Abstract
BCL11A and BCL11B are transcriptional regulators important for lymphopoiesis and previously associated with hematopoietic malignancies. Ablation of the mouse Bcl11b locus results in failure to generate double-positive thymocytes, implicating a critical role of Bcl11b in T-cell development. However, BCL11B is also expressed in CD4+ T lymphocytes, both in resting and activated states. Here we show both in transformed and primary CD4+ T cells that BCL11B participates in the control of the interleukin-2 (IL2) gene expression following activation through T-cell receptor (TCR). BCL11B augments expression from the IL2 promoter through direct binding to the US1 site. In addition, BCL11B associates with the p300 coactivator in CD4+ T cells activated through TCR, which may account for its transcriptional activation function. These results provide the first evidence that BCL11B, originally described as a transcriptional repressor, activates transcription of a target gene in the context of T-cell activation.
Introduction
BCL11B is a C2H2 zinc finger protein initially identified as a mediator of the transcriptional repression function of COUP-TF nuclear receptors.1 However, it has been found to also directly bind and repress transcription driven by GC-rich response elements.2 Studies with germ-line knockout mice have demonstrated that BCL11B is required for specification of the corticospinal motor neurons3 and T-cell development.4 Specifically, Bcl11b-/- mice have a block at DN3 stage of T-cell development with impaired VB to DB recombination at the T-cell receptor β (TCRB) locus and a decreased number of α/β T cells.4 Thymocytes of Bcl11b-/- mice are highly susceptible to apoptosis, which was found to be a consequence of the thymocytes failure to proliferate rather than a direct defect in apoptosis.4 On the other hand, loss of heterozygosity at the Bcl11b locus in adult mice has been associated with generation of thymic lymphomas and skin tumors.5,6 Taken together, these results suggest that BCL11B has a complex biologic function. It has been previously demonstrated for other transcriptional regulators with complex biologic function, such as c-Myc, that depending on the cellular context, they can generate conflicting biologic outcomes.7-9
In addition to expression in thymus, Bcl11b is expressed in peripheral lymphoid organs including spleen and lymph nodes.10 Bcl11b is expressed in peripheral blood lymphocytes,10 mature naive and activated CD4+ T lymphocytes (this report), as well as in the human CD4+ T-cell line Jurkat.11,12 Taking into consideration that Bcl11b is expressed in CD4+ T cells both in resting and activated states, we investigated whether BCL11B plays a role in transcriptional control of IL2 gene expression. The IL2 gene is one of the first genes whose expression is induced immediately after activation of CD4+ T cells through TCR/CD28 and plays a central role in T-cell proliferation and homeostasis.13 In this report, we demonstrate through several lines of evidence that BCL11B regulates expression of the IL2 gene by direct binding to the US1 site in the IL2 promoter and by interaction with the coactivator p300 in response to TCR/CD28 stimulation. To our knowledge, these studies describe for the first time a cellular target gene regulated by BCL11B. In addition, our data reveal a new molecular role for BCL11B, that of a transcriptional activator in the context of T-cell activation.
Materials and methods
Plasmids
The reporter construct -585 IL2 promoter-luciferase, containing the proximal 585 bp of the murine IL2 promoter, was a kind gift from Dr Michael Bell.14 IL2 promoter constructs -190, -210, -243, -254, -290, -390, and -508 were generated by polymerase chain reaction (PCR) and cloned in pGL3 Vector (Promega, Madison, WI). IL-2-243 mutA and mutAB promoter constructs were generated using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). Generation of Flag-Bcl11b construct was described previously.11,12 The construct corresponds to the BCL11B splice isoform 2 or Ritβ (GeneID 64919). Flag-Bcl11b was subcloned in the bicistronic murine stem cell virus (MSCV) vector (Clontech, Palo Alto, CA) (MSCV-BCL11B), in which the expression of Flag-Bcl11b is driven under the LTR control. All the constructs were verified by sequencing.
Antibodies and reagents
Anti-BCL11B (B26-44) polyclonal antibodies were described previously.12 Additional anti-BCL11B antibodies were purchased from Bethyl Laboratories (Montgomery, TX). The mouse anti-Flag (M2) and antiactin antibodies were purchased from Sigma (St Louis, MO). The anti-human CD3 (OKT), anti-mouse CD3, anti-human CD28, and anti-mouse CD28 are from eBioscience (San Diego, CA). IL-2 was obtained from the AIDS Research and Reference/Reagent Program (National Institutes of Health, Germantown, MD).
Cell lines and transduction
Jurkat cells were obtained from ATCC (American Type Culture Collection, Manassas, VA) and grown as per ATCC recommendations. Mouse splenic CD4+ T cells were purified from single cell suspensions using CD4 (L3T4) MACS Microbeads (Miltenyi Biotech, Auburn, CA) following manufacturer's protocol. The purity of the sorted cells was determined to be at least 98% by fluorescence-activated cell-sorter scanner (FACS) analysis using fluorochrome conjugated anti-CD4 antibodies. The sorted cells were grown in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 50 U/mL penicillin, 50 μg/mL streptomycin, 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), nonessential amino acids, 1 mM sodium pyruvate, and 50 μM β-mercaptoethanol. CD4+ T cells were stimulated with cross-linked anti-CD3 (10 μg/mL) and soluble anti-CD28 (2.5 μg/mL) antibodies for 48 hours and then infected with recombinant retroviruses. After infection cells were cultivated in media supplemented with 100 U/mL recombinant human IL-2 and cultured for 4 days. For the second activation, the cells were resuspended into fresh media and treated with phorbol myristate acetate (PMA) and ionomycin or anti-CD3 and anti-CD28, as indicated in the figure legends.
Recombinant retroviruses were generated in Phoenix A or E cells (a kind gift from Dr Gary Nolan). The retroviral supernatants were collected and supplemented with 8 μg/mL polybrene. Jurkat and mouse CD4+ T cells were infected by spinoculation and then sorted with a high-speed cell sorter device (BD Bioscience, Palo Alto, CA) and grown as populations.
Transient transfections and luciferase assays
107 MSCV and MSCV-BCL11B Jurkat cells were transfected by electroporation with 10 μg Firefly luciferase reporter plasmid and 0.5 μg Renilla luciferase as previously described.12 The next day, the cells were equally aliquoted and treated with 10 μg/mL plate-bound anti-human CD3 antibody and 2.5 μg/mL anti-human CD28 antibody, or 50 ng/mL PMA and 1 μM ionomycin. The cells were harvested 24 hours after transfection, and the reporter assays were conducted as previously described.12 Three to 5 independent experiments were conducted in each case, and the quantifications represent means ± SD.
Reverse transcription and real-time PCR—quantitative RT-PCR (qRT-PCR)
Stimulated cells (5 × 106) were harvested, and RNA was isolated using the NucleoSpin RNA kit (Clontech). cDNA was synthesized starting with 1 to 5 μg of total RNA using SuperScript (Invitrogen, Carlsbad, CA), and quantitative PCR was conducted starting with 1/20 to 1/10 of the reverse transcription (RT) reaction using 2 × Sybergreen mix in an iCycler real-time PCR machine (BioRad, Hercules, CA). Negative controls in which the reverse transcriptase was omitted from the RT reaction were conducted in each case. The primers for real-time PCR were chosen such as to cross at least 1 intron, extend products under 200 bp and with no formation of primer dimers. Information on the primer sequences is available upon request. The relative abundance of each message was calculated by normalization to actin in 3 independent experiments and calculated as: 2-(Ct gene - Ct actin), where Ct represents the threshold cycle for each transcript.
Chromatin immunoprecipitation assays (ChIPs)
ChIP assays were conducted as previously described.12 The primer sequence for the human IL2 promoter is 5′-Attcacatgttcagtgtagttt-3′ and 5′-Gtgaaatccctctttgttaca-3′. The primer sequence for the mouse Il2 promoter is 5′-aggaaaatttgtttcatacag-3′ and 5′-tcttcagcatgggaggcaat-3′.
Gene knockdown by small interfering RNA (siRNA)
BCL11B-specific and control nontargeting siRNAs were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA) and Dharmacon (Lafayette, CO). Jurkat cells (5 × 105) were electroporated with 350 pmols siRNA using the Ambion reaction buffer and the BTX Electro Square Porator ECM 830 (Harvard Apparatus, Holliston, MA). A second electoporation was conducted the following day with reporter plasmids as described “Transient transfections and luciferase assays.” Twenty-four hours after transfection the cells were treated with PMA and ionomycin for 7 hours and then harvested for reporter assays and immunoblot analysis.
Results
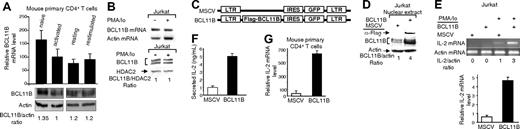
BCL11B is expressed in primary and transformed CD4+ T lymphocytes, both in resting and activated states
Previous data have demonstrated that BCL11B is expressed in spleen, as well as in peripheral blood lymphocytes.10 We therefore investigated the expression of BCL11B in murine splenic CD4+ T lymphocytes, under naive, activated, and resting states both by qRT-PCR and Western blot analysis. The results demonstrate that BCL11B is expressed at similar levels in activated, resting, and restimulated CD4+ T lymphocytes, however, at slightly higher levels in naive cells, as evaluated both by measuring the message levels (Figure 1A, top), as well as the protein levels (Figure 1A, bottom). As BCL11B is known to be expressed in the CD4+ T-cell line Jurkat,11,12 we investigated the expression of BCL11B upon activation of these cells with PMA and ionomycin, which are used to mimic activation through TCR/CD28.15 Our results show that BCL11B is present both in resting and activated Jurkat cells at similar levels (Figure 1B). The 2 bands correspond to the 2 previously described splice isoforms of BCL11B.6,12 The splice isoform 1 or Ritα contains exons 1 to 4, while the splice isoform 2 or Ritβ is identical to the isoform 1, except it lacks exon 3, which encodes 60 amino acids. It has not been established yet if these 2 splice isoforms play different roles in T cells.
Sustained BCL11B expression enhances the level of endogenous IL-2 message in activated CD4+ T cells
Given the fact that BCL11B is expressed in CD4+ T cells both in resting and activated states (Figure 1A-B), we explored a potential role of BCL11B in events occurring early after T-cell activation, such as induction of IL2 gene expression.16 For this purpose, Jurkat cells were stably transduced with retroviruses expressing GFP (MSCV) and Flag-BCL11B-IRES-GFP (MSCV-BCL11B) (Figure 1C). For these experiments we used the splice isoform 2 (Rit1β), lacking exon 3.6 Populations of cells were sorted based on expression of GFP, expanded, and tested for the expression of Flag-BCL11B (Figure 1D) and IL-2 message levels. Activated MSCV-BCL11B cells had significantly higher levels of IL-2 mRNA than cells transduced with vector only, as demonstrated by RT-PCR (Figure 1E, top) and qRT-PCR (Figure 1E, bottom). Ectopic expression of BCL11B alone was not sufficient to induce IL2 gene expression, as no IL-2 was detected in the absence of PMA/ionomycin treatment (Figure 1E, top). ELISA (enzyme-linked immunosorbent assay) analysis of the media from activated MSCV-BCL11B and MSCV Jurkat cells revealed that MSCVBCL11B cells produced more IL-2 compared to MSCV (Figure 1F).
To confirm these results in primary cells, we transduced CD4+ T splenic lymphocytes with MSCV-BCL11B or MSCV retroviruses. Following stimulation with PMA/ionomycin, expression of the IL2 gene was evaluated by qRT-PCR. Enhanced expression of BCL11B resulted in a significant increase in the IL-2 mRNA after stimulation of the cells with PMA/ionomycin (Figure 1G). To exclude the possibility that the enhanced level of IL-2 in MSCVBCL11B-transduced CD4+ T cells is due to reduction in the number of T regulatory cells, we investigated the percentage of Foxp3-positive cells in MSCV-BCL11B and MSCV-transduced cells. Foxp3-expressing cells were found at similar frequencies in the 2 populations of transduced cells (Figure S1, available at the Blood website; see the Supplemental Figure link at the top of the online article). These results indicate that BCL11B enhances IL-2 message levels both in Jurkat and primary CD4+ T cells, and this requires T-cell activation.
BCL11B up-regulates the levels of IL-2 mRNA in transformed and primary CD4+ T lymphocytes. (A, top) Expression of BCL11B evaluated by qRT-PCR in primary CD4+ T cells in the following conditions: freshly isolated (naive), stimulated for 24 hours with anti-CD3/anti-CD28 (activated), rested for 72 hours following the anti-CD3/anti-CD28 treatment (resting), or rested and restimulated with 50 ng/mL PMA and 1 μM ionomycin for 4 hours (restimulated). The relative abundance of BCL11B message was normalized to actin and calculated as described in “Reverse transcription and real-time PCR—quantitative RT-PCR (qRT-PCR).” (Bottom) Western blot analysis of BCL11B in primary CD4+ T cells treated in the conditions shown in the top panel; BCL11B levels were quantified by densitometric analysis and normalized to actin. (B) BCL11B expression evaluated by RT-PCR (top) and Western blot analysis (bottom) in Jurkat cells grown in the presence or absence of PMA (50 ng/mL) and ionomycin (1 μM) for 4 hours. BCL11B levels were quantified by densitometric analysis and normalized to HDAC2. (C) Schematic representation of the MSCV and MSCV-BCL11B retroviral constructs. LTR represents long terminal repeat; IRES, internal ribosomal entry site. (D) Western blot analysis demonstrating expression of Flag-BCL11B in retrovirally transduced Jurkat cells. (E) Levels of IL-2 mRNA evaluated by RT-PCR (top) and qRT-PCR (bottom) before (bottom) and after (both subpanels) treatment for 4 hours with PMA/ionomycin in MSCV and MSCV-BCL11B Jurkat cells. (F) Secretion of IL-2 determined by ELISA in media from MSCV and MSCV-BCL11B Jurkat cells after treatment with PMA/ionomycin for 16 hours. (G) Levels of IL-2 mRNA in MSCV and MSCV-BCL11B primary mouse CD4+ T cells after retroviral transduction and sorting of populations of GFP-positive cells. Cells were stimulated with PMA/ionomycin for 5 hours and the levels of IL-2 message were evaluated by qRT-PCR. The relative abundance of IL-2 message was normalized to actin and calculated as described in “Reverse transcription and real-time PCR—quantitative RT-PCR (qRT-PCR).” Three to 5 experiments were conducted, and the quantifications represent means ± SD.
BCL11B up-regulates the levels of IL-2 mRNA in transformed and primary CD4+ T lymphocytes. (A, top) Expression of BCL11B evaluated by qRT-PCR in primary CD4+ T cells in the following conditions: freshly isolated (naive), stimulated for 24 hours with anti-CD3/anti-CD28 (activated), rested for 72 hours following the anti-CD3/anti-CD28 treatment (resting), or rested and restimulated with 50 ng/mL PMA and 1 μM ionomycin for 4 hours (restimulated). The relative abundance of BCL11B message was normalized to actin and calculated as described in “Reverse transcription and real-time PCR—quantitative RT-PCR (qRT-PCR).” (Bottom) Western blot analysis of BCL11B in primary CD4+ T cells treated in the conditions shown in the top panel; BCL11B levels were quantified by densitometric analysis and normalized to actin. (B) BCL11B expression evaluated by RT-PCR (top) and Western blot analysis (bottom) in Jurkat cells grown in the presence or absence of PMA (50 ng/mL) and ionomycin (1 μM) for 4 hours. BCL11B levels were quantified by densitometric analysis and normalized to HDAC2. (C) Schematic representation of the MSCV and MSCV-BCL11B retroviral constructs. LTR represents long terminal repeat; IRES, internal ribosomal entry site. (D) Western blot analysis demonstrating expression of Flag-BCL11B in retrovirally transduced Jurkat cells. (E) Levels of IL-2 mRNA evaluated by RT-PCR (top) and qRT-PCR (bottom) before (bottom) and after (both subpanels) treatment for 4 hours with PMA/ionomycin in MSCV and MSCV-BCL11B Jurkat cells. (F) Secretion of IL-2 determined by ELISA in media from MSCV and MSCV-BCL11B Jurkat cells after treatment with PMA/ionomycin for 16 hours. (G) Levels of IL-2 mRNA in MSCV and MSCV-BCL11B primary mouse CD4+ T cells after retroviral transduction and sorting of populations of GFP-positive cells. Cells were stimulated with PMA/ionomycin for 5 hours and the levels of IL-2 message were evaluated by qRT-PCR. The relative abundance of IL-2 message was normalized to actin and calculated as described in “Reverse transcription and real-time PCR—quantitative RT-PCR (qRT-PCR).” Three to 5 experiments were conducted, and the quantifications represent means ± SD.
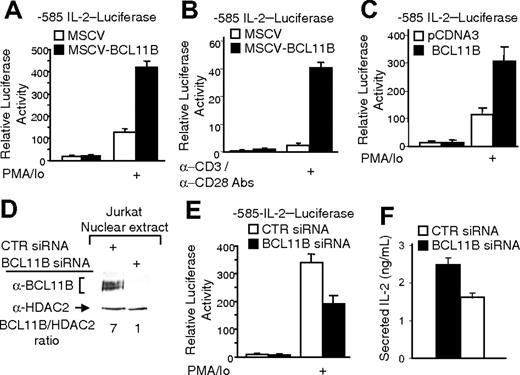
BCL11B augments activation of the IL-2 promoter in activated CD4+ T cells
To test the hypothesis that the higher levels of IL-2 message in MSCV-BCL11B Jurkat cells resulted from increased IL-2 gene expression rather than from regulation of message stability, we conducted transient transfections and reporter assays with the -585 IL-2 promoter-luciferase construct, comprising 585 bp of the IL-2 promoter upstream of the mRNA start site.14 We chose this region of IL-2 promoter as it contains most of the sites known to be involved in the induction of IL2 gene expression.17 Similar to our results on endogenous IL-2 message levels, luciferase activity was higher in activated MSCV-BCL11B cells compared to MSCV (Figure 2A). In the absence of stimulation with PMA/ionomycin, there was no significant difference in the reporter assays in MSCV and MSCV-BCL11B cells and, as expected, the reporter activity was very low (Figure 2A).
We then wanted to determine if the up-regulation of the IL-2 promoter occurred under physiologic conditions. Thus, MSCVBCL11B and control MSCV Jurkat cells were activated with cross-linked anti-CD3 and soluble anti-CD28 antibodies. In these conditions we observed a 10-fold higher induction of the promoter in MSCV-BCL11B cells compared to MSCV control (Figure 2B). To exclude the possibility that the enhanced expression of IL-2 gene in MSCV-BCL11B cells is due to the fact that the transfectants carry altered expression of endogenous genes that affect IL-2 gene expression, we conducted experiments in which we measured the IL-2 promoter activity in cells transiently expressing BCL11B, and the results were similar with stable transductants (Figure 2C).
These data taken together demonstrate that BCL11B activates expression from IL-2 promoter and activation occurs upon T-cell activation through TCR/CD28.
BCL11B regulates IL-2 promoter activity. (A-B) MSCV-BCL11B and MSCV Jurkat cells were electroporated with the -585 IL-2 promoter-Firefly luciferase and CMV-Renilla luciferase constructs. Twenty-four hours after transfection, the cells were treated with PMA/ionomycin for 8 hours (A) or grown for 8 hours in the presence of plate-bound anti-CD3 and soluble anti-CD28 antibodies (B). The luciferase activity was measured using the Promega luciferase dual system. (C) Jurkat cells were electroporated with pCDNA3::BCL11B or pCDNA3 vectors and -585 IL-2 promoter-Firefly luciferase and CMV-Renilla luciferase constructs. Luciferase activity was measured as in panels A and B. (D) Western blot analysis of BCL11B after transfection of Jurkat cells with BCL11B specific (lane 2) or nontargeting (lane 1) siRNAs. HDAC2 was used as loading control. (E-F) Jurkat cells were transfected with the -585 IL-2 promoter-Firefly luciferase, CMV-Renilla luciferase constructs, and BCL11B specific or nontargeting siRNAs. (E) Luciferase activity was measured after PMA/ionomycin treatment for 8 hours; control cells lacked PMA/ionomycin treatment. (F) Secretion of IL-2 was determined by ELISA in media after treatment with PMA/ionomycin for 16 hours. The quantifications represent means of 3 independent experiments ± SD.
BCL11B regulates IL-2 promoter activity. (A-B) MSCV-BCL11B and MSCV Jurkat cells were electroporated with the -585 IL-2 promoter-Firefly luciferase and CMV-Renilla luciferase constructs. Twenty-four hours after transfection, the cells were treated with PMA/ionomycin for 8 hours (A) or grown for 8 hours in the presence of plate-bound anti-CD3 and soluble anti-CD28 antibodies (B). The luciferase activity was measured using the Promega luciferase dual system. (C) Jurkat cells were electroporated with pCDNA3::BCL11B or pCDNA3 vectors and -585 IL-2 promoter-Firefly luciferase and CMV-Renilla luciferase constructs. Luciferase activity was measured as in panels A and B. (D) Western blot analysis of BCL11B after transfection of Jurkat cells with BCL11B specific (lane 2) or nontargeting (lane 1) siRNAs. HDAC2 was used as loading control. (E-F) Jurkat cells were transfected with the -585 IL-2 promoter-Firefly luciferase, CMV-Renilla luciferase constructs, and BCL11B specific or nontargeting siRNAs. (E) Luciferase activity was measured after PMA/ionomycin treatment for 8 hours; control cells lacked PMA/ionomycin treatment. (F) Secretion of IL-2 was determined by ELISA in media after treatment with PMA/ionomycin for 16 hours. The quantifications represent means of 3 independent experiments ± SD.
A decrease in the level of endogenous BCL11B by siRNA reduces the level of expression from IL-2 promoter in activated CD4+ T cells
The results described demonstrate that higher expression of BCL11B significantly enhances the IL-2 promoter activity. To test whether endogenous BCL11B participates in regulation of expression from IL-2 promoter, we employed siRNA technology to decrease the level of endogenous BCL11B in Jurkat cells (Figure 2D). The decrease in endogenous BCL11B resulted in a significant reduction of expression from IL-2 promoter, demonstrating that endogenous BCL11B is involved in regulation of expression from IL-2 promoter (Figure 2E). In addition, the levels of secreted IL-2 in the culture media were decreased (Figure 2F).
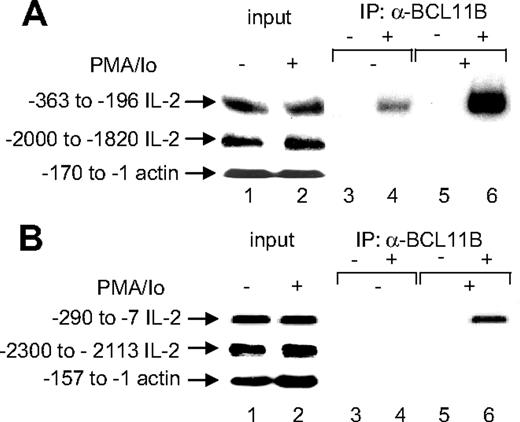
Endogenous BCL11B associates with the IL-2 promoter in Jurkat and primary CD4+ T lymphocytes
We then asked whether BCL11B associates with the IL-2 promoter by conducting ChIP assays. Immunoprecipitation of the cross-linked chromatin from Jurkat cells with anti-BCL11B antibodies, but not with IgG, enriched significantly the IL-2 promoter within a region between -363 and -196, but not in the genomic region located between -2000 and -1820 upstream of IL-2 mRNA start site, or actin promoter (Figure 3A, lanes 3-6). These experiments were conducted in Jurkat cells in the absence of T-cell activation (Figure 3A, lanes 3 and 4) or in Jurkat cells treated with PMA/ionomycin (Figure 3A, lanes 5 and 6). Interestingly, in activated cells the association of BCL11B with the IL-2 promoter was enhanced as compared to resting conditions (Figure 3A, compare lanes 6 and 4). This was not due to up-regulation of BCL11B in response to PMA/ionomycin treatment, as the level of expression of BCL11B in Jurkat cells was not significantly affected by PMA/ionomycin treatment (Figure 1B).
As Jurkat cells are transformed, we wanted to address the question in regard to the association of endogenous BCL11B with the IL-2 promoter in primary CD4+ T cells, in resting and activated states. Immunoprecipitation of the cross-linked chromatin with anti-BCL11B antibodies, but not with IgG, significantly enriched the IL-2 promoter from activated primary CD4+ T cells (Figure 3B). The enrichment was specific, as the genomic region located between -2300 and -2113 upstream from the IL-2 mRNA start site or actin promoter were not enriched (Figure 3B). These results demonstrate that endogenous BCL11B is associated with the IL-2 promoter in primary CD4+ T cells after activation, similar to what was observed in Jurkat cells.
Endogenous BCL11B associates with the IL-2 promoter in Jurkat and primary CD4+ T cells. Immunoprecipitation (IP) of chromatin from Jurkat cells (A) or primary CD4+ T cells (B) stimulated with PMA/ionomycin (lanes 5 and 6) or no stimulation (lanes 3 and 4) using anti-BCL11B (lanes 4 and 6) or nonspecific (lanes 3 and 5) antibodies. Eluted DNA was analyzed by PCR using human (A) or mouse (B) IL-2 promoter-specific primers or actin promoter primers covering the indicated regions. In each case, lanes 1 and 2 represent input from untreated and treated cells, respectively.
Endogenous BCL11B associates with the IL-2 promoter in Jurkat and primary CD4+ T cells. Immunoprecipitation (IP) of chromatin from Jurkat cells (A) or primary CD4+ T cells (B) stimulated with PMA/ionomycin (lanes 5 and 6) or no stimulation (lanes 3 and 4) using anti-BCL11B (lanes 4 and 6) or nonspecific (lanes 3 and 5) antibodies. Eluted DNA was analyzed by PCR using human (A) or mouse (B) IL-2 promoter-specific primers or actin promoter primers covering the indicated regions. In each case, lanes 1 and 2 represent input from untreated and treated cells, respectively.
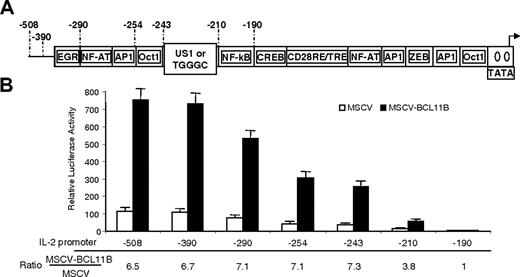
BCL11B augments expression of IL-2 promoter through a region located between -243 and -190
To test whether BCL11B augments expression of the IL-2 promoter through a specific site, we generated several deletion mutants of IL-2 promoter (Figure 4). Deletion of the region between -243 and -210 resulted in a significant reduction in the augmentation of IL-2 promoter activity by BCL11B (Figure 4), while removal of the region between -508 and -243 had no effect on BCL11B-mediated augmentation of IL-2 promoter activity (Figure 4). In addition, deletion of the region between -210 and -190 resulted in an approximately 3-fold reduction of the augmentation mediated by BCL11B on IL-2 promoter, making the levels of the reporter in MSCV-BCL11B and MSCV cells approximately equal (Figure 4).
These results suggest that BCL11B acts on the IL-2 promoter through sites located between -243 and -190.
BCL11B binds and activates the IL-2 promoter through the US1 site
The region of IL-2 promoter between -243 and -210, named the upstream site 1 (US1), is less characterized compared to other regions of the IL-2 promoter.18 We used electrophoretic mobility shift assays (EMSAs) to investigate the binding of BCL11B to the US1 region. As a negative control, we used a DNA fragment covering the IL-2 promoter region from -290 through -243, which contains NF-AT, AP1, and Oct1 binding sites, and whose deletion did not impact the enhancement of IL-2 promoter activity by higher levels of BCL11B (Figure 4). Purified recombinant BCL11B bound the US1 site, but not the negative control (Figure 5A). In addition, we conducted EMSAs with nuclear extracts from Jurkat cells treated with PMA/ionomycin to determine whether endogenous BCL11B binds to the US1 site. Our results show that endogenous BCL11B is present in a complex with the US1 site, as demonstrated by the fact that this complex was specifically shifted by an anti-BCL11B antibody, but not by a nonspecific antibody (Figure 5B, lane 3 versus lane 2).
To narrow down the region(s) within the US1 site required for BCL11B binding, we conducted EMSAs with 2 DNA fragments: the first covering the 5′ distal, and the second covering the 3′ proximal half of the US1 site. BCL11B bound only the proximal half of the US1 site (Figure 5C).
We then conducted mutational analysis of the proximal US1 site to identify the bases required for BCL11B binding (Figure 5D). For this purpose, we mutated every 2 bases to AA, because BCL11B does not bind sites with such mutations.2 Eight mutants were generated, M1 to M8 (Figure 5D, top). As expected, the wild-type (WT) cold competitor inhibited the binding of BCL11B to the US1 proximal site (Figure 5D, bottom, compare WT to no competitor). Mutants M1 to M3 and M6 also inhibited BCL11B binding to the proximal US1 site (Figure 5D, lower panel, compare M1, M2, M3, and M6 to no competitor). Mutants M4 and M5, covering the GGGC of the TGGGC motif, previously demonstrated to be bound by an unknown transcription factor or complex in response to T-cell activation,19 had little effect on BCL11B binding (Figure 5D, bottom, compare M4 and M5 to no competitor), suggesting that these bases are important for BCL11B binding. In addition, mutants M7 and M8 inhibited BCL11B binding slightly more than M4 and M5, however, significantly less robustly than wild-type, suggesting that these bases also contribute to the binding of BCL11B to US1 proximal site (Figure 5D, compare M7 and M8 to no competitor). We therefore named the first site corresponding to mutants M4 and M5, site A, and the second site corresponding to mutants M7 and M8, site B (Figure 5D, top).
IL-2 promoter activation by BCL11B requires the region between -243 and -190. (A) Schematic representation of the IL-2 promoter deletion mutants. (B) Luciferase reporter assays from MSCV-BCL11B (▪) and MSCV (□) Jurkat cells transfected with the IL-2 promoter deletion mutants. Ratios of luciferase activity between MSCV-BCL11B and MSCV Jurkat cells for each deletion mutant are shown. Reporter assays were conducted after treatment of the cells with PMA/ionomycin for 8 hours. The quantifications represent means of 3 independent experiments ± SD.
IL-2 promoter activation by BCL11B requires the region between -243 and -190. (A) Schematic representation of the IL-2 promoter deletion mutants. (B) Luciferase reporter assays from MSCV-BCL11B (▪) and MSCV (□) Jurkat cells transfected with the IL-2 promoter deletion mutants. Ratios of luciferase activity between MSCV-BCL11B and MSCV Jurkat cells for each deletion mutant are shown. Reporter assays were conducted after treatment of the cells with PMA/ionomycin for 8 hours. The quantifications represent means of 3 independent experiments ± SD.
We then wanted to assess the contribution of the mutated sites to the transcriptional regulatory function of BCL11B in T cells in reporter assays. For this purpose we generated reporter constructs in which we mutated the US1 site bases shown by EMSAs to be critical for BCL11B binding. We generated 2 mutant reporter constructs: mutA, in which site A was mutated; and mutAB, in which we mutated both site A and B (Figure 5E, top). In addition to evaluating these mutants in relation to wild-type, we also compared them to a reporter construct containing -210 IL-2 promoter, which lacks the US1 site. Consistent with the EMSA results, the reporter activity in mutAB dropped to the level of -210 IL-2 promoter, indicating that the enhancement of IL-2 promoter activity by BCL11B is mediated through sites required for BCL11B binding (Figure 5E). BCL11B-mediated transcriptional enhancement was only partially reduced within mutA (Figure 5E), which is consistent with the EMSA results (Figure 5D).
Thus, BCL11B functionally binds and activates the IL-2 promoter through a region located in the proximal half of the US1 site.
cAMP elevation reduces augmentation of IL-2 promoter activity by BCL11B
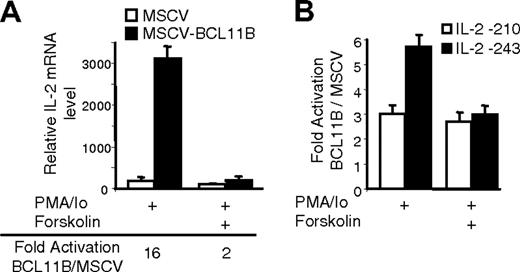
As mentioned, mutants M4 and M5 cover the previously characterized TGGGC motif bound by an unknown transcription factor or complex in response to T-cell activation in a manner sensitive to forskolin.19 We therefore investigated whether forskolin influenced transcriptional augmentation of the IL2 gene expression by BCL11B.
Our results demonstrated that forskolin treatment blocked augmentation of the IL2 gene expression by BCL11B (Figure 6A). To demonstrate that forskolin blocks BCL11B-mediated IL-2 promoter activation by disrupting BCL11B activity on the US1 site, we conducted reporter assays with 2 IL-2 promoter deletion mutants, containing or not the US1 site. BCL11B-mediated activation of the IL-2 promoter construct containing the US1 site was 2-fold higher than that of the mutant lacking the US1 site (Figure 6B). When the cells were pretreated with forskolin, BCL11B-mediated activation of both IL-2 promoter deletion mutants was almost equal, regardless of the presence or absence of the US1 site (Figure 6B). In addition, by conducting EMSA we found that the formation of the US1-BCL11B complex was weaker when Jurkat cells were treated with forskolin (Figure 5B, lower panel).
BCL11B binds and activates the IL-2 promoter through the US1 site. (A) EMSA using purified recombinant GST-BCL11B (lane 3) or GST (lane 2) and US1 (ii) or NFAT-AP1-Oct DNA probes (i). Lane 1 is free probe. (B) EMSA using nuclear extracts from Jurkat cells and the US1 probe. In the top panel the US1 probe was incubated with 5 μg nuclear extracts from Jurkat cells treated with PMA and ionomycin (lanes 1-3) and anti-BCL11B (lane 3) or nonspecific antibodies (lane 2). BCL11B-US1 complexes are indicated by the black arrow, and the antibody-supershifted BCL11B-US1 complex is indicated by the white arrow. In the bottom panel the US1 probe was incubated with extracts from cells treated with PMA/ionomycin or forskolin plus PMA/ionomycin. Treatments were conducted as described in Figure 6. (C) EMSA using purified GST-BCL11B (lanes 3) or GST (lanes 2) and proximal (ii) or distal (i) US1 DNA fragments. Lane 1 is free probe. (D) Mutational analysis of the BCL11B binding site. Top panel shows the sequence of the BCL11B binding site. Underlined are the bases mutated in each mutant (M1-M8). Middle panel shows EMSAs with purified recombinant GST-BCL11B, 32P-labeled BCL11B binding site probe plus excess of unlabeled WT or mutant DNA competitors (M1-M8). The bottom panel represents the quantification of the relative intensity of the corresponding bands. Maximal binding of the probe was determined in the sample lacking competitor DNA. For simplicity EMSAs in panels A-D present only protein-DNA complexes. (E) Reporter assays from MSCV-BCL11B Jurkat cells transfected with the following IL-2 promoter-luciferase constructs: wild-type -210 (lane 1), wild-type -243 (lane 2), mutant A -243 (lane 3), and mutant AB -243 (lane 4) (see top panel). In each case cells were treated with PMA/ionomycin for 8 hours before harvesting. Three experiments were conducted, and the quantifications represent means ± SD.
BCL11B binds and activates the IL-2 promoter through the US1 site. (A) EMSA using purified recombinant GST-BCL11B (lane 3) or GST (lane 2) and US1 (ii) or NFAT-AP1-Oct DNA probes (i). Lane 1 is free probe. (B) EMSA using nuclear extracts from Jurkat cells and the US1 probe. In the top panel the US1 probe was incubated with 5 μg nuclear extracts from Jurkat cells treated with PMA and ionomycin (lanes 1-3) and anti-BCL11B (lane 3) or nonspecific antibodies (lane 2). BCL11B-US1 complexes are indicated by the black arrow, and the antibody-supershifted BCL11B-US1 complex is indicated by the white arrow. In the bottom panel the US1 probe was incubated with extracts from cells treated with PMA/ionomycin or forskolin plus PMA/ionomycin. Treatments were conducted as described in Figure 6. (C) EMSA using purified GST-BCL11B (lanes 3) or GST (lanes 2) and proximal (ii) or distal (i) US1 DNA fragments. Lane 1 is free probe. (D) Mutational analysis of the BCL11B binding site. Top panel shows the sequence of the BCL11B binding site. Underlined are the bases mutated in each mutant (M1-M8). Middle panel shows EMSAs with purified recombinant GST-BCL11B, 32P-labeled BCL11B binding site probe plus excess of unlabeled WT or mutant DNA competitors (M1-M8). The bottom panel represents the quantification of the relative intensity of the corresponding bands. Maximal binding of the probe was determined in the sample lacking competitor DNA. For simplicity EMSAs in panels A-D present only protein-DNA complexes. (E) Reporter assays from MSCV-BCL11B Jurkat cells transfected with the following IL-2 promoter-luciferase constructs: wild-type -210 (lane 1), wild-type -243 (lane 2), mutant A -243 (lane 3), and mutant AB -243 (lane 4) (see top panel). In each case cells were treated with PMA/ionomycin for 8 hours before harvesting. Three experiments were conducted, and the quantifications represent means ± SD.
These results taken together demonstrate that BCL11B binds to the US1 site directly, and several bases in this site, including the GGGC of the previously characterized TGGGC motif, are crucial for its binding. This finding, together with the sensitivity to forskolin of the BCL11B-mediated transcriptional augmentation of IL-2 gene expression, suggests the possibility that BCL11B may be the factor or one of the factors found previously to bind the IL-2 promoter through the TGGGC site in a forskolin-sensitive manner.19
BCL11B interacts with the p300 coactivator after stimulation of CD4+ T cells
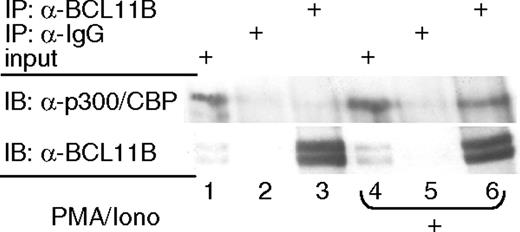
It has been demonstrated that NF-kB, NF-AT, AP-1, and CREB interact with the p300 coactivator, forming an enhanceosome on the IL-2 promoter.20 Therefore, we investigated whether BCL11B also interacts with the p300 coactivator, participating in the recruitment of the coactivator complex to the IL-2 promoter. The presence of p300 in the immunoaffinity purified BCL11B complexes, but not in the IgG complexes, was demonstrated by Western blot analysis, suggesting therefore that endogenous BCL11B associates with p300 (Figure 7). This association was detected only in activated lymphocytes, but not under resting conditions (Figure 7).
These results show that in activating conditions BCL11B associates with p300, which is therefore likely to mediate its transcriptional activation function.
Discussion
Here we describe a new molecular function for BCL11B, as well as the identification of IL-2 as a target gene regulated by BCL11B. Specifically, we demonstrate through several lines of evidence that following activation of CD4+ T lymphocytes through TCR/CD28 signaling, BCL11B functions as a transcriptional activator and participates in the activation of IL-2 gene expression: (1) We show that enhanced expression of BCL11B augments IL-2 mRNA levels both in transformed and primary CD4+ T cells. (2) In addition, we prove that this activation occurs at the level of IL2 gene transcription. (3) We demonstrate that endogenous BCL11B plays a role in the IL-2 promoter regulation, as knockdown of BCL11B by siRNA diminishes the IL-2 promoter activation. (4) We demonstrate that endogenous BCL11B associates with the IL-2 promoter in both Jurkat and primary CD4+ T lymphocytes. (5) We prove that BCL11B participates in the activation of IL-2 promoter through sites located between -243 and -190 and show that BCL11B directly binds to the US1 site located within this region. (6) We show that BCL11B associates with the p300 coactivator in cells activated through TCR/CD28, which may account for its transcriptional activation function.
Forskolin blocks augmentation of IL-2 promoter activity by BCL11B. (A) IL-2 mRNA levels in MSCV (□) and MSCV-BCL11B (▪) Jurkat cells stimulated for 4 hours with PMA/ionomycin and pretreated or not with 10 μM forskolin for 30 minutes before stimulation. IL-2 mRNA levels were evaluated by qRT-PCR and normalized by actin in each sample. (B) Ratios of luciferase reporter assays between MSCV-BCL11B and MSCV Jurkat cells transfected with -210 IL-2 promoter (□) or -243 IL-2 promoter (▪). The cells were stimulated with PMA/ionomycin for 8 hours and pretreated or not with 10 μM forskolin for 30 minutes.
Forskolin blocks augmentation of IL-2 promoter activity by BCL11B. (A) IL-2 mRNA levels in MSCV (□) and MSCV-BCL11B (▪) Jurkat cells stimulated for 4 hours with PMA/ionomycin and pretreated or not with 10 μM forskolin for 30 minutes before stimulation. IL-2 mRNA levels were evaluated by qRT-PCR and normalized by actin in each sample. (B) Ratios of luciferase reporter assays between MSCV-BCL11B and MSCV Jurkat cells transfected with -210 IL-2 promoter (□) or -243 IL-2 promoter (▪). The cells were stimulated with PMA/ionomycin for 8 hours and pretreated or not with 10 μM forskolin for 30 minutes.
BCL11B associates with the p300 coactivator in activated Jurkat cells. Immunoprecipitation (IP) with anti-BCL11B (lanes 3 and 6) or nonspecific (lanes 2 and 5) antibodies and Western blot (IB) analysis with anti-p300 and anti-BCL11B antibodies. Nuclear extracts were prepared from Jurkat cells treated (lanes 4-6) or not (lanes 1-3) with PMA/ionomycin. Lanes 1 and 4 represent input.
BCL11B associates with the p300 coactivator in activated Jurkat cells. Immunoprecipitation (IP) with anti-BCL11B (lanes 3 and 6) or nonspecific (lanes 2 and 5) antibodies and Western blot (IB) analysis with anti-p300 and anti-BCL11B antibodies. Nuclear extracts were prepared from Jurkat cells treated (lanes 4-6) or not (lanes 1-3) with PMA/ionomycin. Lanes 1 and 4 represent input.
BCL11B and the closely related BCL11A are C2H2 zinc finger proteins that have been previously found to elicit transcriptional repression.1,2,11,12,21 Recent studies revealed that BCL11B acts as a transcriptional repressor in resting lymphocytes through association with the NuRD complex and requires MTA1 for this activity.12 In this study we show that BCL11B behaves as a transcriptional activator after stimulation of CD4+ T lymphocytes. Therefore, our new findings suggest that BCL11B may function both as a transcriptional repressor and as a transcriptional activator. This apparently opposite behavior is not unusual, and it has been documented for other C2H2 zinc finger transcription factors, such as YY1 and Ikaros.22,23 In the case of BCL11B, one explanation may reside in the nature of the complexes with which BCL11B associates. The interaction of BCL11B with coactivators (p300) or corepressors (NuRD complex) may be modulated by posttranslational modifications of BCL11B in response to different signaling pathways, including those downstream of TCR activation. Further studies are required to test this hypothesis and identify the modifications and the specific signals that trigger them. Interestingly, BCL11B was not found to be associated with the IL-2 promoter in primary naive cells, but only in activated cells, though the levels of BCL11B were slightly higher in naive cells. One possibility is that the association of BCL11B with the IL-2 promoter is extremely reduced in conditions in which the IL-2 promoter is highly compacted and silenced, possibly due to the fact that the US1 site is not exposed. Alternatively, BCL11B-NuRD complex may be present on the IL-2 promoter in naive cells and may participate in the silencing of IL-2 promoter, but due to the highly compacted chromatin of the IL-2 promoter in naive cells, it is more difficult to immunoprecipitate this fraction of the BCL11BNuRD complex. Another possibility is that as a result of TCR activation, BCL11B may be phosphorylated, which possibly induces a conformational change favoring its association with the IL-2 promoter and coactivator complex. It also is possible that another transcription factor activated in response to TCR activation recruits BCL11B to the IL-2 promoter. Once recruited, BCL11B binds directly to the US1 site, likely in a complex with the other transcription factors.
Here we found that BCL11B directly participates in activation of IL2 gene expression. IL-2 is required for proliferation of T and B lymphocytes and natural killer (NK) cells and plays a central role in regulation of the immune responses.13 In addition, IL-2 is involved in activation-induced cell death (AICD),24 contributing in this way to maintenance of T-cell homeostasis during the late stages of antigen-specific T-cell response.25 The importance of IL-2 in the immune system is confirmed by the fact that IL-2-deficient mice show a severe impairment of the immune system.26 IL-2 is the first cytokine whose expression is induced immediately after CD4+ T-cell activation through TCR signaling.13 Briefly, calcinerurin, a calcium/calmodulin-dependent serine/threonine phosphatase, and PKCθ are activated. The primary target for calcineurin is the nuclear factor of activated T cells (NF-AT), which is dephosphorylated and translocated to the nucleus, where it binds to the IL-2 promoter.27 However, NF-AT is not sufficient, as the transcriptional control of IL-2 gene expression is combinatorial and other transcription factors are required for full activation.17,28,29 AP-1 (Fos/Jun) and NF-kB are also crucial for IL-2 gene expression (reviewed in Serfling17 ), but their induction requires a second costimulatory signal provided by CD28 receptor.30,31 Recently, other transcription factors, including OCT1/OCT2,28,32 CREB,29 MEF2,33 EGR,34,35 and KLF2,36 were found to be involved in transcriptional control of IL2 gene expression. Schnurri-3 (kappa recognition component [KRC]) was shown recently to interact with c-Jun and serve as a coactivator to augment AP-1 activity and IL2 gene transcription.37 BCL11B now adds to the list of transcription factors that regulate expression of the IL2 gene.
BCL11B was found to activate IL2 gene expression by directly binding to its promoter through the US1 site, located between -243 and -210, and less characterized compared to other regions of IL-2 promoter.18 Using mutational analysis we found that the sequence of the BCL11B binding site within the US1 site is 5′-GGGCTAANNCGAC-3′, in which the Cs and Gs are required for binding. Interestingly, most of the bases required for BCL11B binding to the US1 site are conserved in the human and mouse IL-2 promoter.18 In addition, these results are in agreement with the previous data describing the consensus BCL11 binding site as GC-rich.2
What is the molecular mechanism by which BCL11B activates the US1 site in the IL-2 promoter? Previously, it has been reported that p300 acts as a coactivator for several transcriptional regulators, including NF-kB, NF-AT, CREB, and AP-1.20,38,39 These transcription factors cooperatively interact with p300 to form a multiprotein complex named enhanceosome on targeted promoters, such as those of IL2, IFNB, TCRA, TNFA, BLF1, and IL6 genes.20,40-44 We found that BCL11B interacts with p300, and the interaction occurs preferentially after T-cell activation. Our data suggest therefore that BCL11B may be part of the enhanceosome formed on the IL-2 promoter after activation of T cells. The interaction of these transcription factors, including BCL11B, with the same coactivator seems not to be redundant, as individual binding of each transcription factor to the IL-2 promoter is not sufficient for sustained IL2 gene expression.45
Our data suggest that the US1 site is activated by BCL11B only in the specific context of the IL-2 promoter. The US1 site cloned alone upstream of the IL-2 basal (IL-2-70) or SV40 promoters was not sufficient for up-regulation of the reporter activity in conditions of sustained BCL11B expression (data not shown). However, mutations of the BCL11B binding site in the context of the -243 IL-2 promoter affected the BCL11B-mediated promoter activation. One attractive explanation may be that binding of BCL11B alone to US1 site is not sufficient for the recruitment of the p300 coactivator to the IL-2 promoter. In addition, it is possible that US1 site is part of a composite element in the IL-2 promoter, similar to what has been demonstrated for RE/AP1 or NF-AT/AP1 composite elements.46-48
Additional results not shown here suggest that binding of US1 site is not the only mechanism by which BCL11B augments IL-2 gene expression in CD4+ T lymphocytes. We found that BCL11B enhances activation of NF-kB transcription factor downstream of TCR/CD28 signaling (V.B.C. and D.A., manuscript in preparation). These findings explain why the -210 IL-2 promoter reporter had an increased activity as a result of BCL11B overexpression, in spite of the fact that it does not contain a US1 site. However, the region between -210 and -190 contains an NF-kB binding site.
Previous results demonstrated that elevation of cAMP levels through forskolin treatment causes inhibition of IL-2 gene expression through an unidentified transcription factor or complex that binds the US1 site.19 We found that activation of IL-2 promoter mediated by BCL11B is sensitive to cAMP elevation resulting from forskolin treatment. Therefore, our results suggest the possibility that BCL11B may be the factor or one of the factors found previously to bind the IL-2 promoter through the US1 site in a forskolin-sensitive manner.19 It has been demonstrated that increased cAMP levels are associated with development of Th2 effector function and inhibition of Th1-like phenotype. A critical event in this process is the inhibition of expression of IL-2 and other Th1 cytokine genes by cAMP elevation.19,49-52 Therefore, BCL11B may be one of the transcription factors that, in response to cAMP elevation, loses the ability to participate in the IL-2 promoter activation, which consequently results in inhibition of Th1 and promotion of Th2 phenotype.
Prepublished online as Blood First Edition Paper, June 29, 2006; DOI 10.1182/blood-2006-05-021790.
Supported by grants from the National Institutes of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) (K01-AR-02914) (D.A.) and NIH T32-HL-07194 (D.I.A.).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We greatly acknowledge Dr Michael Bell (Mayo Clinic, Rochester, MN), Dr Arthur Weiss (University of California, San Francisco), Dr Laurie Glimcher (Harvard University, Boston, MA), Dr Ronald Schwartz (National Institute of Allergy and Infectious Diseases, Bethesda, MD), and Dr Mark Leid (Oregon State University, Corvallis) for constructs, and Dr Gary Nolan (Stanford University, CA) for Phoenix A and E packaging cells. We thank Dr Wei-Ping Zheng (University of Rochester, NY) for advice with retroviral transduction of primary cells. We thank Ms Tehmeena Qamar and Ms Jennifer Gecewicz for technical assistance, Mr Kenneth Class (Wadsworth Center, Albany, NY) for cell sorting, and Ms Debbie Moran for secretarial assistance. We also thank Dr Michelle Lennartz for reading the manuscript and making helpful suggestions.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal