Abstract
Following initiation of coagulation as part of the hemostatic response to injury, thrombin is generated from its inactive precursor prothrombin by factor Xa as part of the prothrombinase complex. Thrombin then has multiple roles. The way in which thrombin interacts with its many substrates has been carefully scrutinized in the past decades, but until recently there has been little consideration of how its many functions are coordinated or directed. Any understanding of how it is directed requires knowledge of its structure, how it interacts with its substrates, and the role of any cofactors for its interaction with substrates. Recently, many of the interactions of thrombin have been clarified by crystal structure and site-directed mutagenesis analyses. These analyses have revealed common residues used for recognition of some substrates and overlapping surface exosites used for recognition by cofactors. As many of its downstream reactions are cofactor driven, competition between cofactors for exosites must be a dominant mechanism that determines the fate of thrombin. This review draws together much recent work that has helped clarify structure function relationships of thrombin. It then attempts to provide a cogent proposal to explain how thrombin activity is directed during the hemostatic response.
Introduction: natural hemostatic substrates of thrombin
Proteinases play crucial roles in biologic processes involved in organism homeostasis. Their actions require tight regulation and coordination. Failure of regulation and coordination leads to disease. The hemostasis system involves activation, regulation, and coordination of numerous proteinases. The complex reactions occur on activated or damaged cells, platelets, and endothelial cells. In normal physiologic conditions hemostasis is exquisitely initiated, controlled, and terminated. Genetic or physiologic perturbations, however, can lead to severe dysfunction, resulting in either hemorrhagic disorders or thromboembolic disease, illustrating the need for tight regulation of these proteolytic reactions.
Central to the functioning of hemostasis are the roles of thrombin. Numerous potential substrates have been identified for thrombin,1,2 suggesting diverse roles. Only those substrates related to hemostasis are considered here. A prime function of thrombin is the conversion of fibrinogen to fibrin. Two fibrinopeptides (FPs) are cleaved, FPA (the 16-residue N-terminal peptide from the Aα chain) and FPB (the 14-residue peptide from the Bβ chain). Cleavage of FPA occurs first, and this forms a fibrin monomer (termed fibrin I) which spontaneously polymerizes to form protofibrils. Cleavage of FPB generates fibrin II protofibrils that undergo lateral aggregation. In the dynamic process of thrombin generation in blood, some of the early generated thrombin feeds back on the cascade system to activate factors V and VIII, enabling more sustained generation of thrombin.3,4 Thrombin activates factors V and VIII by excision of their central B domains. In factor V, R709, R1018, and R1545 are cleaved, leaving an A1-A2 domain fragment, noncovalently associated with an A3-C1-C2 domain fragment. In factor VIII, the corresponding cleavages are R372, R740, R1649, and R1689. This leaves an A1-A2 fragment associated with an A3-C1-C2 fragment. Thrombin cleaves the factor XIII A subunit at R37, releasing an aminoterminal activation peptide, resulting in exposure of its active site C314 and initiation of fibrin stabilization by covalent crosslinking. Thrombin generation occurring in the context of normal hemostasis is located on the surface of activated platelets that form the primary hemostatic plug. The platelet can be activated by a number of agonists. Thrombin specifically activates the platelet by interacting and cleaving protease-activated receptors (PARs).5 Cleavage of the PAR-1 receptor is at R41, revealing a tethered activating hexapeptide that self-associates with one of its extracellular loops, causing receptor-induced signaling. A second platelet activation mechanism involves proteolysis of glycoprotein V (GpV), part of the GpIb-IX-V complex on its surface, with the generation of hyperresponsive platelets.6 Thrombin can activate factor XI by proteolysis at R369 under rather defined conditions requiring anions such as dextran sulfate.7 However, in the presence of activated platelets, a more likely physiologic activation process involves GpIbα.8 Thrombin may also indirectly enhance platelet adhesion. Recently, thrombin has been shown to proteolyze ADAMTS13 (a disintegrin and metalloprotease with thrombospondin type 1 motif) at multiple sites, leading to its inactivation.9 ADAMTS13 is the protease responsible for von Willebrand factor (VWF) processing. Von Willebrand factor is a key adhesive glycoprotein required for platelet adhesion to the site of injury. By inactivating ADAMTS13, thrombin can potentially aid platelet recruitment to the site of vessel injury. All of these reactions of thrombin with its substrates can be considered as procoagulant; that is, clot promoting.
Thrombin also participates in proteolytic interactions as part of endogenous anticoagulant pathways. First, thrombin generated in the vicinity of the endothelial cell surface is trapped in a high-affinity complex with the cell-surface proteoglycan thrombomodulin.10 Once bound as the thrombin-thrombomodulin complex, its substrate specificity is redirected from procoagulant to anticoagulant reactions. Procoagulant reactions are impeded and efficient activation of protein C occurs, by cleavage at R169 and release of an activation peptide. Second, in cleaving R393 of the reactive loop of the plasma SERPIN (serine protease inhibitor) antithrombin, it is trapped in an essentially irreversible complex and cleared from the circulation. This reaction is accelerated by the glycosaminoglycans heparin and heparan sulfate.11 Third, thrombin is similarly inhibited by cleavage of L444 of the SERPIN heparin cofactor II in the presence of the glycosaminoglycan, heparan sulfate, or the galactosaminoglycan, dermatan sulfate.12
The thrombin-thrombomodulin complex also activates a carboxypeptidase, thrombin-activated fibrinolysis inhibitor (TAFI) by proteolysis at R92,13,14 at a rate approximately 1000 times greater than thrombin alone. Activated TAFI is then able to remove lysine residues from fibrin that form favored binding sites for fibrinolytic proteins. Recent work has suggested that TAFI, when activated, may also have a broad antiinflammatory role. It modulates the proinflammatory properties of bradykinin, complement C5a, and thrombin-cleaved osteopontin with similar efficiency as its effect on fibrin.15
These multiple interactions in which thrombin participates raise the question of how it knows which substrate it should interact with and cleave during hemostasis. Does it, on the one hand, randomly cleave all of these substrates, or, on the other hand, is it directed in some way to cleave the substrates in a coordinated manner?
Thrombin generation
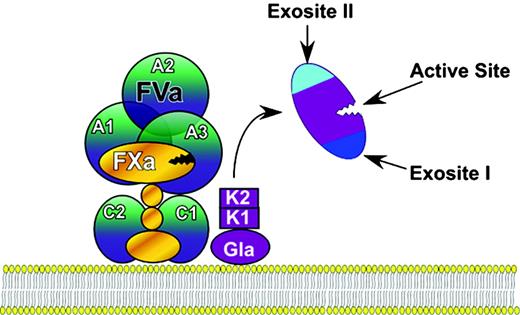
To address this question, it is necessary to consider how the activity of thrombin is controlled by its structure. During its activation by factor Xa, prothrombin is cleaved, first at R320 to generate meizothrombin and then at R271 to generate thrombin and fragment F1 + 2 (an alternative activation pathway is possible but may be less favored kinetically16 and is therefore ignored here). The F1 + 2 fragment contains the Gla and kringle domains, which function to localize prothrombin on membrane surfaces as part of the activation (or “prothrombinase”) complex, that includes factor Va and factor Xa (Figure 1). Once prothrombin is activated, thrombin can escape the complex and is free to attack its substrates. Prothrombin is unique among the activated coagulation proteinases in that it completely loses the domains important for initial recognition interactions when it is activated to its serine proteinase derivative, thrombin. Loss of these domains allows thrombin to diffuse freely to encounter, recognize, cleave, and dissociate from its many substrates. Furthermore, their loss permits exposure of cryptic17 functional regions, the active site, and the charged anion binding regions, termed exosites,18 crucial for extending the range and specificity of thrombin's actions.
Prothrombin activation to thrombin. Prothrombin is first activated as part of the prothrombinase complex to meizothrombin by cleavage at R320. The Gla and kringle (K1 and K2) domains are cleaved at R271, allowing thrombin to diffuse away. Thrombin has 3 regions that are functionally inaccessible in prothrombin and become exposed on activation: the active site, exosite I, and exosite II.
Prothrombin activation to thrombin. Prothrombin is first activated as part of the prothrombinase complex to meizothrombin by cleavage at R320. The Gla and kringle (K1 and K2) domains are cleaved at R271, allowing thrombin to diffuse away. Thrombin has 3 regions that are functionally inaccessible in prothrombin and become exposed on activation: the active site, exosite I, and exosite II.
Thrombin structure and allostery
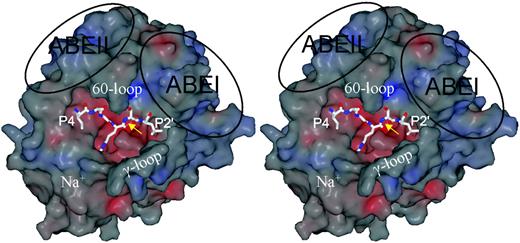
The high-resolution structure of thrombin was considered in detail in a prescient and influential article by Bode et al.18 Thrombin is highly homologous to serine proteases such as chymotrypsin, with a serine residue (S195, chymotrypsin numbering) in an active site grove, along which its substrate must lie (Figure 2). Thrombin has critical features that control specificity, loops, and charged patches surrounding the active site pocket. The 60- and γ-loops frame the canyonlike active site cleft containing S195. The 60-loop is hydrophobic with a structural rigidity provided by 2 adjacent Pro residues (P60b, P60c). It interacts with hydrophobic residues of the substrate, N-terminal to the cleavage site. Deletion mutant studies of the 60-loop have illustrated its role in controlling substrate activation.19,20 The γ-loop is more mobile, hydrophilic, and can make contact with residues C-terminal to the cleavage site.
Of importance for the discussion that follows are the exosites I and II (depicted ABEI and ABEII in Figure 2). Exosite I is centered on residues K36, H71, R73, R75, Y76, R77a, and K109/110, and exosite II includes residues R93, K236, K240, R101, and R233. The structural basis of the exosites and how they interact with cofactors has been reviewed comprehensively elsewhere21 and will not therefore be considered in detail here.
Another important loop contains a site for Na+ binding (Figure 2). Occupancy of its binding site controls some of the activities of thrombin and is important for the consideration of allostery. Allostery of thrombin is an intriguing and only partially resolved issue that can be considered from several (related) viewpoints. First, Na+ when bound to thrombin (denoted initially as the “fast” as opposed to the unoccupied “slow” form22 ) alters its function, increasing access of small substrates to the active site and favoring certain procoagulant (fibrinogen and PAR-1) rather than anticoagulant substrates. The residues energetically linked to Na+-induced allostery are D189, E217, D222, and Y225.23 Occupancy of its binding site affects the activity of thrombin, but, because Na+ concentration in blood is tightly controlled (140 mM), the site will be similarly occupied under all but the most extreme pathologic circumstances. Second, mutants such as E217K24 and W215A/E217A25 mimic, or adopt the conformation of, the slow form and exhibit greatly reduced procoagulant activities but retain or have inducible anticoagulant activities. Such mutants represent potential therapeutic anticoagulants.26 Four crystal structures of the slow form of thrombin have been proposed and show a collapsed active site pocket, but these differ appreciably in detail.24,25,27,28 Third, and of most importance to the present context, occupancy of exosites I and II may induce allosteric changes to the active site to promote catalysis. This is supported by functional analyses that show linkage between the exosites and the catalytic center,29,30 by mutational analysis of exosite residues of thrombin31 and by enhanced reactivity of inhibitors to thrombin, when it is complexed to thrombomodulin.32 Occupancy of exosite I of thrombin by thrombomodulin did not appear to alter the active site in a crystal structure of the complex,33 a finding that may be explained by use of an active site inhibitor to prevent proteolysis during crystallization. At its extreme, allostery might extend to reciprocal exosite inhibition,34 an idea that has, however, been firmly challenged.29
Substrate recognition
Of the very early actions of thrombin, the cleavage of FPA to form fibrin monomer is critical in determining the fate of the proteinase. Thrombin binds directly to the fibrinogen Aα chain, binding being mediated through part of exosite I (Figure 3A; Table 1) and also the active site cleft.36 Recognition of fibrinogen, as opposed to its isolated polypeptide chains or its proteolyzed N-terminal E domain, involves additional surface residues on the face containing the active site serine, identified by clotting experiments.37-39 Release of FPA is sufficient to induce fibrin polymerization, and, while the release of FPB follows closely that of FPA, it is determined by the relative specificity constants of thrombin for the Aα and Bβ chains, and premature release of FPB does not affect polymerisation.40 Exosite I is also involved in the binding of thrombin to the Bβ chain to cleave FPB (Table 1). Another early action of thrombin is activation of factor V, needed to promote further generation of thrombin. Numerous surface residues on thrombin are involved with residues of exosites I and II and the Na+ loop required for cleavage of R70941 (Figure 3B; Table 1). Factor VIII cleavage activation by thrombin involves a similar wide distribution of residues (Table 1). Cleavage at R372 requires residues in both exosites, while cleavage at R1689 does not require exosite II.42 It should be noted that each of these early reactions of thrombin are direct between it and the substrates, and no cofactors are involved in acceleration of the cleavage reactions. This contrasts with the substrate reactions below, which are all assisted by cofactors.
The structural features of thrombin. A stereo view of the surface thrombin in the standard orientation reveals the main features of the protease. The surface is colored according to electrostatic potential, with red and blue representing negatively and positively charged patches, respectively. In this orientation, the active site is centered so that a peptide substrate will run from the left to the right, from its N- to C-termini. Here, the P4 to P2′ residues of antithrombin are depicted as found in the structure of the Michaelis complex, with the reactive center bond (P1-P1′) indicated by the yellow arrow. The active site cleft is flanked above by the 60-loop, and below by the γ-loop making the cleft unusually deep. The 2 major anion binding exosites (ABEI and ABEII) are indicated by the ovals. The adjacent Na+ binding site is also indicated.
The structural features of thrombin. A stereo view of the surface thrombin in the standard orientation reveals the main features of the protease. The surface is colored according to electrostatic potential, with red and blue representing negatively and positively charged patches, respectively. In this orientation, the active site is centered so that a peptide substrate will run from the left to the right, from its N- to C-termini. Here, the P4 to P2′ residues of antithrombin are depicted as found in the structure of the Michaelis complex, with the reactive center bond (P1-P1′) indicated by the yellow arrow. The active site cleft is flanked above by the 60-loop, and below by the γ-loop making the cleft unusually deep. The 2 major anion binding exosites (ABEI and ABEII) are indicated by the ovals. The adjacent Na+ binding site is also indicated.
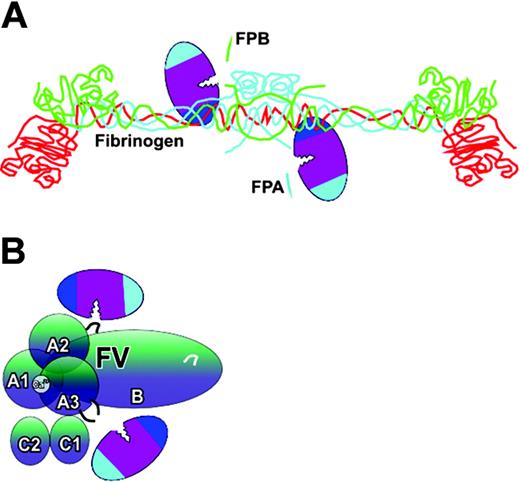
Direct, unassisted interactions of thrombin. Thrombin binds directly to (A) fibrinogen Aα (blue) and Bβ (green) polypeptide chains using exosite I (fibrinogen γ chain is red). This enables the scissile bond to be positioned across the active site cleft. FPA and FPB (depicted as blue and green fragments) are then cleaved. Thrombin also binds directly to factor V (FV) (B). The 3 activation cleavage sites at R709, R1545, and R1018 are recognized using many surface residues, including those of both exosites. Although 2 thrombin molecules are depicted, a single docking event may be sufficient to complete proteolysis of each protein. The schematic of fibrinogen is adapted from Cote et al35 with permission.
Direct, unassisted interactions of thrombin. Thrombin binds directly to (A) fibrinogen Aα (blue) and Bβ (green) polypeptide chains using exosite I (fibrinogen γ chain is red). This enables the scissile bond to be positioned across the active site cleft. FPA and FPB (depicted as blue and green fragments) are then cleaved. Thrombin also binds directly to factor V (FV) (B). The 3 activation cleavage sites at R709, R1545, and R1018 are recognized using many surface residues, including those of both exosites. Although 2 thrombin molecules are depicted, a single docking event may be sufficient to complete proteolysis of each protein. The schematic of fibrinogen is adapted from Cote et al35 with permission.
Proteolytic activation of platelets requires cleavage of PARs. In crystal structures of binding of receptor-based peptides to thrombin, the active site and exosite I were shown to be involved43 (Table 1). The residues on thrombin required for activation of PAR-1 and PAR-4 have been quantitatively assessed using functional analysis and are very similar to those used for recognition of fibrinogen.44,45 There are some differences between PAR-1 and PAR-4, with the former being more dependent on exosite I interaction than the latter.44,46
A restricted common set of surface residues on thrombin around the Na+ loop and active site cleft is important for activity of a number of substrates (in the absence of any cofactor), including antithrombin,37 protein C,39 and factor XIII.47 Use of the same, or a similar, subset of surface residues might suggest competition between these substrates for thrombin; however, it is unlikely that such direct substrate competition will be an important determinant of the direction or fate of thrombin. This is because of the large increases in efficiencies of substrate cleavage obtained in the presence of the cofactors (see next section). In the absence of these cofactors, the substrate reactions are inefficient, and it is unlikely that the unassisted reactions will take place in vivo.
The role of cofactors; variation on a theme
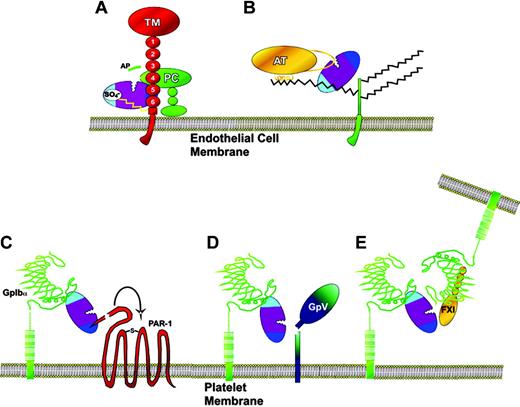
The way in which exosites and cofactors function became evident from their roles in anticoagulant pathways localized on the surface of the endothelium (a recent more general review of the importance of exosites in substrate recognition can be found elsewhere48 ). The protein C anticoagulant pathway is important for the down-regulation of thrombin generation. Thrombin activates protein C, which then proteolytically inactivates factors Va and VIIIa. However, the catalytic efficiency of this activation reaction is very low (kcat/Km, 5.6 × 102 M–1s–1).10 This is increased approximately 1500-fold by thrombomodulin alone and approximately 10 000-fold by thrombomodulin in the presence of a phospholipid membrane (kcat/Km, 5.9 × 106 M–1s–1). Thrombomodulin achieves this increase in efficiency by binding to thrombin at exosite I with very high affinity, binding being mediated by its 5th and 6th epidermal growth factor (EGF)–like domains.33,49 Protein C approaches this complex and associates weakly to thrombomodulin, by an interaction between a positively charged exosite on protein C and the 4th EGF-like domain of thrombomodulin,50 in an orientation that favors its activation. Thrombomodulin thereby functions as a cofactor in the activation of protein C by thrombin, approximating the 2 proteins (Figure 4A) and enabling allosteric changes in thrombin.31,53 Two separate regions of thrombin are required for this reaction: exosite I for thrombomodulin binding, and the active site for protein C activation (Table 1). However, exosite II can also be involved by binding to the chondroitin sulfate moiety present on a fraction of thrombomodulin molecules54 (Figure 4A; Table 1).
The thrombin-thrombomodulin complex is also able to activate TAFI. As described for protein C activation, exosite I is used to bind to the thrombomodulin,39 but exosite II can also again be involved when chondroitin sulfate is attached to the receptor (Table 1).
Antithrombin inhibits thrombin directly in a second anticoagulant pathway. Thrombin recognizes the reactive site loop of antithrombin as a substrate but is trapped during the proteolysis reaction. Antithrombin inhibition of thrombin is relatively inefficient. Catalytic efficiency has the modest value of 6.8 × 103 M–1s–1.55 A fraction of heparin and heparan sulfate chains contain high-affinity sites for antithrombin binding and also provide a template for the efficient diffusion of thrombin. Thrombin is then “approximated” to antithrombin by its binding to an adjacent site on the glycosaminoglycan56,57 (Figure 4B). For this approximation, thrombin uses exosite II to bind to the sulfate groups of the glycosaminoglycan (Table 1). Two sites on thrombin are therefore involved in this cofactor reaction, exosite II and the active site. The outcome of this cofactor participation in the antithrombin/thrombin reaction is an approximate 20 000-fold increase in efficiency, to a kcat/Km of 1.2 × 108 M–1s–1.55
Thrombin is inhibited by another SERPIN, heparin cofactor II, in a reaction accelerated up to 70 000-fold by heparin, heparan sulfate, and dermatan sulfate.58,59 The mechanism is more complex than the heparin-accelerated inhibition of thrombin by antithrombin.12 In addition to a template acceleration reaction mediated by exosite II, the polysaccharide releases a sequestered highly acidic N-terminal tail of heparin cofactor II, which is then able to bind to exosite I and position the thrombin for attempted cleavage of the Leu-Ser reactive center bond (Table 1). This mechanism is supported by crystallographic and mutagenesis studies.60,61
Cofactor-assisted substrate interactions of thrombin on the endothelial cell and platelets. (A) Thrombin is bound to endothelial cell thrombomodulin (TM) by exosite I interaction with EGF-like domains 5 and 6. Exosite II can also interact with any chondroitin sulfate side chain present on the proteoglycan. Protein C (PC) binds to thrombomodulin on EGF-like domain 4, in a position favorable for its activation by thrombin. The high efficiency of activation may arise both from approximation of thrombin and protein C on thrombomodulin, and from exosite-induced conformational change in the active site of thrombin. (B) Thrombin bound via exosite II on endothelial cell heparan sulfate proteoglycan side chains is efficiently inhibited by antithrombin (AT). (C) Thrombin bound to platelet GpIbα via exosite II is brought into proximity to PAR-1 and enhances its activation using exosite I to make contact. (D) The thrombin-GpIbα complex can enhance cleavage of GpV, resulting in hyperactive platelets. (E) The thrombin-GpIbα complex can also activate factor XI (FXI) bound to GpIbα at a separate site in the leucine-rich repeat region. The images of GpIbα interactions have been adapted with permission from Dumas et al51 and Sadler52 (original illustration, Katharine Sutliffe). Copyright 2003 AAAS.
Cofactor-assisted substrate interactions of thrombin on the endothelial cell and platelets. (A) Thrombin is bound to endothelial cell thrombomodulin (TM) by exosite I interaction with EGF-like domains 5 and 6. Exosite II can also interact with any chondroitin sulfate side chain present on the proteoglycan. Protein C (PC) binds to thrombomodulin on EGF-like domain 4, in a position favorable for its activation by thrombin. The high efficiency of activation may arise both from approximation of thrombin and protein C on thrombomodulin, and from exosite-induced conformational change in the active site of thrombin. (B) Thrombin bound via exosite II on endothelial cell heparan sulfate proteoglycan side chains is efficiently inhibited by antithrombin (AT). (C) Thrombin bound to platelet GpIbα via exosite II is brought into proximity to PAR-1 and enhances its activation using exosite I to make contact. (D) The thrombin-GpIbα complex can enhance cleavage of GpV, resulting in hyperactive platelets. (E) The thrombin-GpIbα complex can also activate factor XI (FXI) bound to GpIbα at a separate site in the leucine-rich repeat region. The images of GpIbα interactions have been adapted with permission from Dumas et al51 and Sadler52 (original illustration, Katharine Sutliffe). Copyright 2003 AAAS.
Exosites and cofactors are also involved in procoagulant reactions on the surface of the platelet. GpIbα plays a critical role in initial adhesion reactions between platelets and the subendothelium, mediated through VWF. It also functions as a cofactor in the activation of PARs62,63 and GpV6 (Figure 4C-D). Its cofactor function is similar to the examples already given in the sense that an approximation is achieved. It differs, however, in that the enhanced cleavage arises from substrate receptors merely being in proximity to GpIbα. Crystal structure analyses have demonstrated both exosite I and II binding interactions with GpIbα.51,64 The weight of evidence from functional analyses indicate that exosite II interaction with GpIbα is critical in these cofactor reactions65 (Table 1), but the additional exosite I interactions may play a crosslinking role in platelet activation and signaling.51 Thrombin bound to GpIbα through exosite II is also positioned to activate factor XI on stimulated platelets. Factor XI bound via its Apple 3 domain66 to leucine-rich repeats67 of adjacent GpIbα is recognized by thrombin in part through exosite I interaction68 (Figure 4E; Table 1).
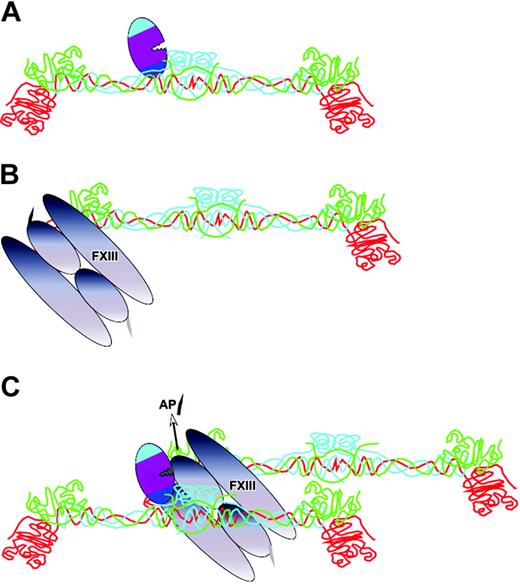
The principle of cofactor-enhanced proteolysis is subject to an interesting variation during factor XIII activation by thrombin. Thrombin activates factor XIII by cleaving a 37-residue activation peptide from the factor XIII A chain. On its own this is not a highly efficient reaction (kcat/Km, 1.4 × 105 M–1s–1).69,70 It has been known for many years that polymerizing fibrin accelerates the activation of factor XIII, but there was no understanding of how this was achieved. Thrombin uses fibrin as a cofactor, but there are unique aspects of this. The cofactor has to be first generated at the site of vascular injury by cleavage of FPA from fibrinogen. It is the subsequent process of polymerization that generates the cofactor. Once thrombin has bound to fibrinogen and cleaved the fibrinopeptides, it remains bound to the fibrin that is formed, in the N-terminal region termed the E domain (Figure 5). It has now been shown that part of exosite I is important for this binding and enhanced activation of factor XIII.47 But how does the needed approximation of factor XIII occur? Factor XIII is known to bind with moderate affinity to the C-terminal D domain of γA/γ′ fibrinogen. This domain engages with complementary sites of the N-terminal E domain to induce polymerization. It is plausible that thrombin is brought to its substrate, factor XIII, by the polymerization process47 (Figure 5), and the catalytic efficiency is thereby increased approximately 80-fold to 1.2 × 107 M–1s–1.69,71
Self-generated cofactor-assisted substrate interactions of thrombin. Once thrombin has cleaved FPA from fibrinogen, fibrin monomer is formed, to which the thrombin remains attached via exosite I interactions at the N-terminal E domain (A). Factor XIII (FXIII) is also loosely bound to fibrin, but at the C-terminal D domain (B). The fibrin monomers will spontaneously polymerize. It has been proposed that this approximates the thrombin and factor XIII (C), resulting in efficient cleavage of the activation peptide (AP).47 Fibrin(ogen) polypeptide chains are colored as in figure 3A.
Self-generated cofactor-assisted substrate interactions of thrombin. Once thrombin has cleaved FPA from fibrinogen, fibrin monomer is formed, to which the thrombin remains attached via exosite I interactions at the N-terminal E domain (A). Factor XIII (FXIII) is also loosely bound to fibrin, but at the C-terminal D domain (B). The fibrin monomers will spontaneously polymerize. It has been proposed that this approximates the thrombin and factor XIII (C), resulting in efficient cleavage of the activation peptide (AP).47 Fibrin(ogen) polypeptide chains are colored as in figure 3A.
Cofactor competition for exosites
The finding that residues on exosite I involved in the fibrin-enhanced activation of factor XIII comprise a discreet subset of those required for the thrombomodulin-enhanced activation of protein C by thrombin, raised the interesting issue of competition between cofactors for exosites.47 If, as suggested in the preceding section, cofactors drive many of the reactions of thrombin, competition between them for the exosites might be expected to determine the fate of thrombin.
A cofactor has a potential choice of either of the exosites with which to interact. How is this choice made? It is known that exosite II interactions are more dependent on salt concentration than are exosite I interactions, suggesting strong ionic interactions. A first consideration of this has been made elsewhere21 on the basis of peptide sequences from cofactors known to bind to the exosites. A number of physiologic and nonphysiologic peptide ligands were considered in terms of the overall properties, acidic, basic, or hydrophobic. Predictably, both exosites required peptides with a high percentage of acidic residues, 50% for exosite I and 72% for exosite II, but the hydrophobic content was somewhat lower for exosite II. The average fractional net negative charge to the hydrophobic fraction was consequently higher for exosite II binding peptides, 2.5 compared with 1.2.
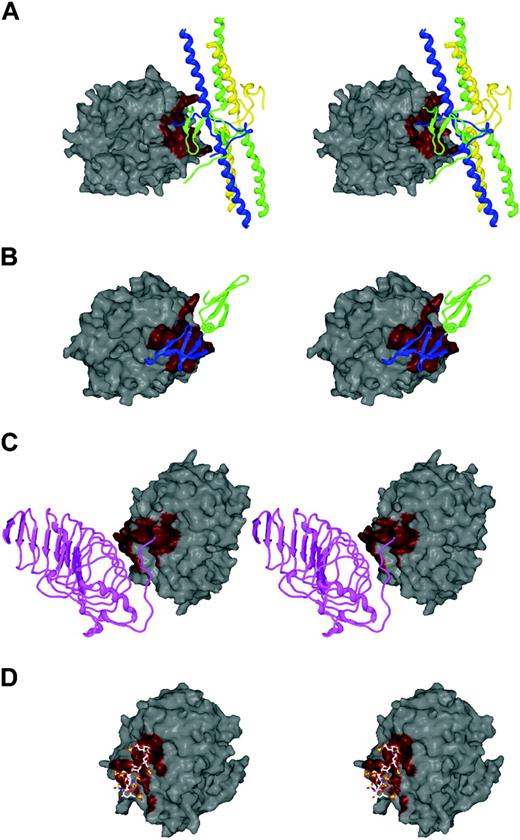
Competition for thrombin exosites is nicely illustrated by recent crystal structures of thrombin complexes with fibrin, thrombomodulin, GpIbα, and heparin.33,36,51,72 In Figure 6A, the structure of fibrin (ribbon, with yellow, green, and blue polypeptide chains) bound to thrombin is shown. The contact region on exosite I is depicted in red. The contact of thrombomodulin (green and blue ribbon EGF domains) with thrombin is shown in Figure 6B, and the red contact region on exosite I can be seen to be overlapping that of fibrin. These structures predict competition between thrombomodulin and fibrin but not its outcome. Because the affinity of thrombin for thrombomodulin is high (∼1 nM) and that of thrombin for fibrin is low (∼1 μM), the former will always be preferred. As shown in Figure 6C, GpIbα (purple ribbon) binds to exosite II (red), using many of the same residues as does heparin (ball and stick) (Figure 6D). Once again, competition will depend in part on relative affinities. Heparin (and presumably heparan sulfate) binds to thrombin with an approximately 1 μM dissociation constant,73 similar to the affinity of thrombin for GpIbα as an isolated peptide65 (it may be greater on the intact platelet74,75 ). The local concentration of heparan sulfate will be much higher over the intact endothelium where the competition is staged, giving it a clear advantage.
Cofactor competition for exosites. The exosite interactions between thrombin and (A) fibrin, (B) thrombomodulin, (C) GpIbα, and (D) heparin have been revealed by recent crystallographic structures. The surface of thrombin is oriented identically in panelsA and B to highlight exosite I, and in panels C and D for exosite II, and the contact surfaces are colored red. It is clear from the stereo depictions that cofactors fibrin (E fragment) and thrombomodulin (EGF domains 5 and 6) share a single binding site on exosite I, and similarly, GpIbα and heparin share a common site on exosite II.
Cofactor competition for exosites. The exosite interactions between thrombin and (A) fibrin, (B) thrombomodulin, (C) GpIbα, and (D) heparin have been revealed by recent crystallographic structures. The surface of thrombin is oriented identically in panelsA and B to highlight exosite I, and in panels C and D for exosite II, and the contact surfaces are colored red. It is clear from the stereo depictions that cofactors fibrin (E fragment) and thrombomodulin (EGF domains 5 and 6) share a single binding site on exosite I, and similarly, GpIbα and heparin share a common site on exosite II.
How thrombin is directed
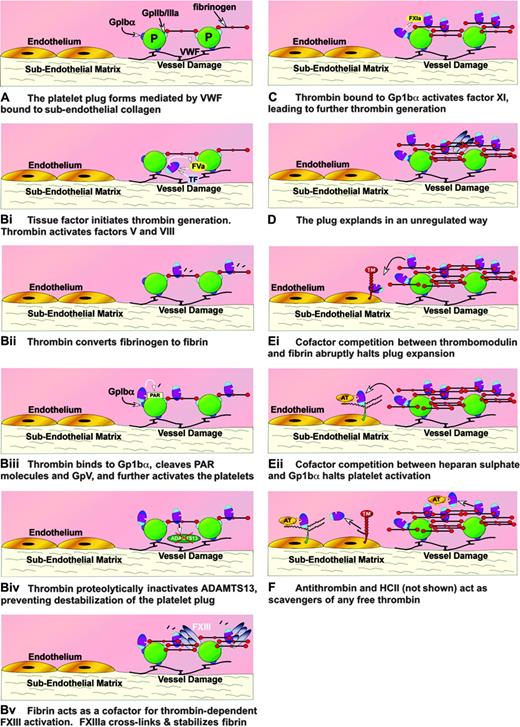
Thus, the many reactions of thrombin therefore involve either direct or cofactor-assisted recognition events involving common exosites. How are these events orchestrated in vivo when the hemostatic plug is being generated? A very early if not initiating event in hemostasis will be the interaction of the platelet with the damaged endothelium, mediated through von Willebrand factor-GpIbα interactions (Figure 7A) and collagen-receptor interactions (not shown). Further, platelets will be recruited to this plug in interactions mediated by adhesive proteins such as fibrinogen with other glycoprotein receptors. Engagement of the glycoproteins results in intracellular signaling and cellular activation. The activated platelet plug is induced to provide a surface of negatively charged phospholipid on which coagulation factors can assemble. Tissue factor (TF) is exposed by vessel injury, and this leads to factor V and factor VIII activation and the generation of the first traces of thrombin on this surface. Thrombin activates more factor V and VIII (Figure 7Bi), aided by the large spread of surface residues used for recognition. The affinity of thrombin for the N terminus of fibrinogen is modest, approximately 7 μM, but the concentration of fibrinogen is high (∼ 15 μM as a monomer), and much (but, crucially, not all) of the thrombin generated will associate with fibrinogen (Figure 7Bii). Recognition of fibrinogen may again be favored by the large number of residues spread over the surface of thrombin that participate in binding. Productive binding results in cleavage of FPA with formation of fibrin monomers. As the fibrin begins to polymerize, it will retain thrombin bound to a subset of those residues in exosite I used for initial recognition. The newly generated polymer will provide the cofactor required for accelerated factor XIII activation. This ensures that factor XIIIa is generated where it is needed, on the fibrin clot surface, where it acts to crosslink the fibrin (Figure 7Bv).
How thrombin is directed. Schematic diagram showing many of the interactions of thrombin over the damaged endothelium as part of the hemostatic response. The direction of thrombin is indicated by use of A, B (i, ii, iii, iv, and v simultaneously), C, D, E (i and ii simultaneously), F. Fibrinogen is shown as a simple domain scheme for simplicity. TF indicates tissue factor.
How thrombin is directed. Schematic diagram showing many of the interactions of thrombin over the damaged endothelium as part of the hemostatic response. The direction of thrombin is indicated by use of A, B (i, ii, iii, iv, and v simultaneously), C, D, E (i and ii simultaneously), F. Fibrinogen is shown as a simple domain scheme for simplicity. TF indicates tissue factor.
Thrombin located on the fibrin clot can migrate, because it is bound through a limited subset of exosite I residues, and its affinity for fibrin is weak. Proximity to GpIbα will enable its binding primarily through exosite II (Figure 7Biii). This glycoprotein will be engaged with VWF, linking the plug to the exposed subendothelium. Thrombin in the vicinity of unattached GpIbα will be able to protect VWF from ADAMTS13 action by proteolysis (Figure 7Biv). This protection, mediated by recognition of ADAMTS13 through exosite I of thrombin,9 prevents destabilization of the plug by ADAMTS13 cleavage of VWF. Thrombin bound to GpIbα through exosite 2 interaction will also be positioned for activation of PARs (Figure 7Biii) (the recognition of which is largely through exosite I) and for GpV cleavage. Complex cell-signaling pathways will be triggered at this stage by proteolysis, nonproteolytic binding of thrombin, and possibly by bivalent thrombin interactions using both exosites.6,64 In situations of extreme hemostatic challenge, factor XI will be activated by thrombin on the platelet surface (Figure 7C) by a mechanism involving both exosites and GpIbα.68 This will enhance downstream thrombin generation. Essentially unregulated and therefore uncontrollable thrombin generation can then occur locally over the damaged endothelium (Figure 7D). Thrombin will continue to be active until the whole platelet plug is surrounded and stabilized with crosslinked fibrin. However, as the clot continues to expand, it will reach areas of intact endothelium. Here it will encounter thrombomodulin (Figure 7Ei). Thrombomodulin has very high affinity for thrombin, much higher than the affinity of thrombin for fibrin. Crucially, both cofactors (fibrin and thrombomodulin) use exosite I and will therefore compete for binding. This competition will only have one outcome, thrombin bound to thrombomodulin. Thrombomodulin will function as a sink, draining and redirecting thrombin activity (from procoagulant to anticoagulant), thereby protecting the intact endothelium from multiple thrombin actions and obstructive thrombus formation.
Heparan sulfate and GpIbα both use exosite II. In areas of damaged vessel with abundant platelet adhesion, binding to GpIbα will be favored. However, over the intact endothelium, the more abundant heparan sulfate will tend to dominate in competition for exosite II and thrombin (bound by exosite II) will be neutralized by approximated antithrombin (Figure 7Eii). A potentially more complicated competitive interaction for GpIbα can be invoked for the second serpin thrombin inhibitor, heparin cofactor II. This inhibitor when bound to its glycosaminoglycan/galactosaminoglycan cofactor will be in a partially activated state with its acidic N-terminal tail displaced and ready to receive thrombin. The latter will be approximated by exosite II contact with sulfate residues in the polysaccharide and by interaction of the N-terminal tail with exosite I. However, in the physiologic context, heparin cofactor II has at best a secondary role in the control of the normal hemostatic clot.76 Should any thrombin break past the functional barrier presented by thrombomodulin, endothelial cell heparan sulfate with bound antithrombin will act as a backup anticoagulant mechanism, with thrombin binding to the glycosaminoglycan side chains through exosite II (Figure 7F). Antithrombin scavenging action may not be restricted to the endothelium. As the hemostatic plug grows into the lumen of the vessel, the thrombin will be associating and dissociating with fibrin, and, because the equilibrium binding affinity is weak, the dissociated thrombin will be a target for free antithrombin. Although efficiency of the thrombin inhibition by direct antithrombin interaction is low, it may be sufficient to limit luminal plug growth. All generated thrombin is eventually cleared from the circulation in complex with either antithrombin, heparan cofactor II, or less specific inhibitors, by receptor-mediated processes.77
Prepublished online as Blood First Edition Paper, June 30, 2005; DOI 10.1182/blood-2005-04-1710.
Supported by grants from the British Heart Foundation and the Medical Research Council.
We thank Dr Jim Crawley for his help in the preparation of the figures.