Abstract
The pre-B-cell receptor (pre-BCR) is thought to signal transcriptional activation of the immunoglobulin light (L) chain gene locus, proceeding to its V-J rearrangement. The pre-BCR signaling pathway for this process is largely unknown but may involve the adaptor protein BASH (BLNK/SLP-65). Here we report that the pre-B leukemia cell lines established from affected BASH-deficient mice rearrange κL-chain gene locus and down-regulate pre-BCR upon PMA treatment or BASH reconstitution. Analyses with specific inhibitors revealed that activation of novel PKC (nPKC) and MEK, but not Ras, is necessary for the rearrangement. Accordingly, retroviral transduction of active PKCη, PKCϵ, or Raf-1, but not Ras, induced the κ gene rearrangement and expression in the pre-B-cell line. Tamoxifen-mediated BASH reconstitution resulted in the translocation of PKCη to the plasma membrane and κ chain expression. These data make evident that the Ras-independent BASH-nPKC-Raf-1 pathway of pre-BCR signaling induces the L-chain gene rearrangement and expression.
Introduction
The rearrangement of immunoglobulin (Ig) genes is a highly regulated process in B-cell development. In pro-B cells, the IgH locus rearranges first with a D-to-JH and then VH-to-DJH recombination. If a μH chain is produced from the rearranged allele and couples with the surrogate light chains (λ5 and Vpre-B) and Ig-α/Ig-β, the resultant surface complex called the pre-B-cell receptor (pre-BCR) is transiently expressed and signals the proliferation of pre-B cells to form a population called large pre-B cells. Pre-BCR also signals the shutoff of RAG1 and RAG2 expression and the down-regulation of itself from the cell surface. The pre-BCR signal is also believed to be responsible for allelic exclusion of the IgH loci. After a few rounds of cycling, large pre-B cells exit the cell cycle and differentiate into small pre-B cells where RAG1/2 is reexpressed and IgL chain (κ or λ) genes undergo V-J rearrangements. Production of IgL coupling with μH leads to the surface expression of IgM that serves as the B-cell receptor (BCR). Thus, pre-BCR signaling is essential for the pro-B- to pre-B-cell transition, which is evidenced in mice and humans carrying mutations in genes encoding components of pre-BCR and showing profound B-cell deficiency.1-3
Biochemical studies using pre-B-cell lines have shown that antibody-mediated crosslinking of pre-BCR induces phosphorylation and/or activation of a set of signaling molecules similar to that induced by BCR crosslinking.4,5 However, it is unknown whether this type of crosslinking represents physiological activation of pre-BCR that promotes B-cell development. Pre-BCR is rather thought to be self-ligated autonomously (without other ligands) and rapidly internalized under physiological conditions.6 Thus pre-BCR-positive cells compose only a minor population in the normal bone marrow. Currently little is known about the molecules involved in the autonomous signaling pathway from pre-BCR, primarily due to the lack of an experimental system. In lieu of such a system, mouse genetics approaches have revealed that the Syk family tyrosine kinases7 and the redundant Src family tyrosine kinases Lyn, Blk, and Fyn8 are essential for the pre-BCR-mediated pre-B-cell development. In addition, the adaptor protein BASH has been shown to be crucial for the pre-BCR function. BASH (also known as BLNK or SLP-65) is phosphorylated by Syk upon BCR ligation and bound with PLCγ2, Vav, Grb2, Btk, and HPK1. BASH is required for intracellular calcium elevation and activation of PLCγ2, MAP kinases including Erk, and NF-κB.9-11 Targeted disruption of BASH resulted in an incomplete block of the pre-BCR-dependent development of pre-B cells, with a marked decrease of B cells and an increase of pre-BCR-positive large pre-B cells.12,13 This developmental block is rendered almost complete by additional mutations in CD19,14 Btk,15 or LAT,16 suggesting the presence of a partially compensatory pathway through these molecules. Incomplete block at the transition from pro-B to pre-B is also observed in PLCγ1+/-PLCγ2-/- mice,17 whereas no such block is seen in mice devoid of all known members of the Vav family (Vav1-3).18
It is reported that transgenic expression of a dominant-negative (DN) mutant of Ras incompletely blocks B-cell development at early pro-B cells,19 or at late pre-B cells,20 probably depending on the expression timing and level of the DN Ras. Conversely, transgenic expression of constitutively active (CA) mutants of Ras or c-Raf-1 results in developmental progression of RAG- or IgH-deficient pro-B cells into small pre-B-like cells in which κ germline transcripts are expressed and also in the L-chain gene rearrangement in the case of IgH-deficient cells.21-23 Taken together, the Ras-Raf signaling pathway inhibits apoptosis of pro-B and late pre-B cells, perhaps through Bcl-2 family proteins, and contributes to their survival.20,21 Whether this function is related to the pre-BCR signaling and also to the induction of L-chain gene rearrangement remains unclear.
Pre-BCR is not essential for the κ gene rearrangement, as it can be detected in a small fraction of pro-B cells from normal as well as pre-BCR-deficient mice.24 However, pre-BCR is thought to induce additional κ germline transcription, which increases the accessibility of the κ chain locus to the V(D)J recombinase and hence the Vκ-Jκ recombination.25-27 Recently, genetic as well as epigenetic requirements for the rearrangement have been proposed and, in particular, monoallelic histone acetylation/methylation and CpG demethylation at the Jκ locus have been shown to coincide with initiation of the Vκ-Jκ rearrangement.28,29 Transcription factors required for the κ germline transcription and recombination, such as E2A, Spi-B, and IRF-4/8, have also been identified.30-32 However, it remains unknown whether and how the pre-BCR signal induces such epigenetic change or engagement by these transcription factors of the κ gene locus.
We have recently reported that pre-B cells from BASH-deficient mice express significantly reduced levels of the κ germline transcript and undergo less extensive κ gene rearrangement despite the increased levels of RAG2, and those from BASH/CD19-double-knockout (DKO) mice exhibited a more severe phenotype.14 Therefore, with occasional redundant backup by CD19, BASH seems to be required for the signal transduction from pre-BCR that induces κ gene rearrangement. However, such in vivo analyses of transgenic or knockout mice are not sufficient to provide a definitive answer as to what molecule signals the κ gene rearrangement, because it is almost impossible to distinguish the de novo induction of rearrangement from survival and/or expansion of preexisting cells undergoing or having undergone the rearrangement. We have reported that a portion of BASH single- and BASH/CD19 double-deficient mice spontaneously develop pre-B-cell acute lymphoblastic leukemia (pre-B ALL) at earlier ages, as others also reported for the former mice.14,33 The leukemic cells are monoclonal and phenotypically resemble the pre-BCR-positive pre-B cells accumulating in the bone marrow of nonaffected mutant mice. By using the novel pre-B-cell lines established from these leukemic cells, we have revealed here the pre-BCR signaling pathway downstream of BASH that induces κ gene rearrangement.
Materials and methods
Cell culture
Pre-B ALL cell lines were established and cultured as described previously.14 BKO84 and DKO35 were the earliest established lines among them and thus used throughout this study. Viable cell number was counted after staining with trypan blue stain 0.4% (Invitrogen, Carlsbad, CA). To examine cell divisions, cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFDA-SE; Molecular Probes, Eugene, OR) and analyzed by flow cytometry. The cells were treated with the following reagents: phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, St Louis, MO; 10 ng/mL unless otherwise noted); bisindolylmaleimide (Calbiochem, San Diego, CA; 2 μM); Go6976 (Calbiochem; 2 μM); FTI-277 (Calbiochem; 1 μM); PD98059 (Calbiochem; 40 μM); and U0126 (Wako, Osaka, Japan; 5 μM). Equal amounts of solvent dimethyl sulfoxide (DMSO; Sigma-Aldrich) were added in the control cultures. The concentration of each inhibitor was determined by titration, and the maximum concentration that does not induce apoptosis of BKO84 cells was used.
MSRE-PCR assays
Methyl-sensitive restriction enzyme-dependent polymerase chain reaction (MSRE-PCR) assay was performed as described previously34 with slight modifications. Genomic DNA was digested with or without HhaI, together with EcoRI to reduce viscosity of the samples, overnight at 37°C and then incubated at 70°C for 20 minutes. The digested genomic DNA (20 ng) was subjected to PCR with primers HhaI-5′: 5′-TGAGGAATGAAGGAACTTCAGGATAGAAAA-3′ and HhaI-3′: 5′-CCTACAGAGTCTCTCATTTTGACATTTTGTGACA-3′, in the following heating cycle: 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute; for 30 cycles. To control the amount of DNA, the sequence 3′ of the recombination signal sequence (RSS) that lies about 25 kb downstream of the Cκ exon was amplified with primers RSS-5′: 5′-CAGTTGAGCTCAGATTTGAGCCCTAATG-3′ and RSS-3′: 5′-TGACTGTTTGCTACTTCAGCTCACTG-3′, in the following heating cycle: 95°C for 1 minute, 58°C for 1 minute, and 72°C for 30 seconds; for 30 cycles. PCR products were separated on 2% agarose gels and visualized by ethidium bromide staining.
Construction of retrovirus vectors
A portion of EGFP in the pMX-IRES-GFP35 was replaced with an extracellular transmembrane domain of rat CD2 (rCD2) to generate pMX-IRES-rCD2. The mouse BASH cDNA14 and the following cDNAs with point mutations were inserted into the pMX-IRES-GFP or the pMX-IRES-rCD2: the CA forms of human PKCη (PKCη A161E), human H-Ras (H-Ras Q61L, H-Ras V12), human Raf-1 (Raf-1 S259D), or human MEK (MEK LA-SDSE)36 and the kinase-dead form of human PKCη (PKCη K384R). These vectors were transfected into PLAT-E packaging cells,35 and on the next day the supernatants were added to the target cells in the presence of 10 μg/mL DOTAP Liposomal Transfection Reagent (Roche, Mannheim, Germany).
Flow cytometry and cell sorting
Cell staining, flow cytometry, and cell-cycle analysis were performed as described.14 Data shown in the figures are representative of at least 3 experiments with essentially the same results. For the experiment shown in Figure 5C, cell sorting was performed on a triple laser MoFlo (Dako-Cytomation, Fort Collins, CA). Cells were stained with 2 μg/mL propidium iodide (PI) before data acquisition, and the cells passing through the PI-negative, GFP-positive gate were collected. For magnetic-activated cell sorting (MACS), κ-positive cells were stained with PE-goat anti-Igκ antibody and anti-PE MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany), and cells infected with virus containing pMX-IRES-rCD2-based vectors were stained with biotin anti-rat CD2 monoclonal antibody (OX34; Cedarlane, Hornby, ON, Canada) followed by Streptavidin-MicroBeads (Miltenyi Biotec). Stained cells were positively selected using MACS MS columns (Miltenyi Biotec) according to the manufacturer's guidelines. More than 90% purity was achieved by this method.
Analysis of κ gene rearrangement
The κ gene rearrangement was analyzed by PCR with primers FWR3 and Jκ5-intron (Figure 2C) as described previously.14 Double strand breaks at Jκ gene segments were analyzed by using ligation-mediated (LM)-PCR with primers κ03, κ0, and BW-1H (Figure 2C) as described previously,26 except that the PCR products were hybridized with Jκ probe made by PCR with primers Jκ1-upstream (5′-TCGCCTACCCACTGCTCTGTTCCTCTTCAG-3′) and the Jκ5-intron (5′-CTAACATGAAAACCTGTGTCTTACACA-3′). To control the amount of template DNA, the sequence downstream of the RSS was amplified by PCR as described under “MSRE-PCR assays.”
Reverse transcriptase-polymerase chain reaction
Total RNA was extracted from cells using TRI reagent (Sigma-Aldrich). Complementary DNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) and subjected to PCR performed as described in Table 1. Primers for κ germline transcript (κGT-5′ and κGT-3′ in Figure 2C) and HPRT are described previously.14
Immunoprecipitation and Western blot analysis
Immunoprecipitation and Western blot analysis were performed as described previously.10 The following antibodies were used: rabbit anti-Btk (a gift from Dr Kiyoshi Takatsu, The Institute of Medical Science, University of Tokyo, Japan); rabbit anti-PLCγ2 (Q-20; Santa Cruz Biotechnology [SCB], Santa Cruz, CA); rabbit antiphospho-ERK2 (active MAPK, Promega, Madison, WI; or phospho-MAPK, Cell Signaling Technology, Beverly, MA); rabbit anti-ERK2 (SCB); rabbit antiphospho-JNK (active JNK; Promega); rabbit anti-JNK (Cell Signaling Technology); HRP antiphosphotyrosine (PY20; BD Biosciences, San Jose, CA); and HRP goat anti-rabbit IgG (Zymed, South San Francisco, CA; or Jackson ImmunoResearch, West Grove, PA).
Establishment of BKO/BASH-ERtm and BKO/ERtm cell lines
The BASH-ERtm expression vector was constructed as follows. Mouse BASH-coding region was amplified by PCR using a 5′ primer (5′-GGAATTCCACCATGGACAAGCTGAATAAGATAACTGTCCC-3′) and a 3′ primer (5′-CGACGCGTTGAAACCTTCACAGCATATTTCAGTCTCG-3′). A coding sequence of the ERtm, a ligand-binding domain of mouse estrogen receptor (amino acid 281 to 599) containing a G525R mutation,37 was amplified using a 5′ primer (5′-TTCACGCGTCGAAATGAAATGGGTGCTTCAGGAG-3′) and a 3′ primer (5′-GGAATTCTCAGATCGTGTTGGGGAAGCC-3′). These PCR products were digested with EcoRI and MluI (sites indicated by underlines) and then ligated with EcoRI-digested MSCV-IRES-GFP38 to yield BASH-ERtm. As a control, ERtm fragment alone was inserted into the same vector. These vectors were transfected into PLAT-E cells using FuGENE 6 reagent (Roche). After 2 days the supernatants were added to BKO84 cells in the presence of 5 mg/mL polybrene. Infected GFP-positive cells (BKO/BASH-ERtm and BKO/ERtm,) were sorted using by FACSvantage (BD Biosciences).
Immunofluorescence microscopy
BKO/BASH-ERtm or BKO/ERtm cells were incubated for 1 hour with 1 μM 4-hydroxy-tamoxifen (4-OHT; Sigma-Aldrich). Cells were first stained with biotinylated anti-pre-BCR antibody (SL156; BD Biosciences), fixed with 4% paraformaldehyde for 15 minutes at room temperature (RT), and permeabilized by incubation with 0.05% Tween 20 and 3% BSA in PBS for 15 minutes at RT. Then the cells were stained with anti-PKCη (SCB) at 4°C overnight, followed by TRITC-conjugated anti-rabbit IgG (Jackson ImmunoResearch) and allophycocyanin (APC)-conjugated streptavidin (eBioscience, San Diego, CA) for 1 hour at RT. Stained cells were analyzed using a confocal laser scanning microscope (TCS SP2; Leica, Mannheim, Germany) equipped with an HCX PL Apo 63×/1.32 NA oil objective lens. TRITC was excited with a krypton laser (568 nm line), whereas APC was excited using a helium-neon laser (633 nm line). Leica confocal software (TCS SP2, version 2.6.1) was used for image acquisition and processing. Digitalized images were analyzed using Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA).
Results
Characterization of differentiation-competent pre-B-cell lines
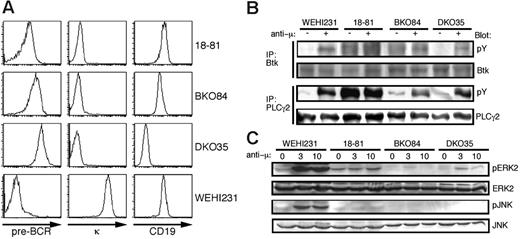
We have established several pre-B-cell lines from bone marrow cells of individual mice affected with pre-B ALL through bulk culture without additive cytokines. In the following study, we mainly used the cell lines BKO84 and DKO35 derived from BASH single- and BASH/CD19-DKO mice, respectively. These cell lines express pre-BCR, but neither the κ nor λ light chains (Figure 1A; data not shown), representing the pre-BCR-positive large pre-B cells developmentally arrested in the knockout mice. Despite a high level of surface pre-BCR expression, and unlike the Abelson murine leukemia virus (A-MuLV)-transformed 18-81 cell line, the basal tyrosine phosphorylation levels of Btk and PLCγ2 were as low in BKO84 and DKO35 cells as in an immature B-cell line WEHI231. However, crosslinking of pre-BCR induced their phosphorylation although the cells lack BASH (Figure 1B). In these cells, ERK2 activity was basally undetectable and only weakly induced by pre-BCR crosslinking compared with 18-81 cells, and JNK activity was undetectable, as in 18-81 cells, regardless of crosslinking (Figure 1C). Thus constitutive pre-BCR signaling appeared to be defective in these cell lines lacking BASH, in contrast to 18-81 cells or another pre-B-cell line as previously reported.4
To test whether these cell lines differentiate in vitro, we treated them with several reagents and found PMA to be a strong inducer of differentiation. After culture with PMA for 2 days, approximately 10% and 0.3% of BKO84 cells became κ or λ positive, respectively, in accord with the physiological ratio of κ- and λ-positive B cells in mice. In addition, overall the cells became small and exhibited down-regulated pre-BCR surface expression (Figure 2A). As short as 2 hours of initial treatment with PMA was sufficient to induce a similar phenotypic change after 2 days, indicating the early commitment to this differentiation (data not shown). PMA treatment resulted in about 2-fold reduction of the gross growth rate of BKO84 cells compared with untreated cells (Figure 2B, left panel), mainly due to the reduced cell division (Figure 2B, right panels). The κ- and λ-positive cells had undergone fewer cell divisions compared with κ- and λ-negative cells after PMA treatment, excluding the possibility that PMA selectively promotes proliferation of cells previously rearranged or committed to rearrange the L-chain genes (Figure 2B, lower right panel). PMA induced a similar differentiation of DKO35 cells and all other pre-B leukemia cell lines derived from BASH- and BASH/CD19-deficient mice but not the A-MuLV-transformed pre-B-cell lines, 18-81 and 38-B9 cells (data not shown).
Characterization of novel pre-B-cell lines generated from BASH-deficient mice. (A) Flow cytometric analysis of the receptor expression on the indicated cell lines. Cells were stained for pre-BCR (left), κ chain (middle), and CD19 (right). (B) The indicated cell lines were stimulated (+) or not (-) with goat anti-mouse μH chain antibody, F(ab′)2 fragment (anti-μ, 10 μg/mL) for 1 minute. Cell lysates were subjected to immunoprecipitation (IP) with anti-Btk or anti-PLCγ2 antibody, and the precipitates were subjected to Western blot analysis using antiphosphotyrosine antibody (pY). The same filters were reprobed with anti-Btk or anti-PLCγ2 antibody as indicated to confirm the protein load. (C) The indicated cell lines were stimulated with anti-μ (10 μg/mL) for 0, 3, and 10 minutes. Cell lysates were subjected to Western blot analysis using antiphospho-ERK2 (pERK2) and antiphospho-JNK (pJNK) antibodies. The same filters were reprobed with anti-ERK2 and anti-JNK antibodies.
Characterization of novel pre-B-cell lines generated from BASH-deficient mice. (A) Flow cytometric analysis of the receptor expression on the indicated cell lines. Cells were stained for pre-BCR (left), κ chain (middle), and CD19 (right). (B) The indicated cell lines were stimulated (+) or not (-) with goat anti-mouse μH chain antibody, F(ab′)2 fragment (anti-μ, 10 μg/mL) for 1 minute. Cell lysates were subjected to immunoprecipitation (IP) with anti-Btk or anti-PLCγ2 antibody, and the precipitates were subjected to Western blot analysis using antiphosphotyrosine antibody (pY). The same filters were reprobed with anti-Btk or anti-PLCγ2 antibody as indicated to confirm the protein load. (C) The indicated cell lines were stimulated with anti-μ (10 μg/mL) for 0, 3, and 10 minutes. Cell lysates were subjected to Western blot analysis using antiphospho-ERK2 (pERK2) and antiphospho-JNK (pJNK) antibodies. The same filters were reprobed with anti-ERK2 and anti-JNK antibodies.
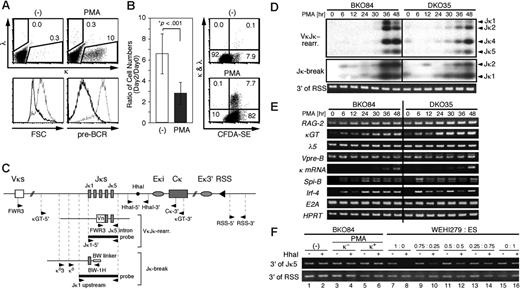
PMA induces κ germline transcription and gene rearrangement in the pre-B-cell lines. (A) BKO84 cells were cultured with PMA (10 ng/mL) or solvent alone (-) for 2 days and then stained simultaneously for κ and λ chains and pre-BCR and analyzed by flow cytometry for the expression of κ and λ chains (top). Numbers indicate percentages of the total live cells gated by light scatters. The dose-response analysis showed that the induction of κ-positive cells reached plateau with 1 ng/mL PMA (data not shown). Also shown are the forward scatter (FSC) profile (lower left) and the pre-BCR expression (lower right) of untreated (dotted line) and PMA-treated (thick line) cells. (B) (Left) BKO84 cells were cultured with PMA (10 ng/mL) or solvent alone (-) for 2 days, and then the number of live cells was counted by trypan blue dye exclusion. Bar graph shows the mean ratio (± SD) of the cell number on day 0 to that on day 2 in 10 independent experiments. *Determined by Student unpaired t test. (Right) BKO84 cells were labeled with CFDA-SE, cultured with PMA (10 ng/mL) or solvent alone (-) for 60 hours, and then stained for κ and λ chain and analyzed by flow cytometry. Numbers indicate percentages of the total live cells gated by light scatters. (C) Schematic representation of Ig κ gene locus (top), an example of the rearranged locus (middle), and an example of aJκ-associated break ligated with the linker (bottom), with relative positions of the primers and the probes used in this study. Eκi and Eκ3′ indicate κ intronic and 3′ enhancers, respectively. (D) Purified genomic DNA from indicated cell lines treated with PMA for the indicated time (in hours) was analyzed for κ gene rearrangements by PCR (top) and for Jκ-associated DNA double strand breaks by LM-PCR (middle) followed by Southern blot hybridization. Primers and probes indicated in panel C were used for these assays. The estimated locations of the amplified bands corresponding to each VκJκ rearrangement (top) or each linker-ligated Jκ break (middle) are indicated on the right of the panels. The amount of genomic DNA used for these assays was monitored by PCR amplifying the 3′ region of RSS that is not compromised after the κ gene rearrangements. (E) Reverse transcriptase-PCR analyses of the indicated transcripts and mRNA. The PCR products were analyzed by agarose gel electrophoresis followed by EtBr staining. (F) BKO84 cells were cultured with PMA for 2 days, then sorted for κ positivity (κ+) or κ negativity (κ-) or cultured without PMA (-). Genomic DNAs from these fractions, or DNAs from WEHI279 and ES cells mixed at the indicated ratio, were treated with (+) or without (-) HhaI, together with EcoRI, and subjected to PCR with primers HhaI-5′ and HhaI-3′ indicated at the top of panel C. The amount of genomic DNA used for this assay was monitored as in the bottom of panel D.
PMA induces κ germline transcription and gene rearrangement in the pre-B-cell lines. (A) BKO84 cells were cultured with PMA (10 ng/mL) or solvent alone (-) for 2 days and then stained simultaneously for κ and λ chains and pre-BCR and analyzed by flow cytometry for the expression of κ and λ chains (top). Numbers indicate percentages of the total live cells gated by light scatters. The dose-response analysis showed that the induction of κ-positive cells reached plateau with 1 ng/mL PMA (data not shown). Also shown are the forward scatter (FSC) profile (lower left) and the pre-BCR expression (lower right) of untreated (dotted line) and PMA-treated (thick line) cells. (B) (Left) BKO84 cells were cultured with PMA (10 ng/mL) or solvent alone (-) for 2 days, and then the number of live cells was counted by trypan blue dye exclusion. Bar graph shows the mean ratio (± SD) of the cell number on day 0 to that on day 2 in 10 independent experiments. *Determined by Student unpaired t test. (Right) BKO84 cells were labeled with CFDA-SE, cultured with PMA (10 ng/mL) or solvent alone (-) for 60 hours, and then stained for κ and λ chain and analyzed by flow cytometry. Numbers indicate percentages of the total live cells gated by light scatters. (C) Schematic representation of Ig κ gene locus (top), an example of the rearranged locus (middle), and an example of aJκ-associated break ligated with the linker (bottom), with relative positions of the primers and the probes used in this study. Eκi and Eκ3′ indicate κ intronic and 3′ enhancers, respectively. (D) Purified genomic DNA from indicated cell lines treated with PMA for the indicated time (in hours) was analyzed for κ gene rearrangements by PCR (top) and for Jκ-associated DNA double strand breaks by LM-PCR (middle) followed by Southern blot hybridization. Primers and probes indicated in panel C were used for these assays. The estimated locations of the amplified bands corresponding to each VκJκ rearrangement (top) or each linker-ligated Jκ break (middle) are indicated on the right of the panels. The amount of genomic DNA used for these assays was monitored by PCR amplifying the 3′ region of RSS that is not compromised after the κ gene rearrangements. (E) Reverse transcriptase-PCR analyses of the indicated transcripts and mRNA. The PCR products were analyzed by agarose gel electrophoresis followed by EtBr staining. (F) BKO84 cells were cultured with PMA for 2 days, then sorted for κ positivity (κ+) or κ negativity (κ-) or cultured without PMA (-). Genomic DNAs from these fractions, or DNAs from WEHI279 and ES cells mixed at the indicated ratio, were treated with (+) or without (-) HhaI, together with EcoRI, and subjected to PCR with primers HhaI-5′ and HhaI-3′ indicated at the top of panel C. The amount of genomic DNA used for this assay was monitored as in the bottom of panel D.
Next, the PMA-induced Vκ-Jκ gene rearrangement was directly examined by PCR (Figure 2C, VκJκ-rearr.). The rearrangement became evident 36 hours after the initiation of PMA treatment in both BKO84 and DKO35 cells (Figure 2D, top). This was accompanied by the induction of DNA broken ends associated with Jκ segments, which was confirmed by LM-PCR (Figure 2C, Jκ-break), indicating a de novo induction of a recombination reaction (Figure 2D, middle). Reverse transcriptase-PCR analysis revealed that mature κ mRNA became detectable after 36 hours and abundant by 48 hours (Figure 2E). The κ germline transcript was detectable before and increased during PMA stimulation, reaching a plateau at around 30 hours in both cell lines. Rag2 mRNA was significantly expressed before and slightly increased after PMA treatment. These data suggest that the κ gene rearrangement was induced through an up-regulation of the level of κ germline transcription but not of Rag2 mRNA in these cell lines. Expression of Spi-B and IRF-4 mRNA markedly increased early in the culture, suggesting their involvement in the process of κ gene rearrangement as proposed previously.31,32 Although E2A has been shown to be critically involved in the κ gene rearrangement,30 the E2A mRNA level was unchanged throughout this culture, in line with the previous data showing that E2A mRNA expression level is rather constant during early B-cell development up to the small pre-B-cell stage.39 Despite the down-regulation of pre-BCR on the cell surface, the mRNA levels for the surrogate light chains, λ5 and Vpre-B, were constant throughout the stimulation period. This suggests protein internalization and/or degradation as a mechanism for the pre-BCR down-regulation, as proposed previously.6
These data indicate that the cells lines BKO84 and DKO35 are developmentally arrested at the pre-BCR-positive pre-B-cell stage due to a signaling defect and that they undergo differentiation in vitro upon PMA treatment in a manner strikingly similar to pre-BCR-dependent differentiation in vivo. Thus, these cell lines appear to serve as an excellent in vitro model of early B-cell development to study a pre-BCR signaling mechanism.
It has been shown that CpG demethylation of κ locus DNA is correlated with its rearrangement. The methyl-sensitive HhaI site located downstream of Jκ5 (Figure 2C), which persists after any Vκ-Jκ rearrangement, has often been used to demonstrate the demethylation of the κ locus.34,40 We examined the demethylation of this HhaI site in BKO84 cells using MSRE-PCR assay.34 In mature B-cell line WEHI279, PCR amplification of the sequence containing the internal HhaI site was abolished after HhaI digestion of the template DNA, indicating the largely demethylated status of this site (Figure 2F, lanes 7 and 8). By contrast, the HhaI site was resistant to digestion in embryonic stem (ES) cells, indicating its fully methylated status (Figure 2F, lanes 15 and 16). The assay using the mixture of DNAs from both cell lines as a template demonstrated that this assay is sensitive enough to detect a 2-fold decrease of DNA methylation at the HhaI site (Figure 2F, lanes 9 to 14). In BKO84 cells, the HhaI site was resistant to digestion before and after PMA treatment for 2 days and even in the sorted κ-positive cells after the PMA induction (Figure 2F, lanes 1 to 6). This indicates that the methylation status of the HhaI site was unchanged during the induction of the κ rearrangement, and thus demethylation of this site is not mandatory for the rearrangement in BKO84 cells.
Differentiation of the pre-B-cell lines by BASH reconstitution
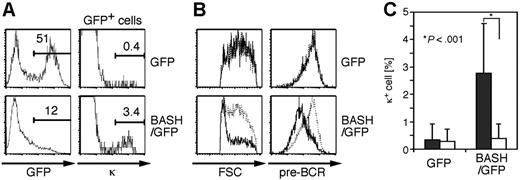
To ascertain whether BASH is required for the pre-BCR induction of the L-chain gene rearrangements, we reconstituted BKO84 cells with BASH via a retroviral vector carrying the GFP gene as an infection marker (pMX-BASH-IRES-GFP [BASH/GFP]). Compared with the mock (GFP)-infected cells, the frequency of BASH/GFP-infected cells was low after 2 days (3.3-fold ± 1.5-fold lower on average [n = 12]), but a significant portion of such cells became κ positive 4 days after the infection (Figure 3A). The κ-positive cell proportion of the BASH-transduced cells varied considerably (1% to 8% in BKO84 and 25% to 33% in DKO35) among experiments but was significantly increased compared with that of nontransduced cells or GFP-transduced cells (Figure 3C). Most of the BASH-reconstituted cells became smaller in size and expressed a reduced level of pre-BCR (Figure 3B, bottom) in contrast with the mock-infected cells (Figure 3B, top). Essentially the same results were obtained with DKO35 as well as other BASH-deficient pre-B ALL cell lines (data not shown). These data indicate that BASH is necessary for pre-BCR-signaled execution of the differentiation program, including κ gene rearrangement and pre-BCR down-regulation in these cell lines, and also that the pre-BCR on these cells is autonomously active in signaling once BASH is reconstituted.
Involvement of novel PKC in the signal inducing κ gene rearrangement
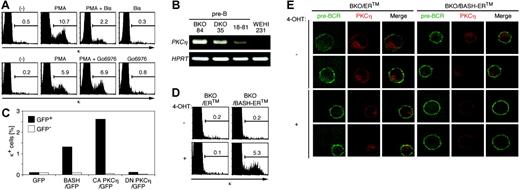
The results shown in the preceding section indicate that BKO84 and DKO35 cell lines should be particularly useful to identify the signaling pathways downstream of BASH that induce the respective events associated with pre-BCR-mediated differentiation. Because even a brief treatment with PMA induced the differentiation, we examined the possible involvement of protein kinase C (PKC) in the signaling pathway by using specific inhibitory reagents. As shown in Figure 4A, PMA-induced κ expression on BKO84 cells was strongly inhibited by bisindolylmaleimide, an inhibitor of conventional PKC (cPKC) and novel PKC (nPKC) (Figure 4A, top), but not by Go6976, an inhibitor of cPKC alone (Figure 4A, bottom). This result indicates that PMA-induced κ gene rearrangement was likely to be mediated by nPKC. PKCη, a member of the nPKC subfamily, is expressed in pro-B cells but down-regulated in small pre-B cells, and its overexpression in a pro-B-cell line resulted in cell-cycle arrest.41 This suggests the involvement of PKCη in the transition from pro-B cells to small pre-B cells. PKCη mRNA was expressed in BKO84, DKO35, and 18-81 but not in a B-cell line WEHI231 (Figure 4B). Thus we examined the effect of a lipid second messenger-independent CA mutant (A161E) or an ATP binding-defective DN mutant (K384R) of PKCη after retroviral transduction into BKO84 cells. Four days after infection, a significant portion of CA-PKCη-transduced cells expressed the κ chain (4.3% ± 2.0% of transduced versus 0.25% ± 0.18% of nontransduced cells, P < .001, n = 11), but essentially none of the cells transduced with the DN-PKCη (0.32% ± 0.15% of transduced versus 0.15% ± 0.10% of nontransduced, n = 6) or the empty (GFP) vector did so (Figure 4C; other data not shown). Essentially the same results were obtained using DKO35 as well as other BASH-deficient pre-B ALL cell lines (data not shown). In addition, transduction of CA form (A159E) of PKCϵ, the closest relative of PKCη in the nPKC family, induced κ chain expression on BKO84 cells (3.5% and 2.0% of transduced versus 0.16% and 0.07% of nontransduced cells). This result indicates that active PKCη and PKCϵ can substitute for BASH in pre-BCR signaling and is sufficient to propagate the signal from BASH that induces κ gene rearrangement. PKCδ and PKCθ might also play a redundant role, because they are expressed in BKO84, DKO35, and 18-81 cell lines (data not shown).
BASH reconstitution of BASH-deficient pre-B-cell lines promotes κ chain expression, down-regulates pre-BCR expression, and reduces cell size. BKO84 cells were infected with retroviruses containing pMX-IRES-GFP (GFP) or pMX-BASH-IRES-GFP (BASH/GFP) and analyzed by flow cytometry 4 days after the infection. (A) Expression of GFP in total live cells (left panels) and of κ chain on the GFP-positive cells (5000 gated cells; right panels). Numbers above the horizontal bars in the histograms indicate the percentages of the gated population. (B) FSC of and pre-BCR expression on the gated GFP-positive (bold line) and GFP-negative (dotted line) cells. (C) The frequency of κ-positive cells among GFP-positive (▪) or GFP-negative cells (□) in BKO84 cells 4 days after infection with GFP or BASH/GFP retroviruses. Bar graph indicates mean value (± SD) of 18 independent experiments. *Determined by Student unpaired t test.
BASH reconstitution of BASH-deficient pre-B-cell lines promotes κ chain expression, down-regulates pre-BCR expression, and reduces cell size. BKO84 cells were infected with retroviruses containing pMX-IRES-GFP (GFP) or pMX-BASH-IRES-GFP (BASH/GFP) and analyzed by flow cytometry 4 days after the infection. (A) Expression of GFP in total live cells (left panels) and of κ chain on the GFP-positive cells (5000 gated cells; right panels). Numbers above the horizontal bars in the histograms indicate the percentages of the gated population. (B) FSC of and pre-BCR expression on the gated GFP-positive (bold line) and GFP-negative (dotted line) cells. (C) The frequency of κ-positive cells among GFP-positive (▪) or GFP-negative cells (□) in BKO84 cells 4 days after infection with GFP or BASH/GFP retroviruses. Bar graph indicates mean value (± SD) of 18 independent experiments. *Determined by Student unpaired t test.
To address whether BASH signaling leads to PKCη activation, we established a BKO84 cell transfectant (BKO/BASH-ERtm) stably expressing a fusion protein consisting of BASH and a mutated hormone-binding domain of mouse estrogen receptor (ER) in which BASH function is basically suppressed but restored upon binding with 4-OHT. A similar BASH fusion protein was shown to transduce signals, leading to Ca2+ flux and activation of NF-κB and AP-1 in μH chain-reconstituted pro-B cells upon 4-OHT treatment.42 After treatment with 4-OHT for 4 days, about 5% of BKO/BASH-ERtm cells expressed κ chain on their surface, whereas BKO84 cells carrying a control vector (BKO/ERtm) did not (Figure 4D). Using this system, we examined localization of endogenous PKCη by immunostaining followed by confocal laser scanning microscopy. After 1 hour treatment with 4-OHT, cytoplasmic PKCη was translocated to the plasma membrane in the BKO/BASH-ERtm cells, which is known as a hallmark of receptor-mediated activation of PKC, but not in the control BKO/ERtm cells. This suggests that BASH recruits PKCη to the pre-BCR signalosome at the plasma membrane and activates it for downstream signaling.
Active Raf-1, but not active Ras or MEK, induces κ gene rearrangement
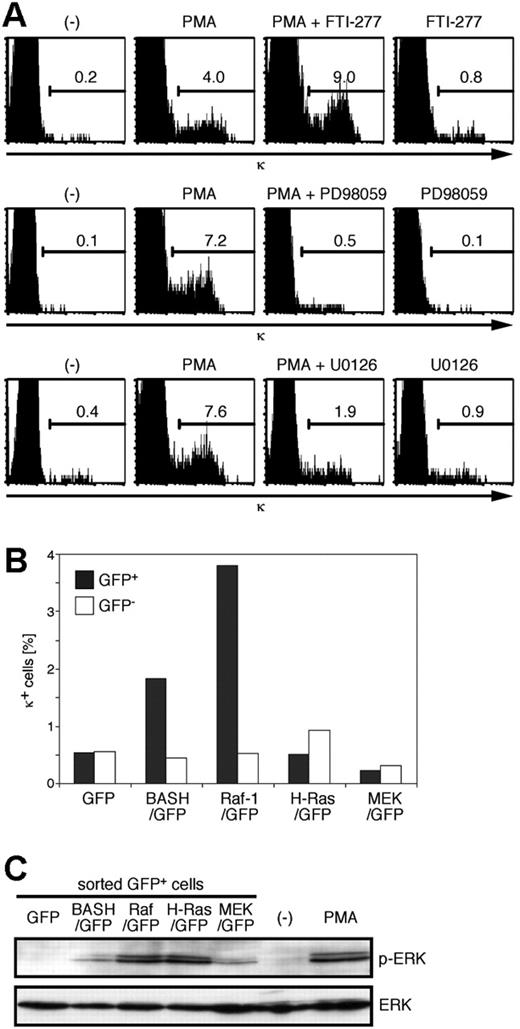
PMA is also known to activate the Ras guanine-releasing protein (GRP) family of nucleotide exchange factors, which proceeds to Ras activation.43 It has been shown that BCR stimulation activates Ras-GRP3 44 and that active Ras may induce L-chain gene locus activation and rearrangement.21,22 To examine whether Ras is required for the PMA-induced κ gene rearrangement, we treated BKO84 cells with Ras-specific farnesyltransferase inhibitor FTI-277 in addition to PMA. As shown in Figure 5A (top), FTI-277 did not suppress the induction of κ-positive cells by PMA but, on the contrary, augmented it. FTI-277 alone only marginally induced κ-positive cells. This result indicated that Ras activation is not necessary and rather inhibitory for PMA-induced κ gene rearrangement. By contrast, the MEK inhibitors PD98059 and U0126 clearly inhibited the PMA induction of κ (Figure 5A, middle and bottom).
Therefore, PMA-induced κ gene rearrangement requires the activation of MEK, possibly by Raf.
Activation of PKCη is necessary and sufficient for κ chain induction in the pre-B-cell lines. (A) BKO84 cells were cultured for 2 days with PMA, bisindolylmaleimide (Bis), Go6976, alone or in combination, or solvent alone (-), as indicated, and analyzed for κ chain expression by flow cytometry. (B) Reverse transcriptase-PCR analysis of PKCη and HPRT mRNA expression in BKO84, DKO35, 18-81, and WEHI231 cells. (C) BKO84 cells 4 days after infection with the indicated retroviral vectors were analyzed by flow cytometry. The bar graph indicates the frequency of κ-positive cells among GFP-positive (▪) or GFP-negative (□) cells. (D) BKO/ERtm and BKO/BASH-ERtm cells were treated with 1 μM 4-OHT (+) or solvent alone (-) for 4 days and analyzed by flow cytometry for the expression of κ chain. Numbers indicate percentages of the total live cells gated by light scatters. (E) Plasma membrane translocation of endogenous PKCη by BASH reconstitution. BKO/ERtm and BKO/BASH-ERtm cells were treated with 4-OHT (1 μM) for 1 hour, stained for surface pre-BCR, fixed and permeabilized, and stained for PKCη as described in “Materials and methods.” Images were obtained by confocal laser scanning microscopy and digitally colored in green (pre-BCR) and red (PKCη) and merged (Merge).
Activation of PKCη is necessary and sufficient for κ chain induction in the pre-B-cell lines. (A) BKO84 cells were cultured for 2 days with PMA, bisindolylmaleimide (Bis), Go6976, alone or in combination, or solvent alone (-), as indicated, and analyzed for κ chain expression by flow cytometry. (B) Reverse transcriptase-PCR analysis of PKCη and HPRT mRNA expression in BKO84, DKO35, 18-81, and WEHI231 cells. (C) BKO84 cells 4 days after infection with the indicated retroviral vectors were analyzed by flow cytometry. The bar graph indicates the frequency of κ-positive cells among GFP-positive (▪) or GFP-negative (□) cells. (D) BKO/ERtm and BKO/BASH-ERtm cells were treated with 1 μM 4-OHT (+) or solvent alone (-) for 4 days and analyzed by flow cytometry for the expression of κ chain. Numbers indicate percentages of the total live cells gated by light scatters. (E) Plasma membrane translocation of endogenous PKCη by BASH reconstitution. BKO/ERtm and BKO/BASH-ERtm cells were treated with 4-OHT (1 μM) for 1 hour, stained for surface pre-BCR, fixed and permeabilized, and stained for PKCη as described in “Materials and methods.” Images were obtained by confocal laser scanning microscopy and digitally colored in green (pre-BCR) and red (PKCη) and merged (Merge).
Activation of Raf-1, but not H-Ras or MEK, induces κ chain expression in the pre-B-cell lines. (A) BKO84 cells were cultured for 2 days with PMA, FTI-277, PD98059, and U0126, alone or in combination, or solvent alone (-), as indicated, and analyzed for the expression of κ chain by flow cytometry. The number in each histogram indicates the percentage of total live cells. (B) BKO84 cells 4 days after infection with the indicated retroviral vectors were analyzed for the expression of κ chain by flow cytometry. The bar graph indicates the frequency of κ-positive cells among GFP-positive (▪) or GFP-negative (□) cells. (C) BKO84 cells 2 days after infection (as in panel B) were sorted for GFP-positive cells. The whole-cell lysates of the sorted GFP-positive cells or the cells treated with PMA (100 ng/mL) for 5 minutes as a positive control were subjected to Western blot analysis using antiphospho-ERK2 (top lane) or anti-ERK2 (bottom lane).
Activation of Raf-1, but not H-Ras or MEK, induces κ chain expression in the pre-B-cell lines. (A) BKO84 cells were cultured for 2 days with PMA, FTI-277, PD98059, and U0126, alone or in combination, or solvent alone (-), as indicated, and analyzed for the expression of κ chain by flow cytometry. The number in each histogram indicates the percentage of total live cells. (B) BKO84 cells 4 days after infection with the indicated retroviral vectors were analyzed for the expression of κ chain by flow cytometry. The bar graph indicates the frequency of κ-positive cells among GFP-positive (▪) or GFP-negative (□) cells. (C) BKO84 cells 2 days after infection (as in panel B) were sorted for GFP-positive cells. The whole-cell lysates of the sorted GFP-positive cells or the cells treated with PMA (100 ng/mL) for 5 minutes as a positive control were subjected to Western blot analysis using antiphospho-ERK2 (top lane) or anti-ERK2 (bottom lane).
In accord with these data, retroviral transduction of the CA forms of H-Ras (L61, Figure 5B; V12, data not shown) did not result in the induction of κ-positive cells above the background level in BKO84 cells (0.23% ± 0.25% of H-RasL61-transduced versus 0.38% ± 0.73% of nontransduced cells, n = 12). Remarkably, a CA form of Raf-1 (Raf-1 S259D) clearly induced κ expression in BKO84 cells (1.8% ± 1.2% of transduced versus 0.29% ± 0.45% of nontransduced cells, P < .001, n = 18), as well as in other pre-B ALL cell lines (data not shown), but not the CA form of MEK (MEK LA-SDSE, 0.16% ± 0.16% of transduced versus 0.12% ± 0.13% of nontransduced cells, n = 7) (Figure 5B; data not shown). Expression of the activated Raf-1, H-Ras, and MEK was confirmed by specific phosphorylation of ERK in the infected BKO84 cells (Figure 5C). These data indicate that active Raf-1 alone, but not Ras, MEK, or ERK, is sufficient to substitute for BASH to signal the κ gene rearrangement. They also suggested that MEK is necessary but not sufficient to relay the signal from Raf-1 for the induction of the κ rearrangement, implying requirement of another mediator downstream of Raf-1 (Figure 6D). Pre-BCR down-regulation was not observed in BKO84 cells transduced with CA-Raf-1, suggesting that Raf-1 activation is not sufficient for this event (data not shown).
Finally, we confirmed that retroviral transduction of BASH, CA-PKCη, or CA-Raf-1 induced κ gene rearrangement in BKO84 cells, as shown by PCR to detect the rearranged alleles (Figure 6A, top) as well as by LM-PCR to detect DNA breaks associated with Jκ segments (Figure 6A, middle). The latter indicated the presence of cells undergoing recombination. No amplification was observed with template DNA from nonstimulated (-) or empty vector-transduced (rCD2) BKO84 cells and from mouse tail or ES cells. As shown in Figure 6B, the BKO84 cells transduced with BASH, CA-PKCη, or CA-Raf-1 (rCD2+) contained an increased proportion of apoptotic cells compared with nontransduced cells (rCD2-), which might be due to DNA breaks associated with unsuccessful L-chain gene rearrangements. It also showed that BASH and CA-PKCη induced a modest G1 arrest of cell cycle with reduction of cell size, but CA-Raf did not (Figure 6B), suggesting that the signal inducing cell-cycle arrest is diverged from that inducing the rearrangement upstream of Raf. The extent of κ rearrangement by transduction of BASH, CA-PKCη, or CA-Raf-1 was less than that induced by PMA (Figure 6A), which is consistent with the frequency of the induced κ-positive cells as detected by flow cytometry (Figure 6C). Notably, a marked synergism was observed between either of these signaling molecules and PMA treatment in the induction of κ-positive cells (Figure 6C). A possible mechanism for this is discussed in the following section.
Discussion
Here we have described newly established pre-B-cell lines that are developmentally frozen at the pre-BCR-positive pre-B-cell stage due to the absence of BASH-mediated pre-BCR signaling. Remarkably, reconstitution of BASH in these cell lines induced κ gene rearrangement, as well as pre-BCR down-regulation and cell-size reduction, indicating that autonomous pre-BCR signaling (without additional ligands) is enough to induce such an event. Using these cell lines as an in vitro model for pre-BCR signal-mediated differentiation, we have revealed that CA forms of PKCη, PKCϵ, and Raf-1 can substitute for BASH in the induction of κ gene rearrangement. Considering the numerous reports showing that PKC isotypes activate Raf-1, including one with PKCη activation of Raf-1,45 we propose that the pre-BCR signal is transmitted through the BASH-nPKC-Raf-1 axis to the specific machinery that activates the κ gene locus for V-J recombination (Figure 6D). As mentioned, not only MEK but also other unidentified molecules appear to be necessary to propagate the signal from Raf-1 to activate the downstream machinery.
Jumaa and colleagues have reported similar experiments using IL-7-dependent pre-B-cell lines derived from SLP-65 (BASH) knockout mice. They have shown that retroviral transduction of BASH enhances surface κ chain expression that is induced by IL-7 withdrawal and that this enhancement requires an N-terminal membrane-anchoring motif and a Btk-binding site of BASH.5,33,46 Supposing these results represent the induction of κ gene rearrangement, it would support the idea that BASH mediates PLCγ2 activation through recruiting Btk, as proposed in the BCR signaling,47 which leads to DAG-mediated nPKC activation (Figure 6D). The BASH-mediated signaling that leads to κ gene rearrangement appears to be restricted to the context of the pre-B-cell stage, because Schebesta et al showed, using a 4-OHT-inducible BASH reconstitution system similar to ours, that BASH reconstitution in Igμ-transgenic Pax5-/- pro-B cells induces down-regulation of RAG1/2 expression that would prohibit the rearrangement.42
BASH and active forms of PKCη and Raf-1 induce κ gene rearrangement in synergy with PMA treatment in the pre-B-cell line. (A) BKO84 cells were infected with the retroviruses carrying pMX-IRES-rCD2 (rCD2), pMX-BASH-IRES-rCD2 (BASH/rCD2), pMX-PKCη-IRES-rCD2 (PKCη/rCD2), or pMX-Raf-1-IRES-rCD2 (Raf-1/rCD2). Four days after the infection, rCD2+ cells were sorted by MACS. Genomic DNAs from the sorted samples, from BKO84 cells treated with PMA for 2 days (PMA) or not (-), and from mouse tail (Tail) or ES cells (ES) were subjected to PCR-detecting κ gene rearrangements (top) and LM-PCR-detecting Jκ-associated DNA breaks (middle) followed by Southern blot analysis. The amount of genomic DNA used for these assays was monitored as in Figure 2D (bottom). (B) BKO84 cells were infected as in panel A. Four days after the infection, rCD2+ (left) or rCD2- (middle) cells were separated by MACS, fixed and permeabilized, stained with PI, and analyzed for DNA contents by flow cytometry. The percentages of the cells in sub-G1 (apoptotic), G1, and S/G2/M (percentages of nonapoptotic cells in parentheses) are denoted in each histogram. FSC profiles of the rCD2+ (thick line) and the rCD2- (dotted line) cells within a live lymphocyte gate (defined by FSC/side scatter [SSC] profile) are shown as histograms (right). (C) BKO84 cells were infected with the indicated retroviruses, stimulated with PMA (PMA) or solvent alone (-) from day 3 after infection, and analyzed for κ expression by flow cytometry on day 5. The bar graph indicates the frequency of κ-positive cells among GFP-positive (+) or GFP-negative (-) cells. (D) Pre-BCR signaling pathway regulating the κ gene rearrangement. A hypothetical “docking area” is indicated as a gray part. DAG indicates diacylglycerol; X, a hypothetical scaffold protein for the BASH-nPKC-Raf-1 pathway. See “Discussion” for more details.
BASH and active forms of PKCη and Raf-1 induce κ gene rearrangement in synergy with PMA treatment in the pre-B-cell line. (A) BKO84 cells were infected with the retroviruses carrying pMX-IRES-rCD2 (rCD2), pMX-BASH-IRES-rCD2 (BASH/rCD2), pMX-PKCη-IRES-rCD2 (PKCη/rCD2), or pMX-Raf-1-IRES-rCD2 (Raf-1/rCD2). Four days after the infection, rCD2+ cells were sorted by MACS. Genomic DNAs from the sorted samples, from BKO84 cells treated with PMA for 2 days (PMA) or not (-), and from mouse tail (Tail) or ES cells (ES) were subjected to PCR-detecting κ gene rearrangements (top) and LM-PCR-detecting Jκ-associated DNA breaks (middle) followed by Southern blot analysis. The amount of genomic DNA used for these assays was monitored as in Figure 2D (bottom). (B) BKO84 cells were infected as in panel A. Four days after the infection, rCD2+ (left) or rCD2- (middle) cells were separated by MACS, fixed and permeabilized, stained with PI, and analyzed for DNA contents by flow cytometry. The percentages of the cells in sub-G1 (apoptotic), G1, and S/G2/M (percentages of nonapoptotic cells in parentheses) are denoted in each histogram. FSC profiles of the rCD2+ (thick line) and the rCD2- (dotted line) cells within a live lymphocyte gate (defined by FSC/side scatter [SSC] profile) are shown as histograms (right). (C) BKO84 cells were infected with the indicated retroviruses, stimulated with PMA (PMA) or solvent alone (-) from day 3 after infection, and analyzed for κ expression by flow cytometry on day 5. The bar graph indicates the frequency of κ-positive cells among GFP-positive (+) or GFP-negative (-) cells. (D) Pre-BCR signaling pathway regulating the κ gene rearrangement. A hypothetical “docking area” is indicated as a gray part. DAG indicates diacylglycerol; X, a hypothetical scaffold protein for the BASH-nPKC-Raf-1 pathway. See “Discussion” for more details.
Because pre-B-cell lines used here cause sporadic spontaneous rearrangement of L-chain gene, although the rearranged cells never accumulate in culture, one could formally argue that the induction of κ-positive cells by the BASH reconstitution is a result of cellular selection for such cells. However, the result of LM-PCR assay indicated that BASH, as well as active PKCη and Raf-1, induced de novo κ gene recombination (Figure 6A). To enrich such cells undergoing recombination, which must be arrested in G1 phase of the cell cycle,48 the BASH-PKCη-Raf-1-mediated signal should delete the cells not being rearranged. Although we cannot exclude such a possibility, the result indicating that the induction of κ germline transcription precedes the gene rearrangement in a time course of PMA-treated BKO84 cells (Figure 2D-E) does not favor such a negative selection.
The present data demonstrate that activation of Raf-1, but not Ras, is necessary for the induction of κ gene rearrangement. This implies a Ras-independent mechanism of Raf-1 activation in pre-BCR signal transduction. In this respect, PMA stimulation or introduction of the CA form of PKCδ, a member of nPKC, was shown to activate MEK and ERK in a manner independent of Ras and dependent on Raf in COS1 cells.49 Transgenic expression of CA-Ras in IgH-deficient mice resulted in developmental progression beyond the pre-BCR checkpoint and L-chain gene rearrangement.22 However, this might represent the CA-Ras-mediated survival/expansion of the population of pro-B cells undergoing or having undergone the rearrangement, which can be detected in normal as well as pre-BCR-deficient mice24,50 but not the de novo V-J recombination of the κ locus. Loss-of-function Ras-mutant mice (perhaps H-, K-, N-ras triple-knockout mice) would be necessary to examine whether Ras is physiologically required for the L-chain gene rearrangement in vivo.
Because transduction of CA-Ras in BKO84 cells induced ERK activation (Figure 5C), it is likely that it activated the Raf-MEK pathway. Then why did CA-Ras fail to induce κ gene rearrangement? We propose the hypothesis that the active Ras forms a signalosome involving Raf-1-MEK-ERK, which is distinct from the signalosome where the BASH-nPKC-Raf-1 pathway activates the 2 further downstream pathways that are required for κ gene rearrangement (Figure 6D). The former signalosome and the latter compete with each other for a limited “docking area” at the plasma membrane that allows activation. This explains the negative impact of Ras on PMA-induced κ gene rearrangement (Figure 5A). This may also explain the observed synergism between the PMA stimulation and transduction of BASH, CA-PKCη, or CA-Raf-1 in the induction of the κ gene rearrangement (Figure 6C). Namely, either of these molecules forms a signalosome nucleated by some scaffold molecule (X in Figure 6D) in addition to the PMA-induced, endogenous PKCη-mediated one, and these together predominate at the “docking area” over the PMA-induced, Ras-mediated signalosome that is incompetent for the rearrangement (Figure 6D).
Prepublished online as Blood First Edition Paper, June 22, 2006; DOI 10.1182/blood-2006-05-024968.
Supported by grants from the Ministry of Education, Culture, Sports, Science and Technology in Japan (D.K., R.G.) and the Japan Society for the Promotion of Science (JSPS) (D.K., R.G., M.Y.). M.Y. is a JSPS Research Fellow.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank T. Kitamura for pMX-IRES-EGFP and PLAT-E packaging cells; M. Kubo for pMX-IRES-rCD2; G. Baier, T. Kawakami and Y. Kawakami, W. Kolch, T. Sato, A. Shaw, and E. Nishida for cDNAs encoding PKCη, PKCϵ, Raf-1, H-RasL61, H-RasV12, and MEK, respectively; K. Takatsu for anti-Btk antibody; M. Ogawara for operating MoFlo; Y. Hara for other flow cytometry and confocal laser scanning microscopy; T. Matsuda, H. Jumaa, M. Busslinger, and K. Rajewsky for discussion; A. Oda, J. Nakayama, Y. Imamura, R. Mizuta, T. Mizuno, T. Shimizu, M. Suzuki, E. Shimura, N. Nakano, and other members of the Research Institute for Biological Sciences (RIBS) for help with experiments and for discussions; and T. Saito for continuous encouragements.






![Figure 6. BASH and active forms of PKCη and Raf-1 induce κ gene rearrangement in synergy with PMA treatment in the pre-B-cell line. (A) BKO84 cells were infected with the retroviruses carrying pMX-IRES-rCD2 (rCD2), pMX-BASH-IRES-rCD2 (BASH/rCD2), pMX-PKCη-IRES-rCD2 (PKCη/rCD2), or pMX-Raf-1-IRES-rCD2 (Raf-1/rCD2). Four days after the infection, rCD2+ cells were sorted by MACS. Genomic DNAs from the sorted samples, from BKO84 cells treated with PMA for 2 days (PMA) or not (-), and from mouse tail (Tail) or ES cells (ES) were subjected to PCR-detecting κ gene rearrangements (top) and LM-PCR-detecting Jκ-associated DNA breaks (middle) followed by Southern blot analysis. The amount of genomic DNA used for these assays was monitored as in Figure 2D (bottom). (B) BKO84 cells were infected as in panel A. Four days after the infection, rCD2+ (left) or rCD2- (middle) cells were separated by MACS, fixed and permeabilized, stained with PI, and analyzed for DNA contents by flow cytometry. The percentages of the cells in sub-G1 (apoptotic), G1, and S/G2/M (percentages of nonapoptotic cells in parentheses) are denoted in each histogram. FSC profiles of the rCD2+ (thick line) and the rCD2- (dotted line) cells within a live lymphocyte gate (defined by FSC/side scatter [SSC] profile) are shown as histograms (right). (C) BKO84 cells were infected with the indicated retroviruses, stimulated with PMA (PMA) or solvent alone (-) from day 3 after infection, and analyzed for κ expression by flow cytometry on day 5. The bar graph indicates the frequency of κ-positive cells among GFP-positive (+) or GFP-negative (-) cells. (D) Pre-BCR signaling pathway regulating the κ gene rearrangement. A hypothetical “docking area” is indicated as a gray part. DAG indicates diacylglycerol; X, a hypothetical scaffold protein for the BASH-nPKC-Raf-1 pathway. See “Discussion” for more details.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/8/10.1182_blood-2006-05-024968/4/m_zh80200602380006.jpeg?Expires=1768278063&Signature=JoKORI~33vTdbcH4MvqGRGDK4WlQMDVVwHEVfntOj7rT6Q4FMIQRcpSABxftqpOjOz7nsL8MQtGMkZvIV3pXNxQjvz7lIb84p3O9LOyxEGTy5LDfuHAh7ZlSnteDAGhkP15z1gI0yfh9WAcHBEcBIB-oDt5JA~44NYQkemw4F7u4ipeCd0FWKoK0ySyoZ0BqtTtcGcq8kcaFULxtbMbcGNfV05PYikaX6m5SWB8djIQ9VlIyfexJ1YfJJ2bCv3z-hShyBRtGsnnFnsvWLt4Ud8A1MY8HpqjX2uV6QESzKoE0TJldXjpsL~N1PfrRW1bN8rY6IPNI1NoihaZOC~fmcA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. BASH and active forms of PKCη and Raf-1 induce κ gene rearrangement in synergy with PMA treatment in the pre-B-cell line. (A) BKO84 cells were infected with the retroviruses carrying pMX-IRES-rCD2 (rCD2), pMX-BASH-IRES-rCD2 (BASH/rCD2), pMX-PKCη-IRES-rCD2 (PKCη/rCD2), or pMX-Raf-1-IRES-rCD2 (Raf-1/rCD2). Four days after the infection, rCD2+ cells were sorted by MACS. Genomic DNAs from the sorted samples, from BKO84 cells treated with PMA for 2 days (PMA) or not (-), and from mouse tail (Tail) or ES cells (ES) were subjected to PCR-detecting κ gene rearrangements (top) and LM-PCR-detecting Jκ-associated DNA breaks (middle) followed by Southern blot analysis. The amount of genomic DNA used for these assays was monitored as in Figure 2D (bottom). (B) BKO84 cells were infected as in panel A. Four days after the infection, rCD2+ (left) or rCD2- (middle) cells were separated by MACS, fixed and permeabilized, stained with PI, and analyzed for DNA contents by flow cytometry. The percentages of the cells in sub-G1 (apoptotic), G1, and S/G2/M (percentages of nonapoptotic cells in parentheses) are denoted in each histogram. FSC profiles of the rCD2+ (thick line) and the rCD2- (dotted line) cells within a live lymphocyte gate (defined by FSC/side scatter [SSC] profile) are shown as histograms (right). (C) BKO84 cells were infected with the indicated retroviruses, stimulated with PMA (PMA) or solvent alone (-) from day 3 after infection, and analyzed for κ expression by flow cytometry on day 5. The bar graph indicates the frequency of κ-positive cells among GFP-positive (+) or GFP-negative (-) cells. (D) Pre-BCR signaling pathway regulating the κ gene rearrangement. A hypothetical “docking area” is indicated as a gray part. DAG indicates diacylglycerol; X, a hypothetical scaffold protein for the BASH-nPKC-Raf-1 pathway. See “Discussion” for more details.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/8/10.1182_blood-2006-05-024968/4/m_zh80200602380006.jpeg?Expires=1768745289&Signature=nPuZXQSl0C~Rr3YRO2MGGW6mdlN6Anb8GI2GHdDVjaDscAzItKY4p9XuNQuYkDMFUYdfi-bdINA66OJ1DhDhrPcAgwLcJsx-f0Eae9eY2PiLA0aIEYjqcxFHIjYVXQHE8c7D5xW79Q~UFjVK~wkukmb4AslPQmGMKSdpkZTvlPXOPXU43~A0tTUBymsqZG4dHVUDMrhO9eLVZp8D6n3Eld03y2YRbuhTMQ3CjByuPZlNNEToDgyAvi7iAREOZcbpMAJO6T1TxJeQsURyOHM3Q8GWH-8qn53O1C3a0wXwXnmguSv796Yk8Mbc9KVPa9~11T8iHgA2HAoEWAa92f7KTA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)