ADS-1, ADS-3, ADS-4), or the corresponding preimmune sera. Bound antibodies were detected using HRP-conjugated goat antirabbit and goat antimouse polyclonal antisera. Quantification was done on a microplate enzyme-linked immunosorbent assay reader at 450 nm. (B) Specificity of polyclonal antisera against LSECtin by Western blot. Whole cell extracts were obtained from COS-7 cells transiently transfected with full-length LSECtin, the Δ2 LSECtin isoform (LSECtin Δ2), or mock-transfected cells (Mock). Ten micrograms of each whole cell extract was subjected to Western blot using the ADS-1 or ADS-4 polyclonal antisera specific for LSECtin. (C) Whole cell lysates were obtained from immature and mature MDDC, and 10 μg of each extract was subjected to Western blot using the ADS-1 anti-LSECtin polyclonal antiserum either alone (left panel) or in the presence of an excess of the immunizing peptide (right panel). Lysates from mock- and LSECtin-transfected COS-7 cells were included as controls. The position of the LSECtin protein is indicated by an arrowhead, and nonspecific bands are denoted as NS. (D) Immature MDDC were lysed in 1% Brij 98 lysis buffer at 37°C and fractionated by sucrose density gradient centrifugation. The low-density Brij 98-insoluble fractions 2-4 (lanes 2-4, Rafts) and the high-density Brij 98-soluble fractions 6-8 (lanes 6-8, soluble) were separated by 12.5% SDS-PAGE under reducing conditions, and the distribution of LSECtin was determined by immunoblotting. Cytoskeletal-associated Rafts (CAR), obtained by solubilization of the cell pellet with Brij98 + Octyl D-glucoside in lysis buffer, were analyzed in parallel (lane 9, CAR). Ten micrograms of each fraction was subjected to Western blot, and the distribution of LSECtin and ganglioside GM1 in the distinct fractions was determined by immunoblotting with the ADS-1 polyclonal antisera (LSECtin) or cholera toxin-HRP (GM1). The experiment was done on MDDC from two independent donors, and one of them is shown.
Expression of LSECtin in human MDDC. (A) Specificity of polyclonal antisera against LSECtin by enzyme-linked immunosorbent assay. High protein-binding 96-well plates were coated with either purified HIS-LSECtin, purified βgal-HIS, or BSA. After washing, wells were treated with either a monoclonal antibody against the HIS epitope (anti-His), a control monoclonal antibody (antiCD86), polyclonal antisera against LSECtin (ADS-1, ADS-3, ADS-4), or the corresponding preimmune sera. Bound antibodies were detected using HRP-conjugated goat antirabbit and goat antimouse polyclonal antisera. Quantification was done on a microplate enzyme-linked immunosorbent assay reader at 450 nm. (B) Specificity of polyclonal antisera against LSECtin by Western blot. Whole cell extracts were obtained from COS-7 cells transiently transfected with full-length LSECtin, the Δ2 LSECtin isoform (LSECtin Δ2), or mock-transfected cells (Mock). Ten micrograms of each whole cell extract was subjected to Western blot using the ADS-1 or ADS-4 polyclonal antisera specific for LSECtin. (C) Whole cell lysates were obtained from immature and mature MDDC, and 10 μg of each extract was subjected to Western blot using the ADS-1 anti-LSECtin polyclonal antiserum either alone (left panel) or in the presence of an excess of the immunizing peptide (right panel). Lysates from mock- and LSECtin-transfected COS-7 cells were included as controls. The position of the LSECtin protein is indicated by an arrowhead, and nonspecific bands are denoted as NS. (D) Immature MDDC were lysed in 1% Brij 98 lysis buffer at 37°C and fractionated by sucrose density gradient centrifugation. The low-density Brij 98-insoluble fractions 2-4 (lanes 2-4, Rafts) and the high-density Brij 98-soluble fractions 6-8 (lanes 6-8, soluble) were separated by 12.5% SDS-PAGE under reducing conditions, and the distribution of LSECtin was determined by immunoblotting. Cytoskeletal-associated Rafts (CAR), obtained by solubilization of the cell pellet with Brij98 + Octyl D-glucoside in lysis buffer, were analyzed in parallel (lane 9, CAR). Ten micrograms of each fraction was subjected to Western blot, and the distribution of LSECtin and ganglioside GM1 in the distinct fractions was determined by immunoblotting with the ADS-1 polyclonal antisera (LSECtin) or cholera toxin-HRP (GM1). The experiment was done on MDDC from two independent donors, and one of them is shown.
The availability of the anti-LSECtin polyclonal antisera allowed us to address the presence of LSECtin protein in dendritic cells. Analysis of immature and mature MDDC demonstrated the presence of an ADS1-reactive protein band that comigrates with the LSECtin protein generated in COS-7 cells after transient transfection (Figure 3C). The specificity of the recognition was further demonstrated by the fact that the detected band was not observed in the presence of an excess of the immunizing peptide (Figure 3C, right panel). Moreover, and like DC-SIGN,23 a significant percentage of LSECtin molecules were detected in lipid raft-enriched membrane fractions (Figure 3D), suggesting that LSECtin might also act as a signaling molecule. Therefore, LSECtin is expressed in monocyte-derived dendritic cells, where it might contribute to increase their pathogen-recognition capability.
Detection of LSECtin protein during MDDC differentiation and macrophage activation confirmed the mRNA data. The IL-4-dependent in vitro generation of AAMØ resulted in up-regulation of LSECtin, which was not expressed by CAMØ (Figure 4A). On the other hand, LSECtin was barely expressed in monocytes, increased after 48 hours in the presence of GM-CSF and IL-4, and reached maximal levels in immature MDDC (5-day treatment with GM-CSF + IL-4; Figure 4A). Analysis of MDDC from additional donors confirmed the presence of LSECtin protein in all cell extracts (data not shown). The presence of LSECtin was further evaluated by flow cytometry (Figure 4B) and confocal microscopy (Figure 4C), which revealed the presence of variable levels of LSECtin protein on the cell surface of monocyte-derived dendritic cells. Furthermore, flow cytometry revealed high levels of LSECtin on the cell surface of CD13+ CD14+ thymic macrophages, whereas weak levels were found on both thymic myeloid (CD13+ CD14−) and plasmacytoid (BDCA2+ CD123+) dendritic cells (Figure 4D). Therefore, LSECtin is expressed on the cell surface of in vitro differentiated (MDDC) and ex vivo isolated (thymic macrophages) myeloid cells.
LSECtin expression on the cell surface of monocyte-derived dendritic cells and thymic-cell subsets. (A) Cell extracts were obtained from monocytes along the MDDC differentiation pathway by treating them with GM-CSF plus IL-4 for the indicated periods of time. In parallel, extracts were obtained from GM-CSF-generated macrophages after a 48-hour treatment with medium (MDM), IL-4 (AAMØ), or interferon-γ (CAMØ). In all cases, 10 μg of each lysate was subjected to Western blot for detection of LSECtin with the ADS-1 polyclonal antiserum and β-actin expression determined in parallel for loading control purpose. (B) Cell surface expression levels of LSECtin in monocyte-derived dendritic cells from three independent donors as determined with the antiLSECtin SOTO1 monoclonal antibody. The supernatant from the P3 × 63 myeloma was used as negative control (X63). (C) MDDC were allowed to differentiate on glass coverslips, fixed (paraformaldehyde 2%), washed, and incubated with either a polyclonal antiserum against LSECtin (ADS-1) or preimmune serum. After washing, cells were incubated with a 1:500 dilution of Alexa 488-labeled goat anti-rabbit IgG (F(ab′)2) antiserum. Nuclei were counterstained with 4′,6-diamidino-2-fenilindol diclorhidrato (DAPI). Coverslips were mounted in Dako Cytomation fluorescent mounting medium (Dako, Glostrup, Denmark) and representative fields photographed through an HCX PL APO lens (63.0 × 1.40 oil objective), with a 1.400000 numerical aperture on a TCS-SP2-ADBS confocal laser LEICA scanning system (LEICA MICROSYSTEMS GmbH, Wetzlar, Germany). (D) Expression of LSECtin in human thymic cell populations. Human thymic cell preparations were depleted of thymocytes by centrifugation on Ficoll after rosetting with sheep erythrocytes. The resulting population was then stained with either PE-labeled anti-CD123 and FITC-labeled BDCA2 or with FITC-labeled anti-CD14 and PE-labeled anti-CD13 antibodies. The BDCA2+ CD123+ (plasmacytoid DC [pDC]), CD14+ CD13+ (macrophages [MØ]), and CD14−CD13+ (myeloid DC [MyDC]) subsets (left panels) were simultaneously stained with the anti-LSECtin SOTO1 monoclonal antibody followed by incubation with an APC-labeled goat anti-mouse antiserum (right panels).
LSECtin expression on the cell surface of monocyte-derived dendritic cells and thymic-cell subsets. (A) Cell extracts were obtained from monocytes along the MDDC differentiation pathway by treating them with GM-CSF plus IL-4 for the indicated periods of time. In parallel, extracts were obtained from GM-CSF-generated macrophages after a 48-hour treatment with medium (MDM), IL-4 (AAMØ), or interferon-γ (CAMØ). In all cases, 10 μg of each lysate was subjected to Western blot for detection of LSECtin with the ADS-1 polyclonal antiserum and β-actin expression determined in parallel for loading control purpose. (B) Cell surface expression levels of LSECtin in monocyte-derived dendritic cells from three independent donors as determined with the antiLSECtin SOTO1 monoclonal antibody. The supernatant from the P3 × 63 myeloma was used as negative control (X63). (C) MDDC were allowed to differentiate on glass coverslips, fixed (paraformaldehyde 2%), washed, and incubated with either a polyclonal antiserum against LSECtin (ADS-1) or preimmune serum. After washing, cells were incubated with a 1:500 dilution of Alexa 488-labeled goat anti-rabbit IgG (F(ab′)2) antiserum. Nuclei were counterstained with 4′,6-diamidino-2-fenilindol diclorhidrato (DAPI). Coverslips were mounted in Dako Cytomation fluorescent mounting medium (Dako, Glostrup, Denmark) and representative fields photographed through an HCX PL APO lens (63.0 × 1.40 oil objective), with a 1.400000 numerical aperture on a TCS-SP2-ADBS confocal laser LEICA scanning system (LEICA MICROSYSTEMS GmbH, Wetzlar, Germany). (D) Expression of LSECtin in human thymic cell populations. Human thymic cell preparations were depleted of thymocytes by centrifugation on Ficoll after rosetting with sheep erythrocytes. The resulting population was then stained with either PE-labeled anti-CD123 and FITC-labeled BDCA2 or with FITC-labeled anti-CD14 and PE-labeled anti-CD13 antibodies. The BDCA2+ CD123+ (plasmacytoid DC [pDC]), CD14+ CD13+ (macrophages [MØ]), and CD14−CD13+ (myeloid DC [MyDC]) subsets (left panels) were simultaneously stained with the anti-LSECtin SOTO1 monoclonal antibody followed by incubation with an APC-labeled goat anti-mouse antiserum (right panels).
Functional characterization of LSECtin
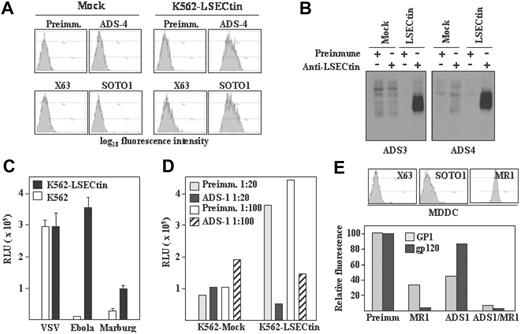
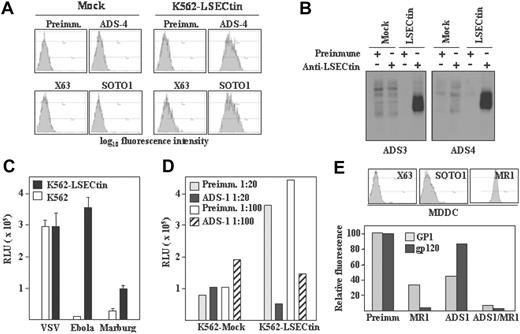
To evaluate the recognition capabilities of LSECtin, stable transfectants were generated in K562 cells (K562-LSECtin), where the lectin was readily detectable on the cell surface either by flow cytometry (Figure 5A) or by immunoprecipitation of biotin-labeled cell surface proteins (Figure 5B) with polyclonal antisera (ADS-3, ADS-4) or the anti-LSECtin SOTO1 monoclonal antibody. Whereas K562-LSECtin and mock-transfected cells exhibited similar binding of VSV, expression of full-length LSECtin conferred K562 cells the ability to bind viral particles pseudotyped with Ebola or Marburg virus GP (Figure 5C). The viral binding to full-length LSECtin was almost completely abrogated in the presence of the ADS-1 polyclonal antiserum (Figure 5D), confirming the specificity of the interaction. In agreement with these results, K562-LSECtin cells specifically bound soluble Ebola virus glycoprotein GP1 (see subsequently). LSECtin also displayed Ebola GP1-binding ability when expressed on in vitro generated dendritic cells. Although HIV gp120 binding to MDDC was exclusively inhibited by anti-DC-SIGN antibodies, Ebola GP1 binding to MDDC was partially dependent on LSECtin (Figure 5E). In fact, complete abrogation of GP1 binding to MDDC was only observed in the presence of anti-LSECtin antiserum (ADS-1) and anti-DC-SIGN antibodies (MR1) (Figure 5E). Therefore, LSECtin displays pathogen-recognition capability when expressed on hematopoietic cells.
Pathogen-recognition ability of LSECtin. (A) Generation of LSECtin stable transfectants. K562 cells were transfected with an empty vector (Mock) or with an LSECtin expression vector (LSECtin) and grown in G418-containing selective medium. LSECtin expression was determined by indirect immunofluorescence with a polyclonal antiserum against LSECtin (ADS-4) or the SOTO1 anti-LSECtin monoclonal antibody. Preimmune polyclonal antiserum (Preimm.) and the supernatant from the P3 × 63 myeloma (X63) were used as controls. (B) Mock-transfected K562 cells (Mock) or K562-LSECtin cells stably transfected with full-length LSECtin (LSECtin) were surface-labeled with a water-soluble and membrane-impermeable biotin derivative (EZ-linked Sulfo-NHS-LC-Biotin; Pierce, Rockford, IL), washed, lysed, and immunoprecipitated with polyclonal antisera against LSECtin (left panel, ADS-3; right panel, ADS-4) or preimmune serum. Precipitated material was resolved by SDS-PAGE under reducing conditions and subjected to Western blot with HRP-streptavidin. (C) Binding of Ebola and Marburg pseudovirus to LSECtin-transfected cells. Mock-transfected and K562-LSECtin transfectants were challenged with vesicular stomatitis virus (VSV), Marburg or Ebola virus GP pseudotypes, and cells were assayed for luciferase expression 48 hours postinfection. (D) Inhibitory effect of the ADS-1 anti-LSECtin polyclonal antiserum on the Ebola pseudovirus binding to LSECtin. The experiment was performed like in D, but cells were preincubated for 10 minutes at room temperature with the preinmune or ADS-1 polyclonal antibodies before viral addition. (E) Binding of Ebola virus GP1 and HIV gp120 to MDDC. Cells were incubated with the recombinant proteins either in the absence or in the presence of antibodies against DC-SIGN (MR1), LSECtin (ADS-1), or both and binding measured by flow cytometry. Protein binding is measured as relative fluorescence, which indicates the binding observed in each experimental condition relative to the binding detected in the presence of preimmune antiserum (considered as 100). The level of expression of LSECtin and DC-SIGN in the assayed MDDC is illustrated in the upper panel.
Pathogen-recognition ability of LSECtin. (A) Generation of LSECtin stable transfectants. K562 cells were transfected with an empty vector (Mock) or with an LSECtin expression vector (LSECtin) and grown in G418-containing selective medium. LSECtin expression was determined by indirect immunofluorescence with a polyclonal antiserum against LSECtin (ADS-4) or the SOTO1 anti-LSECtin monoclonal antibody. Preimmune polyclonal antiserum (Preimm.) and the supernatant from the P3 × 63 myeloma (X63) were used as controls. (B) Mock-transfected K562 cells (Mock) or K562-LSECtin cells stably transfected with full-length LSECtin (LSECtin) were surface-labeled with a water-soluble and membrane-impermeable biotin derivative (EZ-linked Sulfo-NHS-LC-Biotin; Pierce, Rockford, IL), washed, lysed, and immunoprecipitated with polyclonal antisera against LSECtin (left panel, ADS-3; right panel, ADS-4) or preimmune serum. Precipitated material was resolved by SDS-PAGE under reducing conditions and subjected to Western blot with HRP-streptavidin. (C) Binding of Ebola and Marburg pseudovirus to LSECtin-transfected cells. Mock-transfected and K562-LSECtin transfectants were challenged with vesicular stomatitis virus (VSV), Marburg or Ebola virus GP pseudotypes, and cells were assayed for luciferase expression 48 hours postinfection. (D) Inhibitory effect of the ADS-1 anti-LSECtin polyclonal antiserum on the Ebola pseudovirus binding to LSECtin. The experiment was performed like in D, but cells were preincubated for 10 minutes at room temperature with the preinmune or ADS-1 polyclonal antibodies before viral addition. (E) Binding of Ebola virus GP1 and HIV gp120 to MDDC. Cells were incubated with the recombinant proteins either in the absence or in the presence of antibodies against DC-SIGN (MR1), LSECtin (ADS-1), or both and binding measured by flow cytometry. Protein binding is measured as relative fluorescence, which indicates the binding observed in each experimental condition relative to the binding detected in the presence of preimmune antiserum (considered as 100). The level of expression of LSECtin and DC-SIGN in the assayed MDDC is illustrated in the upper panel.
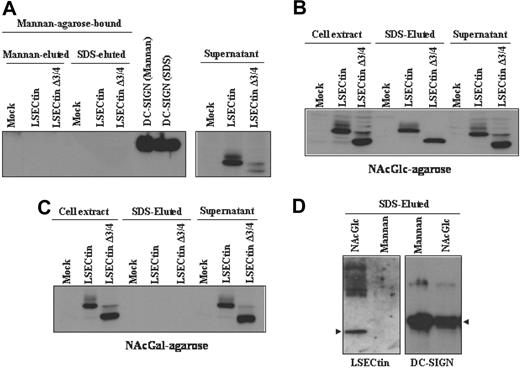
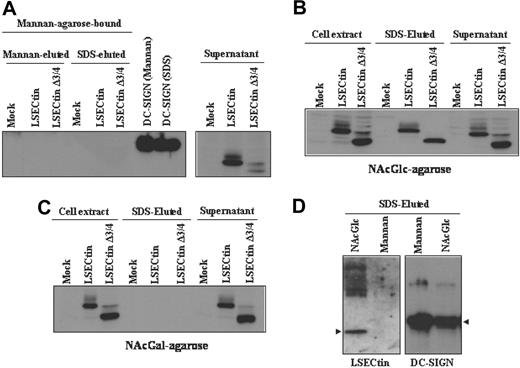
To compare the sugar-specificity of LSECtin with that of DC-SIGN, which acts as an Ebola virus attachment factor,25 we performed sugar affinity experiments by incubation of extracts from DC-SIGN- and LSECtin-transfected K562 cells with Mannan-, N-acetyl-glucosamine-, and N-acetyl-galactosamine-agarose. Both the full-length and the Δ3/4 isoform were not retained by Mannan-agarose, whereas DC-SIGN bound to this matrix (Figure 6A). However, both LSECtin isoforms were specifically retained by N-acetyl-glucosamine-agarose (Figure 6B) but not by N-acetyl-galactosamine-agarose (Figure 6C). LSECtin from MDDC exhibited the same sugar specificity, because it was retained by N-acetyl-glucosamine-agarose but not by Mannan-agarose (Figure 6D). Therefore, and in agreement with the results of pathogen-binding experiments, LSECtin exhibits a sugar-binding specificity that differs from that of the structurally related DC-SIGN lectin on both K562 transfectants and in vitro generated dendritic cells.
Binding specificity of LSECtin in dendritic cells and transfectants. Whole cell lysates from K562 cells stably transfected with full-length LSECtin(LSECtin), the Δ3/4 isoform (LSECtin Δ3/4), DC-SIGN, or mock-transfected (Mock) were loaded onto Mannan- (A), N-acetyl-glucosamine- (NAcGlc, B), or N-acetyl-galactosamine (NAcGal)-agarose (C) and retained proteins eluted with SDS sample buffer (SDS-eluted) and analyzed by Western blot using the ADS-1 anti-LSECtin polyclonal antiserum. As a control, aliquots from nonretained material (supernatant) were analyzed in parallel. (D) Whole cell lysates from monocyte-derived dendritic cells were loaded onto Mannan- or N-acetyl-glucosamine- (NAcGlc)-agarose and retained proteins eluted with SDS sample buffer (SDS-eluted) and analyzed by Western blot using polyclonal antisera against LSECtin (ADS-1) or DC-SIGN (DSG1).
Binding specificity of LSECtin in dendritic cells and transfectants. Whole cell lysates from K562 cells stably transfected with full-length LSECtin(LSECtin), the Δ3/4 isoform (LSECtin Δ3/4), DC-SIGN, or mock-transfected (Mock) were loaded onto Mannan- (A), N-acetyl-glucosamine- (NAcGlc, B), or N-acetyl-galactosamine (NAcGal)-agarose (C) and retained proteins eluted with SDS sample buffer (SDS-eluted) and analyzed by Western blot using the ADS-1 anti-LSECtin polyclonal antiserum. As a control, aliquots from nonretained material (supernatant) were analyzed in parallel. (D) Whole cell lysates from monocyte-derived dendritic cells were loaded onto Mannan- or N-acetyl-glucosamine- (NAcGlc)-agarose and retained proteins eluted with SDS sample buffer (SDS-eluted) and analyzed by Western blot using polyclonal antisera against LSECtin (ADS-1) or DC-SIGN (DSG1).
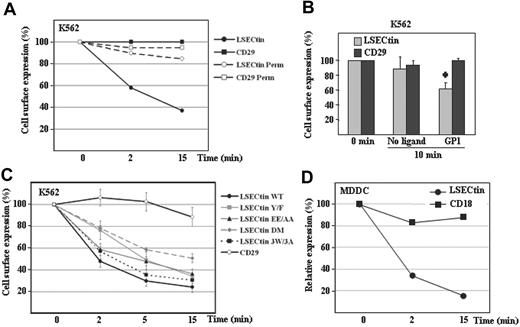
Finally, we evaluated the ability of LSECtin to act as an antigen-capturing receptor by performing internalization experiments with LSECtin-specific ligands. Engagement of LSECtin by SOTO1 led to a rapid loss of the molecule from the cell surface (50% reduction after 2 minutes, 80% after 15 minutes) but did not affect the cell surface levels of CD29 (Figure 7A). To find out whether antibody engagement triggered LSECtin internalization or shedding, a similar experiment was performed using permeabilized cells. No reduction in LSECtin levels was observed when cells were permeabilized before the addition of the secondary fluorescent antibody (Figure 7A), although to a lower extent, the addition of GP1 onto K562-LSECtin cells also resulted in down-regulation of LSECtin without affecting the expression of the CD29 integrin subunit (Figure 7B). Altogether, these results indicate that LSECtin is internalized on ligation on the cell surface and might thus participate in antigen binding and uptake at the early stages of an immune response.
Ligand-induced internalization of LSECtin in transfectants and monocyte-derived dendritic cells. (A) Monoclonal antibody-induced internalization of LSECtin in K562 transfectants. K562-LSECtin cells were incubated with either the SOTO1 (antiLSECtin) or the P5D2 (antiCD29) antibodies for 30 minutes at 4°C. After extensive washing in cold phosphate-buffered saline, cells were transferred to 37°C for the indicated time points, fixed, and the presence of cell surface-bound antibodies detected with FITC-labeled F(ab′)2 anti-mouse IgG. Where indicated, cells were fixed and permeabilized before addition of the fluorescent secondary antibody (Perm). Values represent the fluorescence intensity of the cells at the distinct time points relative to the fluorescence intensity of control cells maintained at 4°C. One representative experiment of two is shown. (B) Ebola GP1-induced internalization of LSECtin. K562-LSECtin cells were either untreated or incubated with Ebola virus GP1-Fc (GP1) for 10 minutes at 37°C, transferred to 4°C, and the expression of LSECtin or the CD29 integrin (negative control) determined by indirect immunofluorescence with SOTO1 or P5D2 monoclonal antibodies and FITC-labeled F(ab′)2 anti-mouse IgG. Values represent the fluorescence intensity of the cells at the distinct time points relative to the fluorescence intensity of control cells maintained at 4°C. Mean ± standard deviation of three independent experiments is shown (*P = 0.001). (C) Cytoplasmic motifs involved in LSECtin ligand-induced internalization. K562 cells were transiently transfected with expression vectors for WT LSECtin or LSECtin mutated at Y6 (LSECtin Y/F), E14E15 (LSECtin EE/AA), W21GRW24VHW27 (LSECtin 3W/3A), or at both Y6 and E14E15 (LSECtin DM). Twenty-four hours later, cells were washed and incubated with the SOTO1 (anti-LSECtin) or the P5D2 (anti-CD29) antibodies for 30 minutes at 4°C. After extensive washing in cold phosphate-buffered saline, cells were transferred to 37°C for the indicated time points (2, 5, 15 minutes), placed on ice, and the presence of cell surface-bound antibodies detected with FITC-labeled F(ab′)2 anti-mouse IgG. Values represent the fluorescence intensity of the cells at the distinct time points relative to the fluorescence intensity of control cells maintained at 4°C. Shown are the mean and standard deviation of three independent experiments. (D) Monoclonal antibody-induced internalization of LSECtin in MDDC. MDDC were incubated with either the SOTO1 (anti-LSECtin) or the TS1/18 (anti-CD18) antibodies for 30 minutes at 4°C. After extensive washing in cold phosphate-buffered saline, cells were transferred to 37°C for the indicated time points and immediately transferred to 4°C. The presence of cell surface-bound antibodies was detected with FITC-labeled F(ab′)2 anti-mouse IgG. Relative expression of each protein was measured by multiplying the percentage of marker-positive cells by their mean fluorescence intensity and is referred to the value obtained for control cells maintained at 4°C (considered as 100). One representative experiment of two is shown.
Ligand-induced internalization of LSECtin in transfectants and monocyte-derived dendritic cells. (A) Monoclonal antibody-induced internalization of LSECtin in K562 transfectants. K562-LSECtin cells were incubated with either the SOTO1 (antiLSECtin) or the P5D2 (antiCD29) antibodies for 30 minutes at 4°C. After extensive washing in cold phosphate-buffered saline, cells were transferred to 37°C for the indicated time points, fixed, and the presence of cell surface-bound antibodies detected with FITC-labeled F(ab′)2 anti-mouse IgG. Where indicated, cells were fixed and permeabilized before addition of the fluorescent secondary antibody (Perm). Values represent the fluorescence intensity of the cells at the distinct time points relative to the fluorescence intensity of control cells maintained at 4°C. One representative experiment of two is shown. (B) Ebola GP1-induced internalization of LSECtin. K562-LSECtin cells were either untreated or incubated with Ebola virus GP1-Fc (GP1) for 10 minutes at 37°C, transferred to 4°C, and the expression of LSECtin or the CD29 integrin (negative control) determined by indirect immunofluorescence with SOTO1 or P5D2 monoclonal antibodies and FITC-labeled F(ab′)2 anti-mouse IgG. Values represent the fluorescence intensity of the cells at the distinct time points relative to the fluorescence intensity of control cells maintained at 4°C. Mean ± standard deviation of three independent experiments is shown (*P = 0.001). (C) Cytoplasmic motifs involved in LSECtin ligand-induced internalization. K562 cells were transiently transfected with expression vectors for WT LSECtin or LSECtin mutated at Y6 (LSECtin Y/F), E14E15 (LSECtin EE/AA), W21GRW24VHW27 (LSECtin 3W/3A), or at both Y6 and E14E15 (LSECtin DM). Twenty-four hours later, cells were washed and incubated with the SOTO1 (anti-LSECtin) or the P5D2 (anti-CD29) antibodies for 30 minutes at 4°C. After extensive washing in cold phosphate-buffered saline, cells were transferred to 37°C for the indicated time points (2, 5, 15 minutes), placed on ice, and the presence of cell surface-bound antibodies detected with FITC-labeled F(ab′)2 anti-mouse IgG. Values represent the fluorescence intensity of the cells at the distinct time points relative to the fluorescence intensity of control cells maintained at 4°C. Shown are the mean and standard deviation of three independent experiments. (D) Monoclonal antibody-induced internalization of LSECtin in MDDC. MDDC were incubated with either the SOTO1 (anti-LSECtin) or the TS1/18 (anti-CD18) antibodies for 30 minutes at 4°C. After extensive washing in cold phosphate-buffered saline, cells were transferred to 37°C for the indicated time points and immediately transferred to 4°C. The presence of cell surface-bound antibodies was detected with FITC-labeled F(ab′)2 anti-mouse IgG. Relative expression of each protein was measured by multiplying the percentage of marker-positive cells by their mean fluorescence intensity and is referred to the value obtained for control cells maintained at 4°C (considered as 100). One representative experiment of two is shown.
Ligand-induced endocytosis is a property shared by lectins involved in antigen capture and presentation and is dependent on the presence of internalization motifs in the cytoplasmic tail.2 To determine the molecular basis for the antibody-induced LSECtin internalization, two potential internalization motifs similar to those present in other lectins (Y6SKW and E14E15) were mutated and assayed for internalization capability. Mutation of either Y6 (to F6) or E14E15 (to A14A15) significantly reduced the ligand-induced internalization of LSECtin (Figure 7C). Although mutation of Y6 exhibited a stronger effect at early time points (2 minutes), disruption of both motifs (LSECtin DM) reduced internalization to a higher extent than each individual mutations (after 5 or 15 minutes) (Figure 7C). By contrast, mutation of the three tryptophan residues within the cytoplasmic tail (W21GRW24VHW,27 LSECtin 3W/3A) had no effect (Figure 7C). Therefore, the ability of LSECtin to be internalized on ligand binding is dependent on the integrity of both Y6SKW and E14E15 motifs in the cytoplasmic tail. LSECtin was found to be internalized at a similar rate in MDDC, because only 20% of LSECtin molecules remain on the cell membrane 15 minutes after antibody engagement (Figure 7D). Therefore, LSECtin is internalized after engagement in both transfectants and monocyte-derived dendritic cells, suggesting its involvement in antigen capture and internalization by myeloid cells.
Discussion
The gene cluster in chromosome 19p13.2 includes the C-type lectin-encoding genes CD23, DC-SIGN (CD209), LSECtin, and DC-SIGNR, with the first three of them arranged adjacent to one another and in the same orientation. In the present report, we demonstrate that LSECtin, originally defined as a liver and lymph node sinusoidal-specific lectin,9 can be induced by IL-4 on human peripheral blood monocytes and is expressed by in vitro generated dendritic cells and alternatively activated macrophages as well as by ex vivo thymic myeloid cells. The IL-4 dependence of LSECtin expression resembles the cytokine responsiveness of the DC-SIGN and CD23 genes, whose expression is either induced or up-regulated by IL-4,10,26 and suggests a common mechanism for their coordinated up-regulation in IL-4-treated monocytes. Moreover, and like DC-SIGN and CD23, LSECtin is internalized on ligand binding. Therefore, DC-SIGN, CD23, and LSECtin comprise a cluster of structurally related lectin genes expressed on myeloid cells regulated by IL-4 and involved in antigen capture. At present, we have no explanation for the discrepancy between the previously reported lack of LSECtin mRNA detection in MDDC9 and the data presented in this article, especially considering that MDDC were generated following an identical protocol. LSECtin expression in MDDC is consistent with its induction in alternatively activated macrophages, which share a number of membrane markers with MDDC,11 and with the presence of LSECtin mRNA in peripheral blood and thymic dendritic cells. Therefore, it can be concluded that LSECtin exhibits a wider cell type distribution than DC-SIGN and CD23 being expressed in both hematopoietic and nonhematopietic cell lineages.
Like in the case of other C-type lectins,27 three distinct alternatively spliced isoforms can be predicted for LSECtin based on the sequences of the amplified mRNAs. The LSECtin gene gives rise to both membrane-bound and a transmembrane-lacking potentially soluble isoform (Δ2 isoform). The relative levels of the distinct LSECtin mRNA species appear to be dependent on the cell type: thymic macrophages exhibit equivalent amounts of LSECtin mRNA coding for full-length and truncated (Δ2 and Δ3/4) molecules, whereas thymic dendritic cells almost exclusively contain LSECtin mRNA for the shorter versions of the molecule. Interestingly, and as in the case of DC-SIGN,27 a significant percentage of LSECtin mRNA species coding for a transmembrane domain-lacking potentially soluble isoform (LSECtin Δ2) have been detected not only in dendritic cells (generated in vitro and isolated ex vivo), but also in alternatively activated macrophages and even in the THP-1 cell line. In fact, and unlike full-length LSECtin, the LSECtin Δ2 isoform can be detected in the supernatant of transfected cells (data not shown), suggesting that it is stable in the extracellular milieu and that it might function as an opsonizing agent. In this regard, a large number of cell surface lectins and lectin-like molecules exhibit potentially soluble isoforms,28 and several soluble lectins display pathogen-recognition capability.29 The soluble form of CD23 is found in serum and is generated by endogenous or exogenous proteases that cleave CD23 from the cell surface26 and has been described as regulating IgE synthesis.30 The availability of the reagents described in the present article will allow us to determine whether LSECtin can be detected in serum and whether soluble LSECtin might also influence the IgE network.
Evaluation of the sugar specificity in precipitation assays has revealed that LSECtin interacts with N-acetyl-glucosamine (NAcGlc) but is not retained by N-acetyl-galactosamine (NAcGal) or Mannan. Although previously described to interact with Mannan,9 our results are in agreement with the lack of Mannan-binding reported by Gramberg et al,19 despite the presence of the EPN motif in calcium-binding site 2 that mediates Mannose/NAcGlc binding in other C-type lectins.31 Thus, LSECtin specificity is distinct from CD23, which binds better to Gal-NAcGal-containing structures than to Gal-NAcGlc-structures,26 and would also differ from DC-SIGN in their ability to be retained by Mannose-containing matrices (Figure 6). Therefore, available data suggest that the three lectins encoded within the 19p13.2 gene cluster are not functionally redundant in terms of sugar-binding specificity. The different carbohydrate specificity of DC-SIGN and LSECtin is further suggested by the distinct aggregation behavior of their corresponding transfectants in K562 cells.32
The nonredundant sugar specificity of the three lectins would also imply that DC-SIGN, CD23, and LSECtin differ in their pathogen-recognition capabilities. In fact, our results and those of others19 indicate that this is indeed the case, because LSECtin does not mediate the binding and uptake of DC-SIGN-interacting pathogens such as HIV, hepatitis C virus,19 Leishmania, Candida, or Aspergillus (data not shown). However, although the failure of K562-LSECtin to capture these pathogens is in agreement with the inability of Mannan-agarose matrix to retain LSECtin, these results should be interpreted with caution, because DC-SIGN-dependent pathogen-recognition activities are greatly dependent on the cell surface level of expression.22 Therefore, an extensive analysis of the sugar-binding specificity of LSECtin should be performed before ruling out its ability to interact with DC-SIGN-interacting pathogens.
Several distinct motifs (tyrosine-based, dileucine-based, glutamic-rich sequences) have been identified as responsible for the internalization of dendritic cell lectins involved in antigen capture for subsequent presentation.33 In the case of LSECtin, its ligand-induced internalization ability is dependent on two internalization motifs (Y6SKW and E14E15) within the cytoplasmic tail of the molecule, which is devoid of a dileucine motif. Analysis of the LSECtin cytoplasmic tail reveals the presence of a tryptophan-rich sequence that conforms to the consensus WXYWXYWXY, where X is a small amino acid and Y is a positively charged residue. Although this sequence resembles motifs found in cytoskeletal and extracellular proteins, its disruption had no effect on LSECtin internalization, which leads us to hypothesize its involvement in signal transduction. The internalization ability of LSECtin fits well with the effector functions of macrophages and dendritic and sinusoidal cells, which constitutively capture extracellular material either for scavenging purposes or for antigen processing and presentation. Along this line, the presence of LSECtin in thymic macrophages and dendritic cells, which are involved in central tolerance induction,34 is suggestive of a role for LSECtin in antigen presentation and tolerance. On the other hand, the expression of LSECtin in MDDC indicates that the molecule could be a potential target molecule in vaccination strategies, a capacity already demonstrated for DEC-205 and DC-SIGN.35,36 If so, determination of the LSECtin specificity might be helpful to understand whether the molecule participates in triggering immune responses (through recognition of PAMPs) or preferentially acts as a tolerance-inducing receptor (through capture of self-antigens).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Ministerio de Educación y Ciencia (grants SAF2005-0021, AGL2004-02148-ALI, and GEN2003-20649-C06-01/NAC) and Fundación para la Investigación y Prevención del SIDA en España (FIPSE 36422/03) to ALC. A.D.S. was supported by a FPI predoctoral grant (BES2004-4405) from Ministerio de Educación y Ciencia (Spain).
Authorship
Contribution: A.D.S. designed the research and performed the experiments; L.A.F., E.G.M., L.M.P., and P.M. performed the research (lipid raft preparation, thymic cell separation, Ebola-binding assays); M.L.T., M.C., M.Z., R.D., and F.B. provided reagents and supervised individual experiments; and A.L.C. supervised research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Angel L. Corbí, Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas, Velázquez 144, 28 006 Madrid, Spain; e-mail: acorbi@cib.csic.es.

![Figure 4. LSECtin expression on the cell surface of monocyte-derived dendritic cells and thymic-cell subsets. (A) Cell extracts were obtained from monocytes along the MDDC differentiation pathway by treating them with GM-CSF plus IL-4 for the indicated periods of time. In parallel, extracts were obtained from GM-CSF-generated macrophages after a 48-hour treatment with medium (MDM), IL-4 (AAMØ), or interferon-γ (CAMØ). In all cases, 10 μg of each lysate was subjected to Western blot for detection of LSECtin with the ADS-1 polyclonal antiserum and β-actin expression determined in parallel for loading control purpose. (B) Cell surface expression levels of LSECtin in monocyte-derived dendritic cells from three independent donors as determined with the antiLSECtin SOTO1 monoclonal antibody. The supernatant from the P3 × 63 myeloma was used as negative control (X63). (C) MDDC were allowed to differentiate on glass coverslips, fixed (paraformaldehyde 2%), washed, and incubated with either a polyclonal antiserum against LSECtin (ADS-1) or preimmune serum. After washing, cells were incubated with a 1:500 dilution of Alexa 488-labeled goat anti-rabbit IgG (F(ab′)2) antiserum. Nuclei were counterstained with 4′,6-diamidino-2-fenilindol diclorhidrato (DAPI). Coverslips were mounted in Dako Cytomation fluorescent mounting medium (Dako, Glostrup, Denmark) and representative fields photographed through an HCX PL APO lens (63.0 × 1.40 oil objective), with a 1.400000 numerical aperture on a TCS-SP2-ADBS confocal laser LEICA scanning system (LEICA MICROSYSTEMS GmbH, Wetzlar, Germany). (D) Expression of LSECtin in human thymic cell populations. Human thymic cell preparations were depleted of thymocytes by centrifugation on Ficoll after rosetting with sheep erythrocytes. The resulting population was then stained with either PE-labeled anti-CD123 and FITC-labeled BDCA2 or with FITC-labeled anti-CD14 and PE-labeled anti-CD13 antibodies. The BDCA2+ CD123+ (plasmacytoid DC [pDC]), CD14+ CD13+ (macrophages [MØ]), and CD14−CD13+ (myeloid DC [MyDC]) subsets (left panels) were simultaneously stained with the anti-LSECtin SOTO1 monoclonal antibody followed by incubation with an APC-labeled goat anti-mouse antiserum (right panels).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/12/10.1182_blood-2006-09-048058/4/m_zh80120701780004.jpeg?Expires=1767954710&Signature=0X1dwx575MXKh8MSEOuh-dt16GS0qlMAEl7b4br-stZFFayBhGIDqPyrWzaJwS2Uk9A3CrhrfQxLwfPNFdxi~3xImIFHr4dd~WYkCeXqycfGgMtJcl9qEvOB6bXpnUONeZFJnegYPzU512I2FNCQP7qDbqh9EeByRFPiDtczAWc-FKN4tB1YDaXpFxTvvyYPvv7MgciH~lWkA2WjqR34a76HwKy33LIORNyh-DV6jJa48u2ELvjnMMFLJa-wSj8xDVicCdzHUxs1707X8uXmdMaTVI~nkpFfj0UeTyu34wPOVT0gZuEFoudj0Gd1WCdaZ3SOPait1DdO-44DpEhieg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. LSECtin expression on the cell surface of monocyte-derived dendritic cells and thymic-cell subsets. (A) Cell extracts were obtained from monocytes along the MDDC differentiation pathway by treating them with GM-CSF plus IL-4 for the indicated periods of time. In parallel, extracts were obtained from GM-CSF-generated macrophages after a 48-hour treatment with medium (MDM), IL-4 (AAMØ), or interferon-γ (CAMØ). In all cases, 10 μg of each lysate was subjected to Western blot for detection of LSECtin with the ADS-1 polyclonal antiserum and β-actin expression determined in parallel for loading control purpose. (B) Cell surface expression levels of LSECtin in monocyte-derived dendritic cells from three independent donors as determined with the antiLSECtin SOTO1 monoclonal antibody. The supernatant from the P3 × 63 myeloma was used as negative control (X63). (C) MDDC were allowed to differentiate on glass coverslips, fixed (paraformaldehyde 2%), washed, and incubated with either a polyclonal antiserum against LSECtin (ADS-1) or preimmune serum. After washing, cells were incubated with a 1:500 dilution of Alexa 488-labeled goat anti-rabbit IgG (F(ab′)2) antiserum. Nuclei were counterstained with 4′,6-diamidino-2-fenilindol diclorhidrato (DAPI). Coverslips were mounted in Dako Cytomation fluorescent mounting medium (Dako, Glostrup, Denmark) and representative fields photographed through an HCX PL APO lens (63.0 × 1.40 oil objective), with a 1.400000 numerical aperture on a TCS-SP2-ADBS confocal laser LEICA scanning system (LEICA MICROSYSTEMS GmbH, Wetzlar, Germany). (D) Expression of LSECtin in human thymic cell populations. Human thymic cell preparations were depleted of thymocytes by centrifugation on Ficoll after rosetting with sheep erythrocytes. The resulting population was then stained with either PE-labeled anti-CD123 and FITC-labeled BDCA2 or with FITC-labeled anti-CD14 and PE-labeled anti-CD13 antibodies. The BDCA2+ CD123+ (plasmacytoid DC [pDC]), CD14+ CD13+ (macrophages [MØ]), and CD14−CD13+ (myeloid DC [MyDC]) subsets (left panels) were simultaneously stained with the anti-LSECtin SOTO1 monoclonal antibody followed by incubation with an APC-labeled goat anti-mouse antiserum (right panels).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/12/10.1182_blood-2006-09-048058/4/m_zh80120701780004.jpeg?Expires=1768325217&Signature=MhkpGD6xlc5k3qmB2Y5H-edoflK43Jg7dmi0uIPJ2GrKc2sRxo5Cidx92RqW66xpK7S1vZa556CmqDNEhe1tMDzw156yjE3X7PAchhHrUWMcFIlLUIkn0IqBmtC~kRae~2tfyt0tIGqS7wNFEjDediF5ILisGXZnFDJi3YLVNY7vCrJ0kHp6UNXUkG0qpi1TwTAwTawn-7IFxSQLPDLOTGdO9UBie4HHNDoouSkhGo9OaBWro8B2l05z9QTCQX10SmmQdKr6AjV2AZRLY85jLMCp0XOZFPkQ~ToGJ-fkKLgVgnbYCRzvbhvBZqQrBTugYS7I8jOAI4TOCx6ZAy2x5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)