Abstract
An acquired gain-of-function mutation in the Janus kinase 2 (JAK2-V617F) is frequently found in patients with myeloproliferative disorders (MPDs). To test the hypothesis that JAK2-V617F is the disease-initiating mutation, we examined whether all cells of clonal origin carry the JAK2-V617F mutation. Using allele-specific polymerase chain reaction (PCR) assays for the JAK2 mutation and for the X-chromosomal clonality markers IDS and MPP1, we found that the percentage of granulocytes and platelets with JAK2-V617F was often markedly lower than the percentage of clonal granulocytes determined by IDS or MPP1 clonality assays in female patients. Using deletions of chromosome 20q (del20q) as an autosomal, X-chromosome–independent clonality marker, we found a similar discrepancy between the percentage of cells carrying JAK2-V617F and del20q. Our results suggest that in a proportion of patients with MPDs, JAK2-V617F occurs on the background of clonal hematopoiesis caused by a somatic mutation in an as-yet-unknown gene.
Introduction
Clonal hematopoiesis has been recognized as a key pathogenetic feature of myeloproliferative disorders (MPDs).1 Recently, an acquired somatic mutation in the JAK2 gene resulting in a valine-to-phenylalanine substitution at position 617 (JAK2-V617F) was described in patients with MPDs.2-6 This discovery implied that the presence of the JAK2-V617F mutation could represent the primary causative lesion in MPDs. However, some of the clinical and genetic data suggested that the role of the JAK2-V617F mutation in the MPD pathogenesis could be more complex and led us to propose 2 alternative models for the role of JAK2-V617F in the clonal evolution of MPDs.5 The first model assumed that JAK2-V617F alone is sufficient to cause MPDs. The second model predicted that MPDs are caused by a mutation in an as-yet-unknown gene that precedes the acquisition of the JAK2-V617F mutation and that JAK2-V617F represents a later event in the disease progression.5 One of the predictions of the first model is that the JAK2-V617F mutation should be present in all cells that represent the MPD clone, whereas according to the second model, only a proportion of clonal cells carry the JAK2-V617F mutation. Here we present data in support of the second model in at least a proportion of patients with MPDs.
Study design
Patients
Collection of patient samples was approved by the local ethics committee. Written consent was obtained from all patients in accordance with the Declaration of Helsinki. We included 66 patients (36 polycythemia vera [PV], 23 essential thrombocythemia [ET], 7 idiopathic myelofibrosis [IMF]). One patient with ET was from the University of Pavia, all other patients were from Basel, Switzerland. The diagnosis of MPD subtypes was assigned using the World Health Organization (WHO) criteria.7
Cell separations, RNA and DNA isolation, microsatellite PCR
All blood samples were processed within 4 hours after collection. Isolation of granulocytes, T lymphocytes, platelets, buccal mucosa, RNA, and DNA, as well as cDNA synthesis, were performed as described.5,8,9 Loss of heterozygosity on chromosome 20q was determined by microsatellite polymerase chain reaction (PCR) with the markers D20S96 and D20S119 derived from the minimal deleted region.10
Detection of JAK2-V617F
The allele-specific PCR for JAK2 genotyping was carried out using 20 ng genomic DNA, 45 nM forward primer JAK2-F, and 22.5 nM each of the allele-specific reverse primers JAK2-R-T and JAK2-R-G (Table S1, available at the Blood website; see the Supplemental Materials link at the top of the online article) in a buffer containing 50 mM KCl, 10 mM Tris pH 8.0, and 1.5 mM MgCl2. Thirty PCR cycles with denaturing at 94°C for 30 seconds, annealing at 61°C for 30 seconds, and extension at 72°C for 30 seconds were applied. The PCR products were analyzed using the 3100 Genetic Analyzer (Applied Biosystems, Carlsbad, CA). The percentage of chromosomes carrying the G>T transversion representing the JAK2-V617F allele (%T) were calculated using the formula: %T = (height of T-peak) / (height of T-peak + G-peak) × 100. The SNaPshot Multiplex Kit (Applied Biosystems, Carlsbad, CA) was used for the detection of the JAK2-V617F mutation with primers shown in Document S2. The calculation of %T was performed as for allele-specific PCR.
Analysis of clonality by X-chromosome inactivation
Detection of single nucleotide polymorphisms in the IDS and MPP1 genes was carried out by allele-specific PCR assays analogous to the JAK2-V617F assay. Primers and PCR conditions are listed in Table S1. Genomic DNA (20 ng) was analyzed to identify female patients heterozygous for IDS and/or MPP1. RNA (2 μg) from granulocytes of heterozygous patients was reverse transcribed in 20 μL reactions and 1 μL of the product was examined for IDS or MPP1 mRNA expression by allele-specific reverse transcriptase (RT)–PCR. The PCR products were analyzed as described for JAK2.
Results and discussion
To test the hypothesis that in some patients with MPDs the JAK2-V617F mutation could be preceded by a disease-causing mutation in an as-yet-unknown gene, we developed assays to compare the proportion of cells carrying JAK2-V617F with the percentage of cells that are of clonal origin (Figure 1). If JAK2-V617F is the disease-initiating event, the percentage of cells that carry JAK2-V617F should correspond to the percentage of clonal cells, as determined by JAK2-independent clonality analysis; for example, X-chromosome inactivation pattern (XCIP) assays. Granulocytes from female patients were reported to be clonal by XCIP assays in the majority of PV and IMF patients,1,11-13 whereas about half of ET patients had polyclonal granulocytes.14-16
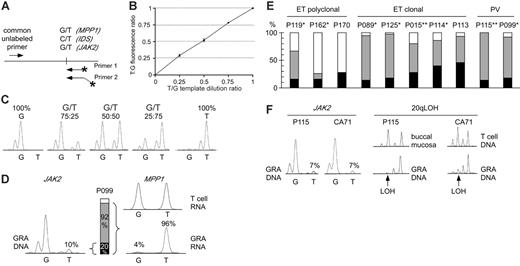
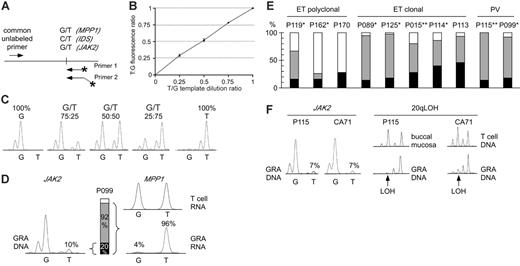
Comparison of clonality and the presence of the JAK2-V617F mutation in MPDs. (A) Design of allele-specific PCR assays for detecting the JAK2-V617F mutation and assessing clonality using the IDS and MPP1 X-chromosomal gene polymorphisms. Horizontal arrows indicate primer positions; fluorescently labeled primers are marked by asterisks. Note that one of the labeled primers is extended by 3 nonhomologous nucleotides to allow separation of the PCR products by size. (B) Analysis of the linearity of the JAK-V617F allele-specific PCR. Quadruplicate reactions were performed on genomic DNAs with known ratios of wild-type and mutant JAK2 alleles. The reactions were performed using homozygous wild-type (G) and homozygous mutant (T) genomic DNA dilutions with increasing proportion of the homozygous mutant DNA. The G and T peak fluorescence ratios were determined and plotted for each genomic DNA dilution. Error bars indicate standard deviation. (C) The chromatograms of 5 DNA samples from panel B are shown. (D) Comparison of clonality determined by X-chromosome inactivation assays with clonality derived from the JAK2-V617F allele-specific PCR in granulocytes (GRA) of one female ET patient (P099). The relative abundance of the mutant JAK2 allele (10%) was calculated by comparing the G and T peak intensities (left). The allelic ratio of 10% T translates to 20% of granulocytes heterozygous for the JAK2-V617F (middle, black bar), or 10% of granulocytes homozygous for the mutation (not shown). Allele-specific PCR assay for the X-chromosomal gene MPP1 is shown at the right. The genomic DNA from patient P099 was heterozygous for a G/T single nucleotide polymorphism (not shown). The relative expression of the 2 MPP1 alleles was determined by comparing the G and T peak intensities obtained by the allele-specific RT-PCR assay in T cells (top chromatogram) and granulocytes (bottom chromatogram). The skewing of expression toward the T allele (96%) translates to the presence of 92% of clonal cells expressing only the T allele (gray bar + black bar), since the remaining 8% of cells that are of polyclonal origin (white bar) express equal amounts of both alleles. (E) Comparison of clonality in patients with ET and PV. Black bars, clonal granulocytes that carry the JAK2-V617F mutation; gray bars, clonal granulocytes that carry the wild-type JAK2 allele; white bars, polyclonal granulocytes. Analysis of T-cell RNA was performed where material was available: *Polyclonal T cells; **Skewed T cells (Figure S2). (F) Comparison of clonality determined by microsatellite PCR for loss of heterozygosity (LOH) on chromosome 20q and clonality defined by the presence of the JAK2-V617F mutation. Allele-specific PCR for JAK2-V617F for 2 patients (P115 and CA71) revealed that only 14% of granulocytes carry the JAK2-V617F mutation (allelic ratio of the T allele = 7%, left panel). In the same 2 patients, LOH (arrows) in the minimal deleted region on chromosome 20q was found in the vast majority of granulocyte DNA (right panel).
Comparison of clonality and the presence of the JAK2-V617F mutation in MPDs. (A) Design of allele-specific PCR assays for detecting the JAK2-V617F mutation and assessing clonality using the IDS and MPP1 X-chromosomal gene polymorphisms. Horizontal arrows indicate primer positions; fluorescently labeled primers are marked by asterisks. Note that one of the labeled primers is extended by 3 nonhomologous nucleotides to allow separation of the PCR products by size. (B) Analysis of the linearity of the JAK-V617F allele-specific PCR. Quadruplicate reactions were performed on genomic DNAs with known ratios of wild-type and mutant JAK2 alleles. The reactions were performed using homozygous wild-type (G) and homozygous mutant (T) genomic DNA dilutions with increasing proportion of the homozygous mutant DNA. The G and T peak fluorescence ratios were determined and plotted for each genomic DNA dilution. Error bars indicate standard deviation. (C) The chromatograms of 5 DNA samples from panel B are shown. (D) Comparison of clonality determined by X-chromosome inactivation assays with clonality derived from the JAK2-V617F allele-specific PCR in granulocytes (GRA) of one female ET patient (P099). The relative abundance of the mutant JAK2 allele (10%) was calculated by comparing the G and T peak intensities (left). The allelic ratio of 10% T translates to 20% of granulocytes heterozygous for the JAK2-V617F (middle, black bar), or 10% of granulocytes homozygous for the mutation (not shown). Allele-specific PCR assay for the X-chromosomal gene MPP1 is shown at the right. The genomic DNA from patient P099 was heterozygous for a G/T single nucleotide polymorphism (not shown). The relative expression of the 2 MPP1 alleles was determined by comparing the G and T peak intensities obtained by the allele-specific RT-PCR assay in T cells (top chromatogram) and granulocytes (bottom chromatogram). The skewing of expression toward the T allele (96%) translates to the presence of 92% of clonal cells expressing only the T allele (gray bar + black bar), since the remaining 8% of cells that are of polyclonal origin (white bar) express equal amounts of both alleles. (E) Comparison of clonality in patients with ET and PV. Black bars, clonal granulocytes that carry the JAK2-V617F mutation; gray bars, clonal granulocytes that carry the wild-type JAK2 allele; white bars, polyclonal granulocytes. Analysis of T-cell RNA was performed where material was available: *Polyclonal T cells; **Skewed T cells (Figure S2). (F) Comparison of clonality determined by microsatellite PCR for loss of heterozygosity (LOH) on chromosome 20q and clonality defined by the presence of the JAK2-V617F mutation. Allele-specific PCR for JAK2-V617F for 2 patients (P115 and CA71) revealed that only 14% of granulocytes carry the JAK2-V617F mutation (allelic ratio of the T allele = 7%, left panel). In the same 2 patients, LOH (arrows) in the minimal deleted region on chromosome 20q was found in the vast majority of granulocyte DNA (right panel).
We established a quantitative allele-specific PCR assay that allows assessing the proportion of granulocytes with the JAK2-V617F mutation (Figure 1A-C). The allelic ratios obtained by this assay represent the proportion of chromosomes with JAK2-V617F. If in a cell population 100% of chromosomes carry the JAK2-V617F mutation, all cells must be homozygous for JAK2-V617F. An allelic ratio of 50% could result either from all cells being heterozygous for the JAK2-V617F mutation, or from a 1:1 mixture of cells homozygous for the mutation and cells without the mutation (Figure 1D). JAK2-V617F was present in 89% of PV patients (32/36), 61% of ET patients (14/23), and 43% of IMF patients (3/7). Some of these patients had allelic ratios below 25%, suggesting that a substantial proportion of granulocytes were negative for JAK2-V617F. To determine whether the granulocytes without the JAK2-V617F mutation are derived from normal (polyclonal) hematopoiesis or from the MPD clone, we examined clonality of granulocytes in female patients using XCIP assays (Table 1). We adapted for allele-specific RT-PCR detection 2 previously described transcriptional clonality assays that are based on frequent exonic polymorphisms in the X-chromosomal genes IDS and MPP1.16-18 These polymorphic markers were demonstrated to be subject to full X-chromosome inactivation in hematopoietic cells by studies of females with normal or clonal hematopoiesis.16-18 In female patients heterozygous for polymorphisms in IDS or MPP1, transcripts of equal quantities from each of the 2 alleles are expected, if the examined cells are polyclonal. A skewing in allelic ratios of more than 70% indicates the predominance of clonal cells in the examined cell population. By these assays, all female PV patients had clonal granulocytes, whereas 6 of 13 ET patients were polyclonal (Table 1). Three of the patients with polyclonal ET were positive for JAK2-V617F. This finding indicates that in polyclonal ET, a small population of clonal V617F-positive cells exists, as previously reported.19 In 2 of 12 female patients with PV and 8 of 13 female patients with ET, less than 25% of the chromosomes in granulocytes carried the JAK2-V617F mutation. Both PV patients and 5 of the 8 ET patients had clonal granulocytes by the IDS and/or MPP1 assays (Table 1, Figure 1E). The low allelic ratio of JAK2-V617F was verified by an alternative assay (SNaPshot; Table S1 and Figure S1). In 8 of 10 patients shown in Figure 1E, we examined clonality in T cells (Table 1 and Supplement 4). In 4 of 6 patients with clonal granulocytes we found polyclonal T cells, excluding constitutive allelic skewing.20-23 Since 4 of 7 informative patients were older than 65, we cannot exclude that in some of them age-related skewing might have contributed to the apparent clonality in granulocytes. However, in a recent report, Levine et al showed that the incidence of neutrophil clonality in patients with ET is the same in age groups below and above 60 years of age and based on statistical analysis reached the same conclusions as presented here.24
To avoid the potential influence of age-related XCIP skewing, we determined granulocyte clonality by assessing deletions on chromosome 20q (del20q), a clonal chromosomal aberration in MPDs.10 We detected del20q in the granulocytes of 4 of 66 patients with MPDs (6%). In 2 of 4 cases (p115 and CA71), the JAK2-V617F mutation was present in only 14% of granulocytes (7% JAK2-V617F allelic ratio), whereas del20q was present in almost 100% of their granulocytes, demonstrating clonality (Figure 1F). This finding further supports our conclusions from the XCIP assays. Moreover, we measured JAK2-V617F in platelet RNA and found allelic ratios closely matching the results obtained with granulocyte DNA (Table 1). If JAK2-V617F is the primary cause of thrombocytosis in patients with ET, the majority of platelets should express the mutant JAK2.
Our results suggest that in a proportion of patients with PV and ET the JAK2-V617F mutation occurs on the background of clonal hematopoiesis caused by an unknown somatic mutation. Thus, the JAK2-V617F mutation represents a genetic defect occurring later in the progression of the disease in these patients. In 2 of these patients, del20q occurred before the JAK2-V617F mutation and could represent such an event.
Prepublished online as Blood First Edition Paper, May 4, 2006; DOI 10.1182/blood-2005-11-009605.
Supported by grants from the Swiss National Science Foundation (310 000-108 006/1), the Swiss Cancer League (OCS-01 411-08-2003), and the Krebsliga beider Basel.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ralph Tiedt and Jürg Schwaller for helpful comments on the manuscript and Mario Cazzola and Francesco Passamonti for DNA from patient CA71.