Abstract
The hematopoietic stem cell (HSC) is a unique cell type found in bone marrow, which has the capacity for both self-renewal and differentiation into all blood lineages. The identification of genes expressed specifically in HSCs may help identify gene products vital to the control of self-renewal and/or differentiation, as well as antigens capable of forming the basis for improved methods of stem cell isolation. In previous studies, we identified a number of genes that appeared to be differentially expressed in murine bone marrow–derived HSCs, using microarray technology. We report here that one of those genes, encoding the murine endothelial protein C receptor (EPCR), is expressed at high levels within the bone marrow in HSCs. Bone marrow cells isolated on the basis of EPCR expression alone are highly enriched for hematopoietic reconstitution activity, showing levels of engraftment in vivo comparable to that of stem cells purified using the most effective conventional methods. Moreover, evaluation of cell populations first enriched for stem cell activity by conventional methods and subsequently fractionated on the basis of EPCR expression indicates that stem cell activity is always associated with EPCR-expressing cells. Based on our findings, we believe EPCR represents the first known marker that `explicitly' identifies hematopoietic stem cells within murine bone marrow.
Introduction
The hematopoietic stem cell (HSC) is a rare cell type found in bone marrow, which has the capacity for both self-renewal and differentiation into all of the specialized cells of the blood.1 Because of the unique biologic properties of HSCs, and their therapeutic utility, there have been intense efforts to fully characterize their phenotypic and functional properties, and to identify molecular mechanisms involved in the control of their function. Due to the low frequency of HSCs in bone marrow, such efforts have necessitated the development of methods for the physical isolation of HSCs. To date, strategies for the purification of HSCs have relied primarily on fluorescence activated cell sorter (FACS)–based protocols that are based on the detection of cell-surface antigens (markers) that are differentially expressed by HSCs (or non-HSCs).2-4 While none of the markers employed in such fractionations singularly enable the purification of stem cells, effective protocols have been developed through the collective use of a number of markers that define an immunophenotypic `address' for HSCs.
The distinctive ability of HSCs to fully reconstitute the hematopoietic system following transplantation suggests that some elements of the gene expression program of these cells are unique. Based on this assumption, we had previously employed genome-wide screening of HSCs to identify genes that were differentially expressed by stem cells when compared with total marrow.5 Here, we report that one of the genes identified in those studies, encoding the murine endothelial protein C receptor (EPCR),6 is expressed at high levels within the bone marrow by HSCs. Bone marrow cells isolated on the basis of EPCR expression alone are highly enriched for hematopoietic reconstitution activity, showing levels of engraftment in vivo comparable to that of stem cells purified using the most effective conventional methods. Moreover, evaluation of cell populations enriched for stem cell activity by conventional methods and subsequently fractionated on the basis of EPCR expression indicates that stem cell activity is always associated with EPCR-expressing cells. As such, EPCR represents the first known marker that `explicitly' correlates with HSC activity within the murine bone marrow.
Materials and methods
Antibodies
Rat anti–mouse EPCR monoclonal antibodies were produced by standard methods, using purified recombinant soluble EPCR as the immunogen, as described previously.7 Antibody was produced by culturing hybridoma cells in serum-free media and isolating the secreted antibody by chromatography on MEP HyperCel (Pall, East Hills, NY). Antibody clone 1560 used in this study was of isotype IgG2b Kappa and blocked protein C activation by thrombin on cells expressing both thrombomodulin and EPCR. Clone 1560 was also found to bind 293 cells transfected and expressing murine EPCR, but not to bind control (untransfected) cells. Fluorescently labeled anti–rat IgG secondary antibodies were obtained from Molecular Probes (Eugene, OR). Antibodies against Sca-1, CD34, c-Kit, and lineage markers (B220, CD3e, Ter119, Gr-1, CD11b) were obtained from Pharmingen (San Diego, CA).
Reverse transcriptase (RT)–polymerase chain reaction
RNA was extracted using a Microprep RNA Isolation Kit (Stratagene, La Jolla, CA) and subjected to first-strand synthesis using random hexamer primed reverse transcription (Amersham-Pharmacia, Piscataway, NJ). Template from this reaction was used to perform multiplex polymerase chain reaction (PCR) containing primers for both GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and EPCR. Primers were designed against the 3′ untranslated region of the mRNA as follows: EPCR top GCTGGTAAGGGAAGGTCGTAGCTCA, EPCR bottom CACTGCTCCCATATTGTTCAACCTCA; GAPDH top TTCCAGTATGACTCCACTCACG, GAPDH bottom GTTCACACCCATCACAAACATG. PCR conditions were as follows: 95°C for 10 minutes followed by 31 cycles of 95°C for 1 minute, 55°C for 1 minute, 72°C for 1 minute. Reactions were run on a 2% agarose gel, and images were captured using a Kodak DC290 Gel Imaging System (Kodak, Rochester, NY).
FACS
Sorting was performed on a triple laser MoFlo (Cytomation, Fort Collins, CO) using Summit software (Cytomation). Hoechst 33342 was excited at 351 nm, and fluorescence emission was detected using 405/BP30 and 570/BP20 optical filters against Hoechst blue and Hoechst red, respectively, and a 555-nm long-pass dichroic mirror (all from Omega Optical, Brattleboro, VT) to separate emission wavelengths. Both Hoechst blue and red fluorescence were acquired on a linear scale. Propidium iodide (PI) fluorescence was measured through a 630BP30 filter after being excited by a 488-nm Argon laser. A live gate was defined as the cells negative for PI, allowing dead cells to be excluded. All analyses were performed using FlowJo software (Treestar, Ashland, OR).
Methylcellulose-based colony-forming assays
CFU assays were performed by sorting 20 000 total marrow stained with EPCR antibody or the relative fraction of 20 000 cells represented by the test population (ie, 19 700 EPCR negative or 280 EPCR intermediate) and placed in MethoCult media containing stem cell factor (SCF), interleukin 3 (IL-3), IL-6 and erythropoietin (Epo) following the manufacturer's protocol (StemCell Technologies, Vancouver, BC, Canada). Colonies were scored following 12 days of culture in a humidified incubator at 37°C with 5% CO2. EPCR-expressing fractions were normalized to the counts obtained from total marrow to determine the percentage of each progenitor type isolated in the tested fraction.
Competitive repopulation assays
Recipient C57BL/6J (CD45.2) mice, 5 to 6 weeks old, were lethally irradiated with 1300 rads in two 650-rad doses 3 hours apart and were transplanted by retro-orbital injection the following day. Donor B6.SJL-PtprcaPep3b/BoyJ (CD45.1) mice expressing the Ly5.1 isoform of the CD45 marker were killed and their femurs and tibiae crushed with a mortar and pestle. Samples for side population (SP) sorting were suspended at a cell density of 4 × 106 cells/mL as counted from 5 μM to 10 μM on a Coulter counter and stained with a concentration of 10 μg/mL Hoechst 33342 dye. Cells were subjected to a ficoll gradient to isolate lymphocytes prior to staining with monoclonal antibodies against surface antigens. Staining was performed using a monoclonal antibody against mouse EPCR (mAb 1560) followed by detection with secondary antibody against rat IgG conjugated to Alexaflour-488 (Molecular Probes). After staining, cells were washed and resuspended in media containing propidium iodide to exclude dead cells. Sorted donor cells were transplanted with 2 × 105 competitor cells from an unirradiated C57BL/6J recipient littermate. Blood samples were taken each month following transplantation, and chimerism was measured by flow cytometry using antibodies specific for each isoform of the CD45 receptor.
Analysis of multilineage engraftment
Blood was taken from animals that underwent transplantation with 1000 EPCRhi cells 6 months after transplantation and stained using monoclonal antibodies against CD45.1 conjugated to phycoerythrin, CD45.2 conjugated to fluorescein, and one of the following biotinylated lineage antibodies: anti-B220 (B lymphocytes), anti-CD3e (T lymphocytes), anti– Gr-1 (granulocytes), anti–Mac-1 (macrophages), or anti-Ter119 (erythroid cells). After washing, samples were stained with streptavidin-labeled allophycoerythrin and analyzed using a FACSCalibur (Becton Dickinson, San Jose, CA). The ratio of CD45.1- to CD45.2-expressing cells within each subset of lineage-positive cells was determined using FlowJo (Treestar, Ashland, OR) analysis software.
Results
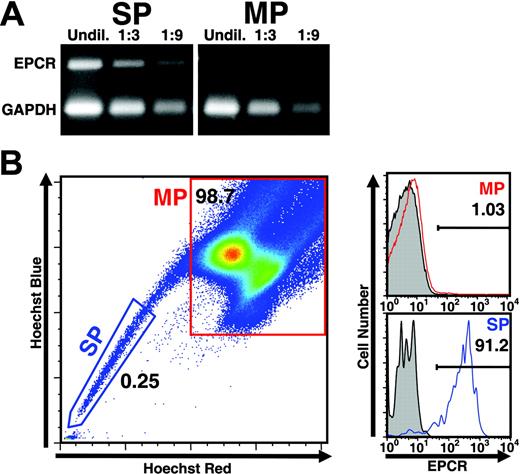
In our previous studies,5 RNA prepared from HSCs purified on the basis of dual wavelength analysis of Hoeschst-stained bone marrow cells8,9 (termed CD34– SP cells) and stem cell–depleted bone marrow cells (termed main population [MP] cells) were analyzed for their respective gene expression profile using Affymetrix microarrays in order to identify differentially expressed genes. From this analysis, the murine EPCR gene6 was identified as one of the most differentially expressed genes, exhibiting at least 30-fold greater expression in SP than in MP cells.5 To confirm the differential expression of the EPCR gene observed by microarray analysis, RT-PCR was performed on RNA samples isolated from either side population (SP) or main population (MP) cells prepared from murine bone marrow. As shown in Figure 1A, whereas EPCR sequences were readily amplified from SP RNA samples, no appreciable signal was detected in reactions with similar quantities of RNA from MP cells. To determine if the specific expression of EPCR mRNA in SP cells resulted in expression of EPCR protein at the cell surface, a rat anti–mouse EPCR monoclonal antibody (mAb 1560) was used to detect EPCR on bone marrow cells also stained with Hoechst dye. As shown in Figure 1B, more than 90% of SP cells stained positively for EPCR, whereas only a small fraction (1%) of MP cells showed evidence of EPCR expression. Interestingly, as shown in Figure 2A, the expression level of EPCR at the cell surface was found to correlate with the extent of dye efflux (ie, cells expressing increasing levels of EPCR manifested increasing dye-efflux activity/decreasing fluorescence). We have previously reported that SP cells exhibiting the highest dye-efflux activity are most enriched for hematopoietic reconstitution activity.8,9
Specific expression of EPCR by SP cells. (A) RNA from sorted side population (SP) or main population (MP) cells was extracted and subjected to multiplex RT-PCR to detect expression of EPCR message. EPCR message was easily amplified from SP cell RNA but was nearly undetectable within a similar quantity of MP RNA (n = 3). (B) C57BL/6 total bone marrow was stained with Hoechst 33342 dye to identify the SP and further stained with mAb 1560, a monoclonal antibody specific for EPCR. Gray histograms denote level of staining produced by isotype control antibody. Red and blue histograms represent specific staining by mAb 1560. EPCR protein is readily detectable at the cell surface of SP cells with very little expression detectable in MP (n = 5).
Specific expression of EPCR by SP cells. (A) RNA from sorted side population (SP) or main population (MP) cells was extracted and subjected to multiplex RT-PCR to detect expression of EPCR message. EPCR message was easily amplified from SP cell RNA but was nearly undetectable within a similar quantity of MP RNA (n = 3). (B) C57BL/6 total bone marrow was stained with Hoechst 33342 dye to identify the SP and further stained with mAb 1560, a monoclonal antibody specific for EPCR. Gray histograms denote level of staining produced by isotype control antibody. Red and blue histograms represent specific staining by mAb 1560. EPCR protein is readily detectable at the cell surface of SP cells with very little expression detectable in MP (n = 5).
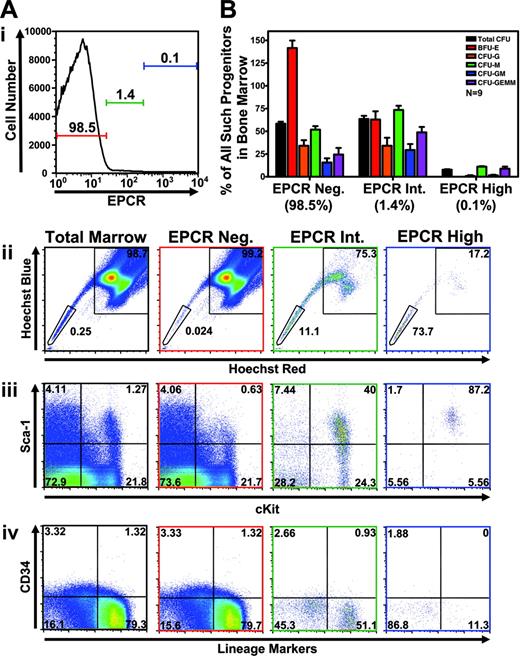
EPCR-expressing marrow correlates with HSC phenotype and shows significant CFU activity. (A) Correlation of expression level of EPCR to HSC phenotype was analyzed by performing multiparameter flow cytometry and gating the indicated intensity of EPCR expression (denoted by colored horizontal bars with percentage of total cells indicated) and replotting gated cells to display the indicated parameters. Frame color of each graph denotes the histogram gate in which plotted cells are found (black frame indicates total bone marrow). Each graph uses the identical set of gates for the specified combination of parameters set using isotype control antibodies. Hoechst profiles are shown with identical SP and MP gates, with top and bottom text indicating percentage of plotted cells falling into MP or SP gates, respectively. Replotting of EPCR-positive cells (gated in the top histogram) to display levels of other markers analyzed (bottom plots) shows that EPCR expression correlates well with enrichment for Sca-1 and c-Kit expression and depletion of CD34 and lineage marker staining. In the experiment shown, nonspecific antibody binding as measured by isotype control using the identical EPCRhi gate accounted for approximately 10% of EPCR-positive cells analyzed (n = 3). Numbers in panels Aii-Aiv represent the percentage of total cells found in each quadrant. (B) Populations gated in panel A were sorted and subjected to methylcellulose-based colony formation assays (CFU) to determine the relative proportion of each progenitor type found within various EPCR-expressing subfractions. Although accounting for only 1.4% of total bone marrow, EPCR intermediate–expressing cells contained a large proportion of colony-forming activity in the marrow. EPCR high–expressing cells, which were most highly enriched for cells exhibiting HSC immunophenotype, showed very little progenitor activity (n = 9). Error bars represent standard deviation.
EPCR-expressing marrow correlates with HSC phenotype and shows significant CFU activity. (A) Correlation of expression level of EPCR to HSC phenotype was analyzed by performing multiparameter flow cytometry and gating the indicated intensity of EPCR expression (denoted by colored horizontal bars with percentage of total cells indicated) and replotting gated cells to display the indicated parameters. Frame color of each graph denotes the histogram gate in which plotted cells are found (black frame indicates total bone marrow). Each graph uses the identical set of gates for the specified combination of parameters set using isotype control antibodies. Hoechst profiles are shown with identical SP and MP gates, with top and bottom text indicating percentage of plotted cells falling into MP or SP gates, respectively. Replotting of EPCR-positive cells (gated in the top histogram) to display levels of other markers analyzed (bottom plots) shows that EPCR expression correlates well with enrichment for Sca-1 and c-Kit expression and depletion of CD34 and lineage marker staining. In the experiment shown, nonspecific antibody binding as measured by isotype control using the identical EPCRhi gate accounted for approximately 10% of EPCR-positive cells analyzed (n = 3). Numbers in panels Aii-Aiv represent the percentage of total cells found in each quadrant. (B) Populations gated in panel A were sorted and subjected to methylcellulose-based colony formation assays (CFU) to determine the relative proportion of each progenitor type found within various EPCR-expressing subfractions. Although accounting for only 1.4% of total bone marrow, EPCR intermediate–expressing cells contained a large proportion of colony-forming activity in the marrow. EPCR high–expressing cells, which were most highly enriched for cells exhibiting HSC immunophenotype, showed very little progenitor activity (n = 9). Error bars represent standard deviation.
To phenotype bone marrow cells expressing increasing amounts of EPCR with regard to the expression of markers commonly used to define murine HSCs,2,8,10 multiparameter flow cytometry was employed to analyze both Hoechst staining as well as antibody staining directed against EPCR, Sca-1, CD34, c-Kit, and “lineage markers.” As shown in Figure 2A, EPCR expression was found to correlate closely with both SP phenotype as well as the traditional KSL34– cell-surface phenotype of HSCs. Cells expressing the highest levels of EPCR were found to be homogeneously positive for Sca-1 and c-Kit and at the same time negative for CD34 and lineage marker staining. These cells were found primarily at the SP tip (73.7%), although a detectable fraction was found in MP (17.2%). To characterize the progenitor activity of EPCR-expressing fractions, methylcellulose assays were performed to quantify the proportion of total progenitor activity found within EPCR depleted, EPCR intermediate, or EPCR highly expressing cells. As shown in Figure 2B, EPCR intermediate–expressing cells contained more than 60% of total colony-forming unit (CFU) progenitor activity in the marrow despite accounting for only 1.4% of total marrow cells, indicating a significant enrichment for progenitors. The EPCR high fraction previously found to be most enriched for cells exhibiting the SP and KSL CD34–negative phenotype displayed only limited progenitor activity in this assay.
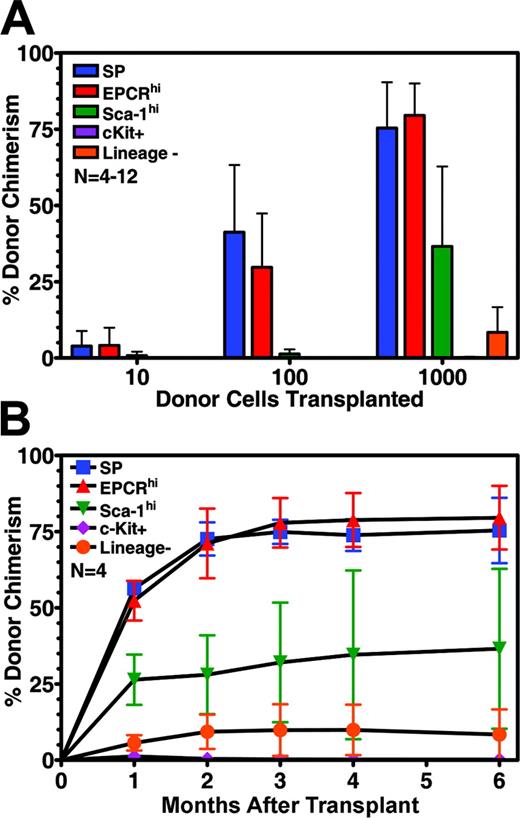
To directly demonstrate that EPCRhi cells possess hematopoietic reconstitution activity, and to quantify that activity relative to stem cell populations enriched by other means, EPCRhi cells and several other purified cell populations were assayed by an in vivo competitive repopulation assay.11 As shown in Figure 3A, cells purified solely on the basis of high levels of expression of EPCR showed potent hematopoietic reconstitution activity, comparable to that of SP cells. Detectable engraftment could be observed after the transplantation of only 10 cells, even in the presence of 200 000 unfractionated bone marrow competitor cells. Based on the level of engraftment observed after transplantation of 100 positive cells, EPCR-positive bone marrow cells appear to be more than 1000-fold enriched for HSC activity. In contrast, cells fractionated on the basis of the expression of common stem cell markers (eg, Sca-1, c-Kit, or lineage negative) showed far weaker engraftment activity. Mice that underwent transplantation with EPCR-positive cells displayed durable hematopoietic reconstitution by donor cells and engraftment kinetics similar to cells purified by dye efflux (Figure 3B). EPCR-positive cells were found to contribute equally to all blood lineages long-term, thus indicating their pluripotent nature (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article).
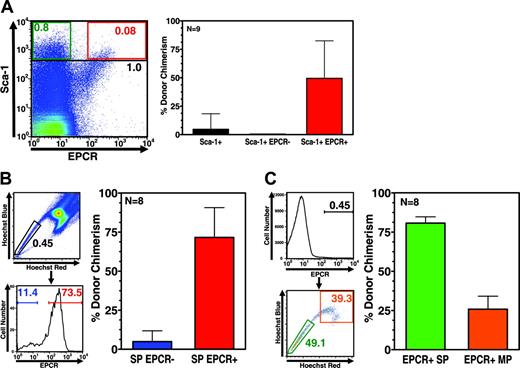
Based on the potent hematopoietic reconstitution activity of EPCR-positive cells, we next asked whether EPCR expression is always associated with the HSC phenotype. To address this, we determined whether the repopulating activity of cell populations enriched for stem cell activity (on the basis of conventional fractionation strategies) is always associated with EPCR-expressing cells, and conversely, whether all EPCR-expressing cells possess repopulating activity. First, Sca-1hi cells were fractionated for EPCR expression, and the resulting EPCR-positive and -negative populations were analyzed for their engraftment capacity in a competitive repopulation assay. EPCR antibody costaining of Sca-1–stained marrow revealed a distinct subfraction of double-positive cells representing approximately 10% of total Sca-1hi cells, which contained all of the reconstitution activity of the Sca-1hi fraction (Figure 4A). In a second experiment, cells were sorted via dye-efflux methodology, and subsequently fractionated for EPCR expression prior to competitive repopulation assays. As shown in Figure 4B, more than 70% of SP cells were EPCR positive and again, only the EPCR-positive subpopulation of SP cells demonstrated potent repopulating ability. In a third experiment, bone marrow cells were first isolated on the basis of EPCR expression, and subsequently fractionated on the basis of dye-efflux activity prior to competitive repopulation assays. Fractionation of EPCR-positive cells via dye efflux revealed significant SP and MP populations (Figure 4C). As expected, the EPCR-positive SP fraction showed potent reconstitution activity; however, the EPCR-positive MP fraction was also found to contain significant engrafting activity (Figure 4C). These results are particularly noteworthy, as they suggest that a significant proportion of the total stem cell activity in bone marrow may not be recoverable via SP isolation alone. Previous studies of the engraftment abilities of SP and MP fractions had suggested that the majority of stem cell activity resided in the SP fraction.8 In light of the current findings, it is possible that our previous studies may have been compromised by the inability to identify and isolate the relevant cells within the MP gate capable of reconstitution.
Hematopoietic reconstitution potential of EPCR-positive cells and other purified stem cell populations. (A) Competitive repopulation experiments showing the contribution to blood chimerism made by indicated numbers of cells purified by different methods (n = 4 to 12 mice per group). (B) Kinetics of 1000 HSC engraftment of mice shown in panel A over the course of study. Error bars represent standard deviation.
Hematopoietic reconstitution potential of EPCR-positive cells and other purified stem cell populations. (A) Competitive repopulation experiments showing the contribution to blood chimerism made by indicated numbers of cells purified by different methods (n = 4 to 12 mice per group). (B) Kinetics of 1000 HSC engraftment of mice shown in panel A over the course of study. Error bars represent standard deviation.
Discussion
Overall, our results demonstrate that high EPCR expression more accurately identifies murine bone marrow–derived HSCs capable of engraftment in vivo than any previous method relying on marker expression or dye efflux. Prior to the studies described here, FACS-based methods for the identification and isolation of HSCs have relied on the fractionation of cells based on staining for proteins whose expression is only relatively stem cell–specific, such as Sca-1, c-Kit, CD34, and differentiated cell antigens (“lineage” markers).2,9,10 Accordingly, those protocols for isolating stem cells required the simultaneous use of multiple reagents and multiparameter FACS capabilities. Recently, a number of new cell-surface markers have been described12,13 as well as a family of receptors whose combinatorial patterns of gene expression provide the basis for the isolation of highly purified HSCs.14 To our knowledge, however, EPCR represents the first marker that enables the isolation of stem cells on the basis of the expression of a single gene product. The purification of stem cells through the isolation of EPCRhi-expressing cells may therefore prove to be a simple and highly effective alternative to currently utilized methods.
EPCR expression accurately identifies HSC activity. (A) Flow cytometry of total bone marrow stained with both Sca-1 and EPCR antibodies showing that EPCR identifies a distinct subfraction of Sca-1hi cells that constitutes approximately 10% of the Sca-1hi population. Black, green, and red text indicates the percentage of total cells found to be Sca-1hi, Sca-1hi EPCR–, and Sca-1hi EPCR+, respectively (left panel). Right panel indicates level of engraftment 4 months after transplantation of 100 cells sorted using the gates shown in the left panel in competition with 200 000 unfractionated bone marrow competitor cells. EPCR expression clearly fractionates all stem cell activity from the Sca-1hi population. (B) Gating logic used to isolate the 500 SP cells, either positive or negative for EPCR expression, that were transplanted in competitive repopulation experiments against 200 000 unfractionated bone marrow competitor cells (left panels). Engraftment of cells sorted 4 months after transplantation showing little engraftment activity by SP cells lacking EPCR (right panels). (C) Gating logic used to isolate 500 EPCR-expressing cells exhibiting either SP or MP dye-efflux phenotype (left panel). Repopulation of sorted cells 4 months after transplantation in competition with 200 000 unfractionated bone marrow cells showing significant engraftment by EPCR-expressing MP cells (right panel). Error bars represent standard deviation.
EPCR expression accurately identifies HSC activity. (A) Flow cytometry of total bone marrow stained with both Sca-1 and EPCR antibodies showing that EPCR identifies a distinct subfraction of Sca-1hi cells that constitutes approximately 10% of the Sca-1hi population. Black, green, and red text indicates the percentage of total cells found to be Sca-1hi, Sca-1hi EPCR–, and Sca-1hi EPCR+, respectively (left panel). Right panel indicates level of engraftment 4 months after transplantation of 100 cells sorted using the gates shown in the left panel in competition with 200 000 unfractionated bone marrow competitor cells. EPCR expression clearly fractionates all stem cell activity from the Sca-1hi population. (B) Gating logic used to isolate the 500 SP cells, either positive or negative for EPCR expression, that were transplanted in competitive repopulation experiments against 200 000 unfractionated bone marrow competitor cells (left panels). Engraftment of cells sorted 4 months after transplantation showing little engraftment activity by SP cells lacking EPCR (right panels). (C) Gating logic used to isolate 500 EPCR-expressing cells exhibiting either SP or MP dye-efflux phenotype (left panel). Repopulation of sorted cells 4 months after transplantation in competition with 200 000 unfractionated bone marrow cells showing significant engraftment by EPCR-expressing MP cells (right panel). Error bars represent standard deviation.
The applicability of the method to the isolation of human HSCs, although of great interest, remains uncertain. Although we have recently identified a rare population of EPCR-expressing cells in human bone marrow, preliminary efforts to document their activity in standard assays for human stem cells/progenitors have been unsuccessful (A.B.B., unpublished results, September-October 2003). Further studies are in progress to address this important issue.
EPCR is a type I transmembrane receptor that augments the activation of protein C (PC) by the thrombin-thrombomodulin complex at the luminal surface of blood vessels during coagulation.15 EPCR is related structurally to the major histocompatibility complex (MHC) class 1/CD 1 family of molecules, most of which are involved in inflammation. Like the CD 1 family, EPCR has a tightly bound lipid (mostly phosphatidylcholine) located in a region corresponding to the antigen-presenting groove of other family members.16 In the mouse, EPCR has been shown to be expressed during embryogenesis primarily within trophoblast giant cells involved in placental development, and in the adult primarily within endothelial cells that line the large vessels.7 Although EPCR expression has been described in human monocytes,17 the expression of EPCR within murine hematopoietic cells had not been reported previously. EPCR expression has also been reported in certain cancer cell lines.18,19
Studies of EPCR knockout mice indicate that EPCR expression is required during embryogenesis, as mice deficient in EPCR do not survive beyond day 10.20 Recent reports, however, have shown that mice expressing EPCR only within trophoblast giant cells can survive to adulthood,21 suggesting that embryonic lethality in the absence of EPCR is not due to a lack of expression by HSCs. EPCR-null adult mice showed modified thrombin and activated protein C (APC) generation but other hematopoietic defects were not reported.21
In light of the EPCR knockout studies, the role, if any, that EPCR plays in stem cell function remains unclear. While it is possible that EPCR's function in stem cells relates to its well-described role in coagulation,15 EPCR has been shown to possess additional biologic functions related to intracellular signaling that may also be of relevance. It has been shown recently that EPCR is required for APC-mediated activation of protease activated receptor 1 (PAR-1; thrombin receptor),22 a member of a family of G protein–coupled receptors that are activated by extracellular cleavage.23 While the physiologic significance of this mode of PAR-1 activation has been questioned,24 cleavage of PAR-1 by APC has been reported to activate the mitogen-activated protein kinase (MAPK) pathway in endothelial cells,22,25 a pathway that is implicated in a variety of biologic processes that could be relevant in the context of HSCs (eg, growth factor signaling, cell cycle control, apoptosis).26 In addition to studies that evaluate the effects of APC or PAR agonists on various stem cell functions, further studies of EPCR conditional knockout animals could lead to a better understanding of the role, if any, of EPCR expression in hematopoietic stem cell function in vivo.
Prepublished online as Blood First Edition Paper, November 22, 2005; DOI 10.1182/blood-2005-06-2249.
Supported by National Institutes of Health grant 5PO-HL54 785 (R.C.M.). C.T.E. holds the Noble Chair in Cardiovascular Research at Oklahoma Medical Research Foundation and is an investigator of the Howard Hughes Medical Institute.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank G. Mostoslavsky, D. Kotton, and G. Murphy for their helpful discussions and critical reading of the manuscript. We also thank Peggy Russell for her excellent technical assistance.