Abstract
Type 1 interferons (IFNs) are induced in vivo, administered therapeutically, and potential targets for amelioration of autoimmune diseases. The cytokines mediate profound antiproliferative effects. Signal transducer and activator of transcription 1 (STAT1)-dependent signaling pathways are required for inhibition of proliferation, and viral infections can elicit high levels of type 1 IFNs as well as total STAT1 protein expression. Thus, a mechanism must be in place to help antigen-specific T cells overcome IFN-induced inhibition of proliferation. The studies reported here demonstrate that total CD8 T-cell proliferation in the presence of IFNs, ex vivo in response to cytokines and in vivo during viral infection, is inhibited through a STAT1-dependent mechanism. In contrast, major proportions of antigen-specific CD8, but not CD4, T cells are rendered less sensitive to this inhibition, express lower endogenous levels of total STAT1, and are selectively proliferating in the presence of type 1 IFN, at key times after viral challenge. Taken together, these novel results show that differential STAT1 expression is used by the immune system to modify cytokine-mediated effects on T-cell expansion and have implications for the consequences of therapeutic intervention in cytokine function.
Introduction
Type 1 IFNs (IFNα and IFNβ) mediate a wide range of biologic effects, including antiviral and antimicrobial defense, immunoregulation, proliferation inhibition, and survival of activated T cells.1 They are induced during a variety of infections, most notably certain viral infections; are used therapeutically to treat cancers, viral infections, and multiple sclerosis1-4 ; and are being developed as vaccine adjuvants.5 There is a growing interest in neutralizing their in vivo functions to help control certain autoimmune diseases, including systemic lupus.6 Gene expression analyses using cell lines have shown that hundreds of genes can be differentially regulated by IFN-α/β,7 and certain biologic effects attributed to these cytokines are paradoxical.8 Thus, there must be sophisticated mechanisms controlling the downstream consequences of exposure to the cytokines.
Receptor binding of IFNα/β elicits phosphorylation of the transcription factors STAT1 and STAT2. Heterodimers of phosphorylated STAT1 and STAT2 can help promote gene expression through the IFN-stimulated responsive elements.9,10 In addition to this complex, dimers of STAT1 can be activated to bind to γ-activated sequence elements. Other STAT family members have been implicated in the signaling activated by type 1 IFNs,11 but the best understood of the IFN-mediated effects are through STAT1.12 This transcription factor is critical for the antiproliferative effects of type I and II IFNs.13,14 STAT1 is also an important intermediary in the activation of many of the biologic responses at early times after in vivo challenge with viruses or treatments with IFNs.15,16 In contrast, other effects of IFNs are inhibited by the presence of STAT1, but revealed under STAT1-negative conditions.8,17 Interestingly, STAT1 expression is induced as a result of STAT1 activation,7 and the protein levels of STAT1 are elevated during viral infection.18 These observations suggest that regulation of STAT1 protein levels may provide a mechanism to naturally modify the effects of type 1 IFNs.
The studies presented here were undertaken to evaluate IFN effects on T-cell proliferation and to investigate how the antigen-specific cells required for defense can escape the growth inhibitory effects mediated by the cytokines in vivo. Responses to lymphocytic choriomeningitis virus (LCMV) infections of mice were characterized because the conditions are associated with well-defined and extended production of type 1 IFNs, as well as striking expansion of antigen-specific CD8 T cells.19-21 Moreover, previous work from our group has shown that at the times overlapping with IFNα/β production, STAT1 protein expression is dramatically elevated.18 The results of our new work demonstrate that STAT1 is an important contributor to the early regulation of nonspecific CD8 T-cell expansion during infection. Moreover, they show that, although STAT1 protein expression is induced in all populations, including CD4 T cells, responding CD8 T-cell subsets have relatively less STAT1 and breakthrough to proliferate at times overlapping with viral replication and the IFNα/β response. These events precede the return of other populations to low STAT1 levels. Thus, the antiproliferative effects exerted by type 1 IFNs may provide conditions that limit nonspecific T-cell expansion early during immune responses, but the required antigen-specific CD8 T cells can curtail these effects by selectively expressing reduced STAT1 levels. The results have profound implications concerning the secondary consequences of manipulating in vivo IFN concentrations and functions.
Materials and methods
Mice and in vivo manipulations
Specific pathogen-free wild-type C57BL/6 mice were purchased from Taconic Laboratory Animals and Services (Germantown, NY). STAT1-deficient mice initially on the 129 background22 were crossed onto the C57BL/6 background for more than 5 generations, and colonies were established in the animal care facilities at Brown University. Experimental groups were age matched, and all mice used in experiments were 8 to 12 weeks of age. Animals obtained outside of Brown University were housed for at least 1 week before use. Handling of mice and experimental procedures were in accordance with institutional guidelines for animal care and use. Experiments were initiated on day 0, with mice either not infected or infected intraperitoneally with 2 × 104 plaque-forming units (PFUs) LCMV Armstrong strain, clone E350. Where indicated, LCMV was measured in a viral plaque assay with vero cells as described.23
Splenic leukocyte preparations and CD8 T-cell enrichment
On indicated days following initiation of experiments, mice were killed and spleens were harvested. Leukocytes were prepared as previously described.23 In certain experiments, CD8 T cells were isolated by negative selection using magnetic-activated cell sorting (MACS) enrichment kits and the program DepleteS on the autoMACS instrument (Milteny Biotec, Auburn, CA). The purity of enriched samples was greater than 90%.
CFSE proliferation analysis
Leukocytes were resuspended to 1 × 107 cells/mL in PBS containing 5% FBS, and CFSE labeling was performed as described24 with some modifications. Briefly, CFSE was rapidly mixed with the cells to a final concentration of 5 μM and incubated for 5 minutes at room temperature. After 2 washes with 10 volumes of PBS-5% FBS, the CFSE-labeled leukocytes were incubated in the absence or presence of cytokines. In all experiments, the concentration of cells was 1 × 106 cells in 200 μL RPMI-10% FBS. Cells were incubated in 96-well plates at 37°C for the times indicated. A final concentration of 20 ng/mL IL-2, IL-7, and IL-15 was used. rhIL-2 was from Cetus Corporation (Berkeley, CA); rmIL-7 and rmIL-15 were purchased from R&D Systems (Minneapolis, MN). Human IFNαA/D, the universal interferon, was used to complete a titration of sensitivity to the factor. Use at either 1 × 103 or 1 × 104 U/mL resulted in maximal inhibition. The mouse IFNαA from PBL Biomedical Laboratories (Piscataway, NJ) was used at 1 × 104 U/mL. Following stimulation, cells were labeled with allophycocyanin (APC)-conjugated anti-CD8 antibody or peridin chorophyll protein (PerCP)-conjugated CD4 antibody (BD Biosciences; Mountain View, CA). CFSE and antibody fluorescence was acquired using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) and CellQuest software. Laser outputs were 15 mW at 488 and 635 nm wavelengths. To isolate CFSE-low and CFSE-high CD8 T cells, splenic leukocytes were isolated at day 5 after LCMV infection, labeled with CFSE, and maintained in culture for 3 days. After the incubation, total CD8 T cells were enriched on magnetic beads and sorted based on CFSE intensity using a Becton Dickinson FACSVantage SE with a 70-μm nozzle and equipped with a laser producing 488 nm excitation (Lifespan Core Research Laboratories, Providence, RI). The purity and viability of the sorted populations were analyzed immediately after sorting.
Flow cytometric analyses
Studies evaluating in vivo BrdU incorporation by CD8 T cells were adapted from the BrdU Flow Kit protocol (BD Biosciences). Briefly, 1 mg BrdU was injected intraperitoneally into mice 2 hours prior to harvest. Splenic leukocytes were isolated as described, surface labeled with PerCP-conjugated CD8 antibody and APC-conjugated H-2Db tetramer binding the immunodominant LCMV peptide GP33-41 (KAVYNFATC) (Beckman Coulter, Immunomics, San Diego, CA), or APC-conjugated CD4 antibody followed by intracellular staining with fluorescein isothiocyanate (FITC)-labeled anti-BrdU and/or phycoerythrin (PE) anti-STAT1. For intracellular STAT1 labeling, a custom-made PE-conjugated STAT1 antibody was used (BD Biosciences), using the solutions and protocol from the BrdU Flow kit to detect nuclear and cytoplasmic STAT1. The approach was developed as a modification of reported staining of human monocytes.25 Studies of cells isolated from the STAT-/- mice demonstrated specificity of staining. Isotype control antibodies were used in all the analyses. The mean fluorescence intensities of the isotype and STAT1-/- controls were lower than the first log of intensity with narrow uniform peaks.
IFN evaluation
Serum was obtained from mice anesthetized with isoflurane (Abbott Laboratories, Abbott Park, IL) as previously described.26 Active IFNα/β was evaluated as protection against vesicular stomatitis virus (VSV)-induced cytopathic effects. Protection was scored at 2 to 3 days after challenge by visual examination for reduction in cytopathic effects. One unit per milliliter of IFNα/β is defined as the dilution at which 50% protection from VSV-mediated lysis occurs. The limit of detection was 8 U/mL. For IFNα enzyme-linked immunosorbent assay (ELISA), the primary antibody was a rat anti-mouse IFNα mAb (F-18; Hycult Biotechnology, Uden, The Netherlands; distributed by Cell Sciences), the secondary antibody a rabbit anti-mouse IFNα polyclonal Ab (PBL Biomedical Laboratories), and the tertiary antibody a horseradish peroxidase-conjugated donkey anti-rabbit F(ab′)2 Ab (Jackson ImmunoResearch Laboratories, Baltimore, PA); the substrate was ABTS Peroxidase Substrate (KPL, Gaithersburg, MD) and the standard, a recombinant mouse IFNαA from PBL Biomedical Laboratories. Colorimetric changes of enzyme substrates were detected at 405 nm wavelength using a SpectraMax 250 reader (Molecular Devices, Sunnyvale, CA). The limit of detection was 1500 pg/mL. For certain experiments, the mouse IFNα ELISA Kit from PBL Biomedical Laboratories was used. The limit of detection for this assay was 31.25 pg/mL.
Cell lysates and Western blot analysis
Total splenic leukocytes, enriched CD8 T cells, and non-CD8 cells, as well as CD8 T cells sorted for high or low CFSE staining were lysed in a solution containing 50 mM Tris HCl pH 7.5, 0.3M NaCl, 0.5% Triton X-100, 2 mM EDTA, 0.4 mM Na3VO4, and a cocktail of protease inhibitors (Roche Diagnostics, Indianapolis, IN). Protein (30 μg) was separated on SDS-PAGE gels (Gradipore 4%-20% LongLife MicroGels; Gradipore, Hawthorne, NY), following the Gradipore protocol for blotting. Monoclonal anti-STAT1 antibody (clone 1) was purchased from BD Transduction Laboratories (Lexington, KY); rabbit polyclonal anti-STAT4 antibody (C-20) obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and rabbit polyclonal anti-β-actin from Abcam (Cambridge, United Kingdom) were used as a loading control. Blot images were acquired in a BioRad (Hercules, CA) GelDoc and analyzed using the public domain National Institutes of Health (NIH) Image program (developed at the US National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/).
Real-time PCR
RNA from sorted CD8 T cells was extracted with the RNAeasy kit from Qiagen (Valencia, CA), using on column digestion with DNAse I (Qiagen). One-step real-time reverse transcriptase-polymerase chain reaction (RT-PCR) was performed with Quantitect Probe RT-PCR kit (Qiagen) following the amplification parameters given in the manufacturer protocol. Sequences of STAT1 primers and FAM-labeled probe were as follows: forward primer, 5′-AAGCGAACTGGATACATCA; reverse primer, 5′-CCGGGACATCTCATCAAAC; and probe, 5′-CAGACCACAGACAACC. Fold changes were calculated relative to the level of β-actin, determined using the QuantiTect Mm_Actb Assay (Qiagen). Reactions were carried out using Applied Biosystems (Foster City, CA) 7300 Real Time PCR system. To calculate the relative expression of target and reference genes, amplification efficiencies were determined.27 Results were plotted as ratio of STAT1 expression between proliferating and nonproliferating cells, and as amplification curves of ΔRn (an indicator of the signal generated by the PCR) against cycle numbers. The CT values, indicating the earliest cycle for detection, are given in the legends.
Results
Role for STAT1 in IFN-mediated inhibition of CD8 T-cell proliferation
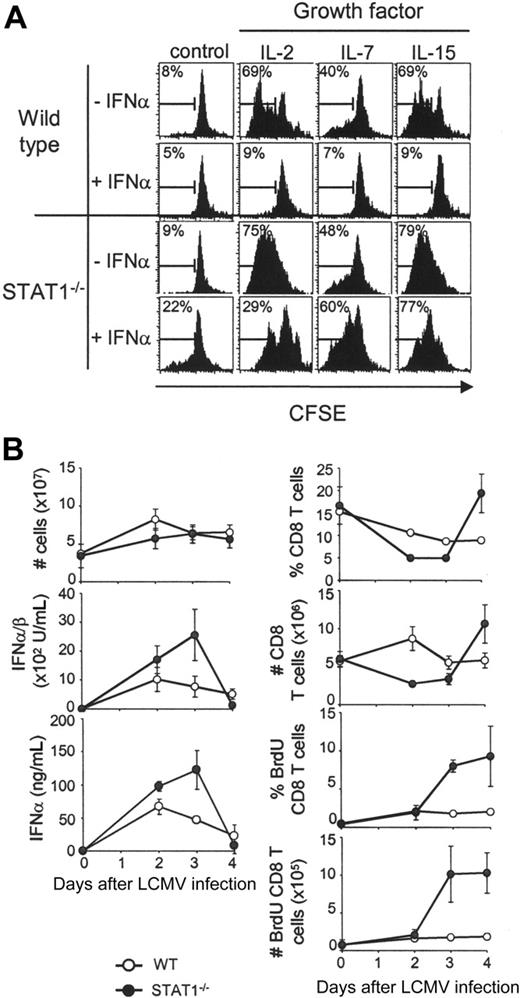
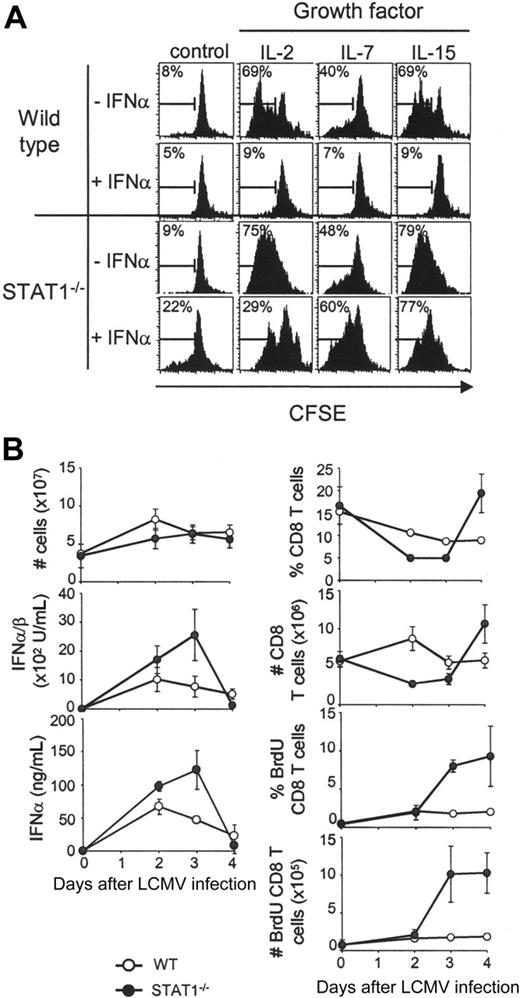
The STAT1 effects on CD8 T-lymphocyte proliferation were evaluated in vitro with responses to exogenous growth factors. Splenic leukocytes derived from STAT1-competent or -deficient mice were labeled with carboxy-fluorescein diacetate, succinimidyl ester (CFSE) and incubated with IL-2, IL-7, or IL-15. All of these cytokines use the common cytokine-receptor γ-chain to induce cell proliferation and have well-described effects on CD8 T-cell proliferation.28-30 Flow cytometric analyses of proliferation, as measured by dilution of CFSE staining, demonstrated that presence of the growth factors resulted in 40% to 70% of the wild-type CD8 T cells dividing after 5 days in culture (Figure 1A). As expected, addition of the type 1 IFN, IFNα, dramatically inhibited this proliferation. Although IL-2, IL-7, or IL-15 also supported the division of STAT1-deficient cells, IFNα failed to inhibit their proliferation (Figure 1A). These results confirm the role of STAT1 as a key mediator in the ability of IFNα to regulate proliferation of CD8 T lymphocytes.
To examine the STAT1-dependent effects on proliferation in vivo, the CD8 T-lymphocyte proliferation elicited in response to LCMV infection was examined. For these experiments, the thymidine analog, BrdU, was injected into STAT1-competent and -deficient mice. This approach marks populations replicating their DNA. Uninfected (day 0) mice or mice infected with LCMV for 1, 2, 3, or 4 days received injections of BrdU 2 hours prior to killing (Figure 1B). These periods of infection overlap with peak type 1 IFN induction. Although overall splenic cell yields were not dramatically altered in wild-type as compared with STAT1-deficient mice and both groups responded to infection with the production of type 1 IFNs, CD8 T cells had deregulated proliferation in STAT1-deficient mice (Figure 1B). In contrast to the minimal expansion of, and low BrdU incorporation by, wild-type CD8 T lymphocytes, there was a greater than 2-fold expansion, and approximately 10% of these cells in the STAT1-deficient mice were labeled with BrdU. The expanding subsets were not specific for the LCMV immunodominant epitopes because they did not bind class I MHC molecules presenting these determinants (data not shown). Thus, STAT1 is critical for inhibiting nonspecific CD8 expansion at early times after challenge in vivo.
Differential regulation of STAT1 protein levels in responding CD8 T cells
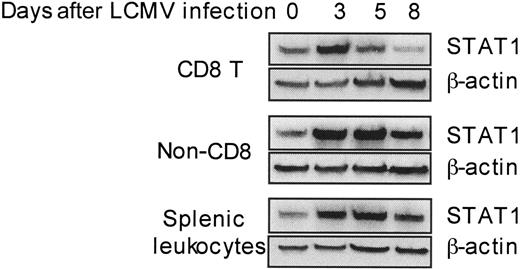
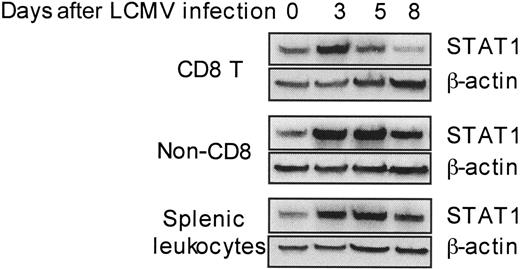
The aforementioned results demonstrate that high type 1 IFNs and STAT1 protein levels can be beneficial in limiting nonspecific T-cell proliferation. However, antigen-specific T cells need to be expanded under these conditions. In uninfected spleens, approximately 10% of the leukocytes are CD8 T cells having a diverse, nonspecific repertoire for antigen. By day 5 after infection, the CD8 T cells comprise up to 15% of the spleen and a low frequency of these can be identified as virus specific. The subset can represent greater than 50% of the leukocytes and is primarily virus specific by day 8 after infection.21,31,32 The day-5 and -6 time points are key because they overlap with periods of CD8 T-cell effector function, declining viral burdens, and type 1 IFN responses.21 Previous studies from our group have demonstrated that STAT1 levels are induced in total splenic leukocytes at days 2 to 5 after LCMV infection.18 To extend this characterization to particular cell subsets, STAT1 levels were determined by Western blot analysis of proteins extracted from CD8 T cells, as compared with non-CD8 and total splenic leukocyte populations isolated from uninfected (day 0) or day 3, 5, or 8 LCMV-infected mice. Remarkably, although STAT1 protein was modulated in all the populations tested (Figure 2), being low on days 0 and 8 and induced to high levels on days 3 and 5 after infection, the kinetics of induction were narrower in CD8 T cells, reaching its peak at day 3 and lower at day 5. The virus (4.7 ± 0.1 log PFU/g on day 5, and 4.2 ± 0.1 log PFU/g on day 6) and type 1 IFNs (50.5 pg/mL serum on day 5 and below detection on day 6 for this experiment [see also Pien et al21 ]) were still detectable at these times. Hence, STAT1 levels are limited in CD8 T cells at times when elevated expression of the transcription factor is maintained in other subsets, and a subset of the population is being activated in the presence of virus and type 1 IFN to mediate viral clearance.
Contribution of STAT1 to regulation of CD8 T-cell proliferation. (A) CD8 T cells from wild-type or STAT1-deficient mice proliferate in response to growth factors in culture, but STAT1 is required for the antiproliferative effects mediated by type 1 IFN. Total splenic populations were prepared from uninfected mice, CFSE-labeled on day 0, and cultured for 5 days in the absence or presence of IFNα (1 × 104 U/mL), with or without IL-2, IL-7, or IL-15 (20 ng/mL each). Flow cytometry histogram plots of gated CD8 T cells are shown. Numbers are proportions of cells dividing as assessed by dilution of CFSE. (B) STAT1 contributes to the negative regulation of early CD8 T-cell proliferation in vivo during LCMV infection. Wild-type and STAT1-deficient mice were infected with LCMV and followed for 4 days after infection, and CD8 T-cell proliferation was measured by BrdU incorporation in vivo. Proportions and numbers of total, as well as BrdU-positive CD8 T cells, are shown. Active IFNα/β in serum was measured by bioassay and serum IFNα by ELISA. All data are means ± SDs. Results for both panels are representative of at least 3 independent experiments.
Contribution of STAT1 to regulation of CD8 T-cell proliferation. (A) CD8 T cells from wild-type or STAT1-deficient mice proliferate in response to growth factors in culture, but STAT1 is required for the antiproliferative effects mediated by type 1 IFN. Total splenic populations were prepared from uninfected mice, CFSE-labeled on day 0, and cultured for 5 days in the absence or presence of IFNα (1 × 104 U/mL), with or without IL-2, IL-7, or IL-15 (20 ng/mL each). Flow cytometry histogram plots of gated CD8 T cells are shown. Numbers are proportions of cells dividing as assessed by dilution of CFSE. (B) STAT1 contributes to the negative regulation of early CD8 T-cell proliferation in vivo during LCMV infection. Wild-type and STAT1-deficient mice were infected with LCMV and followed for 4 days after infection, and CD8 T-cell proliferation was measured by BrdU incorporation in vivo. Proportions and numbers of total, as well as BrdU-positive CD8 T cells, are shown. Active IFNα/β in serum was measured by bioassay and serum IFNα by ELISA. All data are means ± SDs. Results for both panels are representative of at least 3 independent experiments.
Differential dynamic modulation of STAT1 levels in CD8 T cells during LCMV infection. CD8 T, non-CD8 T, and total splenic leukocytes were isolated from uninfected or LCMV-infected mice at the indicated days after challenge. Cell lysates were prepared, and STAT1 protein levels were detected by Western blot with β-actin measurement as a loading control. Data presented are representative of 2 independent experiments. Densitometry analyses of all tests demonstrated that the STAT1 values, normalized based on β-actin, for samples prepared on day 5 from non-CD8 T cells and total splenic leukocytes were at least 160% greater than those prepared from CD8 T cell.
Differential dynamic modulation of STAT1 levels in CD8 T cells during LCMV infection. CD8 T, non-CD8 T, and total splenic leukocytes were isolated from uninfected or LCMV-infected mice at the indicated days after challenge. Cell lysates were prepared, and STAT1 protein levels were detected by Western blot with β-actin measurement as a loading control. Data presented are representative of 2 independent experiments. Densitometry analyses of all tests demonstrated that the STAT1 values, normalized based on β-actin, for samples prepared on day 5 from non-CD8 T cells and total splenic leukocytes were at least 160% greater than those prepared from CD8 T cell.
Differential STAT1 expression in proliferating CD8 T cells
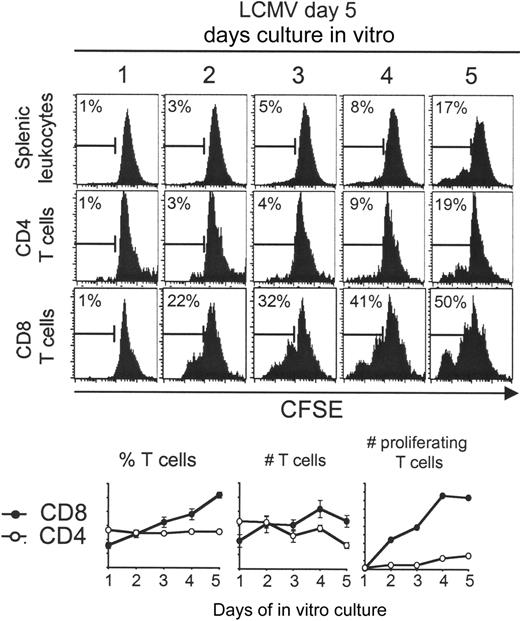
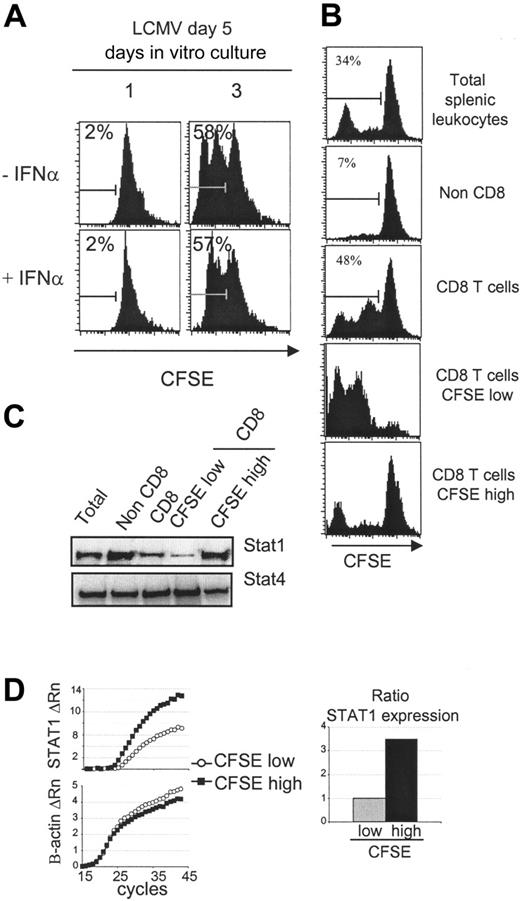
Two different approaches were developed to selectively evaluate STAT1 levels in CD8 T cells induced to proliferate during the infection. The first took advantage of the observation that splenic leukocytes harvested on day 5 after LCMV challenge continue to develop, without addition of exogenous factors, during in vitro culture.33 Cells obtained from uninfected, day 5, or day 8 LCMV-infected mice were labeled with CFSE and evaluated for proliferation at days 1, 2, 3, 4, and 5 after culture. Extensive ex vivo proliferation could only be demonstrated with the cells isolated on day 5 (Figure 3; data not shown). The CD4 T-cell subset had only modest proliferation under these conditions, but the CD8 T-cell subset continued to proliferate such that greater than 30% of the cells had diluted out the CFSE by day 3 after culture, and the recovery of this subset almost doubled. Interestingly, the addition of exogenous IFNα, at the concentrations blocking the cytokine-supported expansion of cells from uninfected mice (Figure 1), was unable to abolish the expansion of the in vivo-activated cells (Figure 4A; data not shown). Thus, the proliferation was relatively insensitive to IFN-mediated inhibition. To assess differences in STAT1 levels between the proliferating and nonproliferating cells, leukocytes isolated on day 5 after infection were CFSE labeled and cultured for 3 days ex vivo. CD8 T cells were prepared by magnetic bead purification and sorted into CFSE-low and -high populations (Figure 4A-B). Western blot analyses of the proteins extracted from the different cell populations showed that the CD8 had less STAT1 than the non-CD8 populations and that the proliferating had very low amounts of STAT1 compared with the nonproliferating CD8 T cells (Figure 4C). A similar difference was obtained analyzing STAT1 mRNA expression: it was lower in the proliferating cells and higher in the nonproliferating CD8 T cells with a difference of 3.5 points (Figure 4D). These results demonstrate that STAT1 is differentially expressed such that proliferating cells have relatively low and nonproliferating cells have relatively high levels. They suggest that selection of T-cell subsets for expansion is dependent on low STAT1 expression.
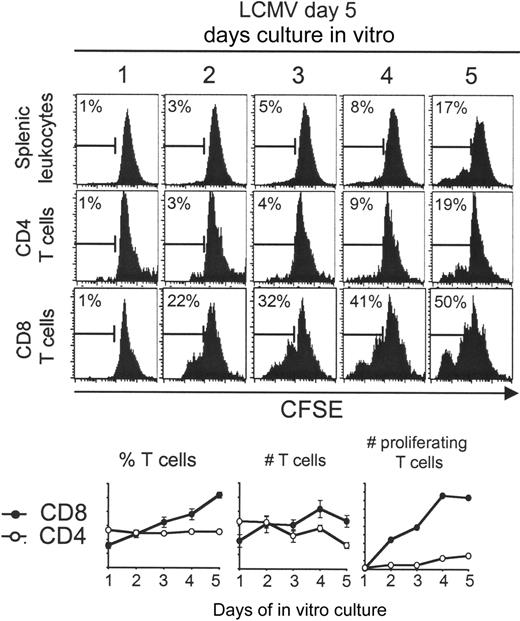
Splenic CD8 T cells derived from day 5 LCMV-infected mice continue to expand ex vivo. Total splenic leukocytes were prepared, CFSE-labeled, and cultured for 1 to 5 days. Histograms of CFSE dilution within electronically gated CD8 and CD4 populations are shown. The marker in the histogram shows percentage of proliferating cells based on CFSE dilution. The averages of different samples had SD between 0% and 2% for the splenic leukocytes, 0% and 5% for the CD4 T cells, and 0% and 4% for the CD8 T cells. Expressed on the graphs are percentages, as well as number of total and proliferating CD4 (○) and CD8 (•) T cells. The bars denote deviations of the means. The experiment has been repeated more than 2 times, obtaining similar results.
Splenic CD8 T cells derived from day 5 LCMV-infected mice continue to expand ex vivo. Total splenic leukocytes were prepared, CFSE-labeled, and cultured for 1 to 5 days. Histograms of CFSE dilution within electronically gated CD8 and CD4 populations are shown. The marker in the histogram shows percentage of proliferating cells based on CFSE dilution. The averages of different samples had SD between 0% and 2% for the splenic leukocytes, 0% and 5% for the CD4 T cells, and 0% and 4% for the CD8 T cells. Expressed on the graphs are percentages, as well as number of total and proliferating CD4 (○) and CD8 (•) T cells. The bars denote deviations of the means. The experiment has been repeated more than 2 times, obtaining similar results.
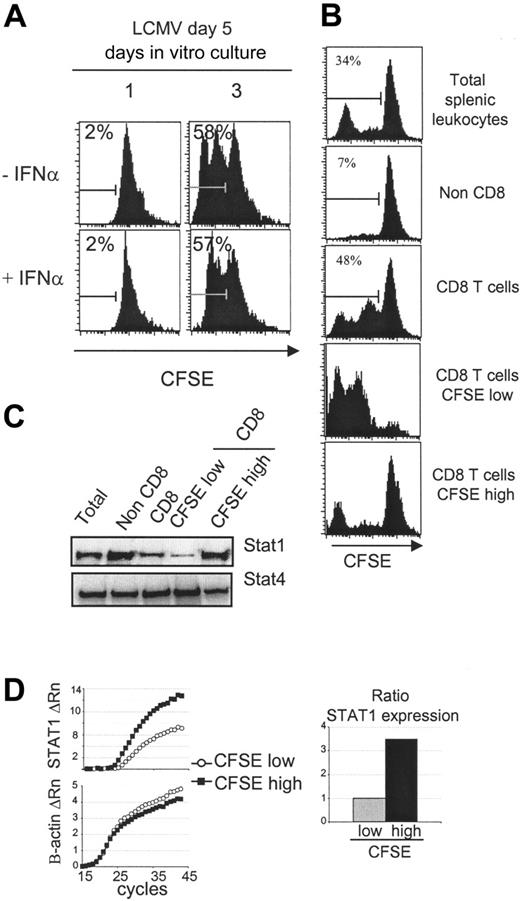
Reduced STAT1 levels in proliferating CD8 T cells expanding ex vivo. (A) Splenic CD8 T cells derived from day 5 LCMV-infected mice cultured ex vivo are insensitive to the antiproliferative effects of IFNα. Total splenic leukocytes were prepared, CFSE-labeled, and incubated 1 or 3 days with or without IFNα (1 × 104 U/mL). Histograms of CFSE dilution within electronically gated CD8 populations are shown. The experiment has been repeated more than 2 times with similar results. The overall averages ± SDs for the combined experiments were, respectively without or with IFNα, 2.2 ± 0.5 and 1.9 ± 0.7 on day 1 and 58.9 ± 3.9 and 59.2 ± 2.0 on day 3. (B) Cultured CD8 T cells were separated into high and low proliferating, based on dilution of CFSE. CD8 T cells were enriched from a 3-day culture of CFSE-labeled splenic populations prepared from day 5 LCMV-infected mice and then sorted into CFSE-low and -high subsets. CFSE staining intensities of the different samples are shown. (C) The STAT1 protein levels are lower in proliferating than nonproliferating CD8 T cells. Western blot analysis of STAT1 levels in the populations prepared in panel B. Western blot analysis of STAT4 was used as loading control. Also tested was β-actin (not shown). This figure is representative of 3 experiments. (D) STAT1 RNA levels are lower in proliferating CD8 T cells. Real-time PCR analysis of STAT1 RNA levels prepared from sorted CFSE-low (○) and CFSE-high (▪) CD8 T cells is shown. The ratio of STAT expression was calculated using amplification efficiencies (1.93 for STAT1 and 1.99 for β-actin). The experiment was repeated twice. The average CT values ± SDs for the low CFSE samples were 29.0 ± 4.4 for STAT1 and 20.9 ± 0.4 for β-actin, and for the high CFSE samples were, respectively, 25.6 ± 1.6 and 22.8 ± 0.6.
Reduced STAT1 levels in proliferating CD8 T cells expanding ex vivo. (A) Splenic CD8 T cells derived from day 5 LCMV-infected mice cultured ex vivo are insensitive to the antiproliferative effects of IFNα. Total splenic leukocytes were prepared, CFSE-labeled, and incubated 1 or 3 days with or without IFNα (1 × 104 U/mL). Histograms of CFSE dilution within electronically gated CD8 populations are shown. The experiment has been repeated more than 2 times with similar results. The overall averages ± SDs for the combined experiments were, respectively without or with IFNα, 2.2 ± 0.5 and 1.9 ± 0.7 on day 1 and 58.9 ± 3.9 and 59.2 ± 2.0 on day 3. (B) Cultured CD8 T cells were separated into high and low proliferating, based on dilution of CFSE. CD8 T cells were enriched from a 3-day culture of CFSE-labeled splenic populations prepared from day 5 LCMV-infected mice and then sorted into CFSE-low and -high subsets. CFSE staining intensities of the different samples are shown. (C) The STAT1 protein levels are lower in proliferating than nonproliferating CD8 T cells. Western blot analysis of STAT1 levels in the populations prepared in panel B. Western blot analysis of STAT4 was used as loading control. Also tested was β-actin (not shown). This figure is representative of 3 experiments. (D) STAT1 RNA levels are lower in proliferating CD8 T cells. Real-time PCR analysis of STAT1 RNA levels prepared from sorted CFSE-low (○) and CFSE-high (▪) CD8 T cells is shown. The ratio of STAT expression was calculated using amplification efficiencies (1.93 for STAT1 and 1.99 for β-actin). The experiment was repeated twice. The average CT values ± SDs for the low CFSE samples were 29.0 ± 4.4 for STAT1 and 20.9 ± 0.4 for β-actin, and for the high CFSE samples were, respectively, 25.6 ± 1.6 and 22.8 ± 0.6.
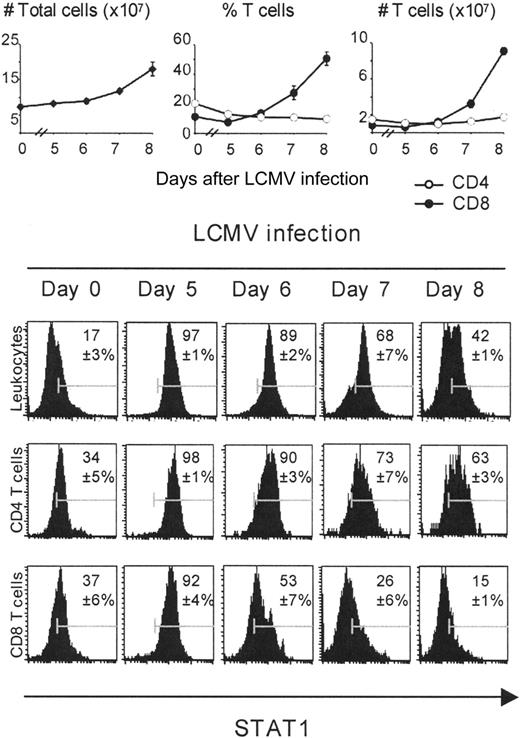
To directly examine STAT1 protein levels within individual cells immediately after isolation, a variety of antibodies were screened for their ability to fluorescently label total STAT1 protein intracellularly. The approach required optimizing conditions for cytoplasmic and nuclear staining of total STAT1 using a phycoerythrin-conjugated anti-mouse STAT1 antibody. Isotype control antibodies and cells derived from STAT1-/- mice were used as negative controls to demonstrate specificity of staining (see “Materials and methods”). The results of staining for STAT1 protein in total leukocyte populations complemented the results of the Western blot analysis and extended them to evaluate expression within individual cells. Low but detectable levels of STAT1 were expressed with relative uniformity in populations from uninfected mice, expression was dramatically elevated with greater than 87% of cells expressing high levels on days 5 and 6 after infection, and decreased on days 7 and 8 (Figure 5). As expected, the CD8 but not the CD4 T-cell subset was expanding in vivo starting on day 6 after infection (Figure 5). Gating for analysis of the CD4 T-cell population demonstrated that the majority of these cells had STAT1 levels similar to those observed in total populations (Figure 5). In contrast, the CD8 T-cell population also had STAT1 being induced but a subset had lower expression on day 6 as compared with total or CD4 T cells, that is, 47% versus 11% and 10%. Moreover, the levels of expression were lower in a proportion of the T cells throughout the peak induction of STAT1 with bright staining at only 53% on day 6, 26% on day 7, and 15% on day 8 as compared with approximately 90%, 70%, and 40% to 60% of the total and CD4 T-cell populations (Figure 5). Thus, 2 different populations of CD8 T cells were developing, one with induced STAT1 levels equivalent to those achieved in total and CD4 T-cell populations and another with lower STAT1 levels.
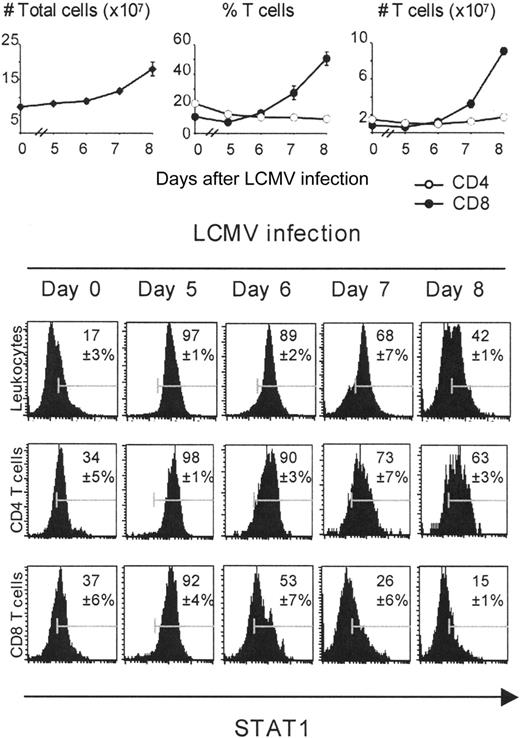
Differential expansion and time course of STAT1 expression on CD4 and CD8 T cells during the course of infection. For these experiments, populations were prepared from uninfected or day 5, 6, 7, and 8 LCMV-infected mice. Splenic leukocytes were counted using a hemocytometer. Percentages of CD4 and CD8 T cells were obtained by FACS analysis. The intracellular staining for expression of STAT1 protein was analyzed by FACS, electronically gating on total splenic leukocytes, CD8 or CD4 T cells. The numbers in the histograms represent percentages of STAT1-positive cells, compared with the isotype controls.
Differential expansion and time course of STAT1 expression on CD4 and CD8 T cells during the course of infection. For these experiments, populations were prepared from uninfected or day 5, 6, 7, and 8 LCMV-infected mice. Splenic leukocytes were counted using a hemocytometer. Percentages of CD4 and CD8 T cells were obtained by FACS analysis. The intracellular staining for expression of STAT1 protein was analyzed by FACS, electronically gating on total splenic leukocytes, CD8 or CD4 T cells. The numbers in the histograms represent percentages of STAT1-positive cells, compared with the isotype controls.
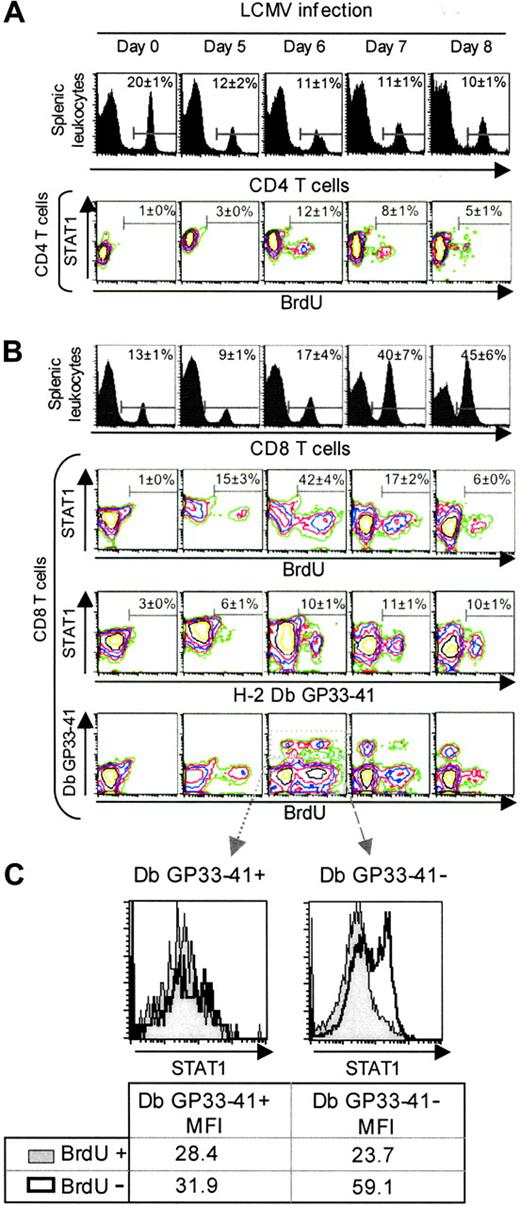
To further explore the subset of the proliferating cells in vivo, splenic leukocytes were harvested from uninfected or LCMV-infected mice who received injections of 5-bromo-2′-deoxyuridine (BrdU) 2 hours prior to killing, and fluorescently labeled with anti-CD8 or anti-CD4, anti-BrdU, and anti-STAT1.31 As expected, the proportions of CD4 T cells were not dramatically increasing on days 5, 6, 7, or 8 after infection (Figure 6A). Only 12% of these cells were incorporating the DNA precursor, BrdU, on day 6, but they were contained within the CD4 T-cell subset having relatively low levels of STAT1. Analysis of the expanding CD8 T cells demonstrated that greater than 40% of these were incorporating BrdU on day 6 after infection, and they were preferentially represented in the higher proportion of STAT1-low cells (Figure 6B). Thus, as compared with total cells and CD4 T cells, CD8 T cells are expanded to higher proportions and numbers during infection, have higher proportions of the low STAT1subset, and the populations replicating their DNA during the 2 hours immediately prior to harvest are found in the low STAT1 subset.
Differential STAT1 expression in antigen-specific CD8 T cells
To examine STAT1 levels within the antigen-specific CD8 T-cell subset, a fluorescently labeled MHC class I Db tetramer conjugated to the LCMV peptide GP33-41 was also used for staining. This LCMV peptide is an immunodominant viral determinant, and Db GP33-41 is bound by T-cell receptors specific for the complex.31 Splenic leukocytes harvested from uninfected or LCMV-infected mice who had received injections of BrdU 2 hours prior to being killed were stained with anti-CD8, anti-BrdU, anti-STAT1, and Db GP33-41. The antigen-specific CD8 T-cell subset was preferentially found in the low STAT1 subset on day 6 after infection, representing about 10% of the CD8 T cells (Figure 6B). Analysis of the Db GP33-41-positive and BrdU-positive or -negative subsets showed that all of the antigen-specific cells were low for STAT1 (Figure 6C). In contrast, analysis of the Db GP33-41-negative subset showed that STAT1-low and -high populations were present (Figure 6C). These results suggest that the greater higher proportions of the STAT1-low, BrdU-positive cells, that is, 40%, contained antigen-specific cells for other immunodominant viral peptides31 or cells in cycle having down-regulated antigen receptors.34 Although the proportions cannot be precisely quantitated, these data conclusively show that the in vivo-proliferating, antigen-specific CD8 T cells display a phenotype characterized by low STAT1 expression.
Discussion
The data presented here show that STAT1 is an important factor in regulating inappropriate CD8 T-cell proliferation in vivo at early times during viral infection. Moreover, they conclusively demonstrate that different cell types are induced to express a range of total STAT1 levels during the course of responses to viral infections. Although all leukocytes have elevated expression of the transcription factor, a subset of dividing, antigen-specific cells has lower levels and reduced sensitivity to IFN-mediated antiproliferative effects. The dichotomy of the magnitude of expression allows antigen-specific CD8 T-cell expansion, even in the presence of type 1 IFN, to defend against infection. Thus, a fine-tuning of cytokine effects by regulation of total levels of intracellular signaling molecules is defined.
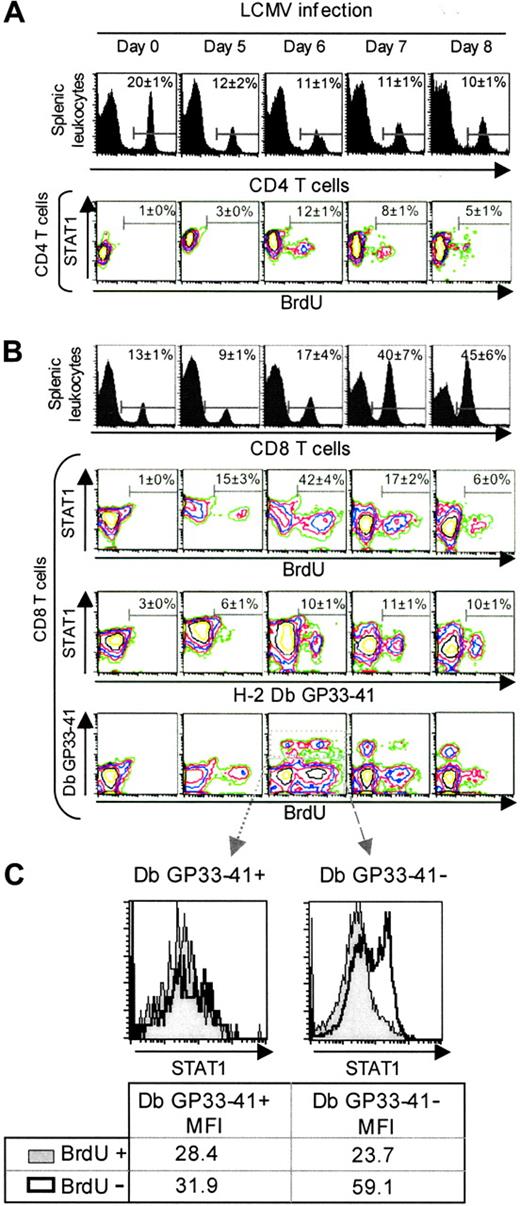
Modulation of STAT1 levels in vivo-reduced levels in antigen-specific proliferating CD8 T cells. For these experiments, populations were prepared from uninfected or day 5, 6, 7, and 8 LCMV-infected mice. BrdU was injected at 2 hours before harvest, and the isolated cells were labeled to evaluated CD4, CD8, BrdU, and STAT1 expression by FACS (A) CD4 T cells experience low levels of proliferation during LCMV infection. Histograms represent the percentage of CD4 T cells in the splenic leukocytes. The contour plots show the staining for STAT1 and BrdU in the electronically gated CD4 T-cell subset. (B) The proliferating and antigen-specific CD8 T-cell subsets are contained within the low STAT1 populations. Histograms represent the percentage of CD8 T cells in the splenic leukocytes. The contour plots show the staining for the electronically gated CD8 T-cell subset. In the first set, STAT1 versus BrdU incorporation, as a marker of in vivo proliferation, is shown. In the second set, STAT1 versus the tetramer Db GP33-41, as a marker of cells specific for the LCMV immunodominant epitope, is shown. In the third set, Db GP33-41 versus STAT1 is shown. Data presented are means ± SDs. (C) STAT1 levels are low in Db GP33-41-positive and -negative expanding CD8 T cells. Histograms represent STAT1 levels in the electronically gated peptide specific or unspecific populations. Numbers show mean fluorescence intensities. These experiments have been repeated more than 3 times.
Modulation of STAT1 levels in vivo-reduced levels in antigen-specific proliferating CD8 T cells. For these experiments, populations were prepared from uninfected or day 5, 6, 7, and 8 LCMV-infected mice. BrdU was injected at 2 hours before harvest, and the isolated cells were labeled to evaluated CD4, CD8, BrdU, and STAT1 expression by FACS (A) CD4 T cells experience low levels of proliferation during LCMV infection. Histograms represent the percentage of CD4 T cells in the splenic leukocytes. The contour plots show the staining for STAT1 and BrdU in the electronically gated CD4 T-cell subset. (B) The proliferating and antigen-specific CD8 T-cell subsets are contained within the low STAT1 populations. Histograms represent the percentage of CD8 T cells in the splenic leukocytes. The contour plots show the staining for the electronically gated CD8 T-cell subset. In the first set, STAT1 versus BrdU incorporation, as a marker of in vivo proliferation, is shown. In the second set, STAT1 versus the tetramer Db GP33-41, as a marker of cells specific for the LCMV immunodominant epitope, is shown. In the third set, Db GP33-41 versus STAT1 is shown. Data presented are means ± SDs. (C) STAT1 levels are low in Db GP33-41-positive and -negative expanding CD8 T cells. Histograms represent STAT1 levels in the electronically gated peptide specific or unspecific populations. Numbers show mean fluorescence intensities. These experiments have been repeated more than 3 times.
The modulation of STAT1 protein levels following viral infection within antigen-specific CD8 T cells is reported here for the first time. The studies show that the ability to proliferate in the presence of type 1 IFNs is a consequence of less total protein rather than protein absence. Although earlier studies with STAT1-deficient cells suggested that the presence or absence of the molecule might be the factor determining the consequences of cytokine exposure,8 the in vivo titration of the levels observed here provides a more biologically conservative mechanism, with the potential to maintain a cell's ability to access antiviral gene targets for defense while limiting the antiproliferative effects of IFN exposure. The titration of expression is more than about 2 logs of fluorescence intensity when evaluated within individual cells by FACS analysis, and intensity changes with the kinetics of infection in all populations (see Figure 5). Consistent with the observed dynamic increases and decreases in expression, careful timing was required to demonstrate the difference, with ex vivo cells expanded in culture, by Western blot analysis (see Figure 4). This may explain why earlier attempts by others have failed to detect reduced STAT1 within total proliferating CD4 T-cell populations by Western blot analysis.35 Alternatively, because the proportions of CD4 T cells with low STAT1 levels were much smaller (Figures 5, 6), the difference in STAT1 levels may provide a mechanism preferentially favoring antigen-specific CD8 T-cell proliferation in the context of high type 1 IFN induction. Current studies in our laboratory are extending the characterization of STAT1 levels to other cell types and advancing the understanding of the pathways regulating STAT1 expression in antigen-specific CD8 T cells.
The results add to and extend earlier work from our group demonstrating that type 1 IFNs promote IFN-γ production by CD8 T cells,21,36 and that the presence of STAT1 negatively regulates the induction of IFN-γ by IFN-α/β.8,18 Taken together, these studies indicate that cells are controlling responses to the cytokines, ranging from inhibition of proliferation to induction of IFN-γ, based on access to intracellular signaling pathways. The new work also provides a biologic context for doing this during a competent immune response. The data complement studies from others showing that the type 1 IFNs, as well as another cytokine using STAT1 for receptor signaling, IFN-γ,9 can act to increase the abundance of CD8 T cells during viral infections and in response to antigen stimulation in culture.37,38 In contrast to the early inhibition of proliferation reported here (Figure 1), these reports demonstrate that another consequence of IFN exposure may act later during the development of an immune response to enhance the numbers of antigen-specific cells. The balance is likely to be one more example of how cell conditioning to express different levels of STAT molecules may change responses to the cytokines in paradoxical ways. Ongoing experiments are evaluating the possible modification of gene targets after IFN treatment of populations conditioned to express various levels of STAT1 in vivo, and the requirements for other STAT molecules in the expression of individual genes (M.P.G. and C.B., unpublished observations). By providing the biologic context for the regulation of STAT expression, these experiments extend and complete the work characterizing how different STAT molecules play particular roles in accessing type 1 IFN effects in T cells.14,39
The results have profound implications concerning the consequences of therapeutic intervention to regulate IFN levels and functions.2-6 They suggest that treatments with IFNs to protect against viral infections and cancer may differentially regulate immune function depending on the cellular levels of total STAT1 protein. If the cytokines are administered under conditions whereby all of the T cells respond with high expression of STAT1, the treatments may interfere with the expansion of antigen-specific cells for defense. However, neutralization of endogenous IFN function to protect against autoimmune diseases may result in conditions promoting the activation and expansion of nonspecific cells as well as limit the antimicrobial defense mechanisms activated by the cytokine. Clearly, learning how to control the levels of STAT1 within cells and using this during cytokine manipulation may help promote appropriate immune responses and allow access to the antimicrobial defense functions as needed.
Prepublished online as Blood First Edition Paper, October 6, 2005; DOI 10.1182/blood-2005-07-2834.
Supported by the National Institutes of Health (grants RO1-AI55677, RO1-CA41268, and F31-GM20760), the Rhode Island Foundation, and a National Defense Science and Engineering Graduate Fellowship.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ken Nguyen for thoughtful insights and suggestions, and Kathryn Doiron for technical assistance.