Abstract
Accurate diagnosis and classification of leukemias are the bases for the appropriate management of patients. The diagnostic accuracy and efficiency of present methods may be improved by the use of microarrays for gene expression profiling. We analyzed gene expression profiles in 937 bone marrow and peripheral blood samples from 892 patients with all clinically relevant leukemia subtypes and from 45 nonleukemic controls by U133A and U133B GeneChip arrays. For each subgroup, differentially expressed genes were calculated. Class prediction was performed using support vector machines. Prediction accuracy was estimated by 10-fold cross-validation and was assessed for robustness in a 100-fold resampling approach using randomly chosen test sets consisting of one third of the samples. Applying the top 100 genes of each subgroup, an overall prediction accuracy of 95.1% was achieved that was confirmed by resampling (median, 93.8%; 95% confidence interval, 91.4%-95.8%). In particular, acute myeloid leukemia (AML) with t(15;17), AML with t(8;21), AML with inv(16), chronic lymphatic leukemia (CLL), and pro–B-cell acute lymphoblastic leukemia (pro–B-ALL) with t(11q23) were classified with 100% sensitivity and 100% specificity. Accordingly, cluster analysis completely separated all 13 subgroups analyzed. Gene expression profiling can predict all clinically relevant subentities of leukemia with high accuracy.
Introduction
The diagnosis and classification of leukemia rely on the simultaneous application of multiple techniques. Cytomorphology and histomorphology are combined with cytochemistry and multiparameter flow cytometry to assign the diagnostic sample to the correct entity. Furthermore, chromosomal analysis, often supplemented by fluorescence in situ hybridization (FISH), and molecular techniques, such as polymerase chain reaction (PCR), is needed to definitively confirm the diagnosis. A comprehensive and standardized algorithm for a diagnostic workflow and an effective and carefully designed combination of methods is essential to guarantee that all the required diagnostic information is gathered.
This huge amount of laboratory assessment is necessary not only to diagnose and classify leukemia samples correctly but to detect biologically homogeneous entities that require specific treatment approaches. Thus, the detailed leukemia classification proposed by the French-American-British (FAB)1,2 Cooperative Group has been improved by thoroughly defined genetic and other characteristics, resulting in the new World Health Organization (WHO) classification.3 This also led to new prognostic markers and even to disease-specific therapeutic approaches. The prime example for this strong link between a comprehensive diagnosis and a disease-specific treatment approach has been the use of all-trans retinoic acid (ATRA) in patients with acute promyelocytic leukemia. Both the correct diagnosis and the efficacy of the specific treatment are based on the presence of the translocation (15;17) and of the corresponding PML/RARA fusion gene.4-6 Although it did not result in the development of a new targeted drug therapy, identifying acute myeloid leukemia (AML) with a complex aberrant karyotype, depending on the age of the patient, is nonetheless highly relevant for the management of the patient. This dismal diagnosis—again, depending on the age of the patient—is the basis for the decision to perform allogeneic stem cell transplantation very early or even to withhold antileukemia therapy.7-10 The recent introduction of imatinib mesylate into the therapeutic management of patients with chronic myeloid leukemia (CML) has revolutionized the treatment strategies for this disease and may change therapeutic concepts for BCR/ABL-positive acute lymphoblastic leukemia (ALL) in the near future.11-16 Again, the basis for the correct diagnosis and the specifically targeted therapy is the presence of the genetic alteration, the t(9;22) translocation. In addition, the BCR/ABL fusion gene is increasingly used to sensitively assess response to therapy by monitoring minimal residual disease (MRD) levels.17 In patients with acute leukemias and chronic lymphatic leukemia (CLL), monitoring of MRD is increasingly used to guide risk-adapted therapy.18-22
To achieve the correct and complete diagnosis in each analyzed sample, a modern, state-of-the-art laboratory must provide significant resources for laboratory equipment, working time, and skilled, experienced personnel. What is needed is a novel diagnostic tool that provides the opportunity to satisfy diagnostic needs while enabling efficient use of resources.
Microarray analysis used to perform gene expression profiling may be the method of choice in this regard23-25 because it allows simultaneous detection of the expression of nearly all human genes in one experimental approach, thereby providing maximal insight into gene regulation and gene alterations in the analyzed sample on the transcriptional level. Although many studies exploited this technique to gain clues to the pathogenesis of a large variety of malignant diseases and to characterize unidentified disease subentities, little focus has been applied to gene expression profiling for diagnostic purposes.26-28
Here, we describe the results of an extensive microarray study on leukemia samples that was performed in parallel with all standard techniques and that resulted in a global, one-step, diagnostic approach. In 937 samples from patients with newly diagnosed leukemia and from nonleukemic controls, we focused on all leukemic subentities clinically relevant with respect to specific treatment approaches and prognostication. Using unsupervised and supervised biostatistical methods based primarily on support vector machines (SVMs), we confirmed and reproduced 12 predefined leukemia subtypes and separated all from each other and from nonleukemic control bone marrow samples with an accuracy of 95.1%. Thus, the single method, gene expression profiling using microarrays, may effectively be applied as a complementary diagnostic method in patients with leukemia and may even substitute for other methods in the future.
Patients, materials, and methods
Patient and control samples
Nine hundred thirty-seven bone marrow and peripheral blood samples from patients with newly diagnosed leukemia and from nonleukemic controls were included in the present analysis. CLL samples consisted of peripheral blood, whereas samples from all other entities in general consisted of bone marrow. Samples were sent to the Laboratory for Leukemia Diagnostics in Munich, Germany, between February 1998 and February 2004 for reference diagnosis from local and national hospitals. The median shipment time was 1 day (range, 0-3 days). All samples underwent standardized processing, including sample central registration,29 preparation, and evaluation by cytomorphology,30 cytochemistry, multiparameter immunophenotyping,31 cytogenetics,32 fluorescence in situ hybridization (FISH), and molecular genetics.33 Stabilized cell lysates were stored at –80°C for a median of 13 months (range, 0-67 months). Diagnoses and other sample and patient characteristics are given in Tables 1 and 2. After 800 samples had been identified for inclusion in the study, another 137 samples were selected according to diagnosis to achieve a distribution among the disease subtypes, thereby ensuring an adequately powered study. In all samples with balanced translocations, the corresponding fusion transcript was verified on the molecular level—that is, PML/RARA for t(15;17), AML1/ETO for t(8;21), CBFB/MYH11 for inv(16) or t(16;16), MLL/and various partner genes for t(11q23) in AML and ALL, and BCR/ABL for t(9;22) in ALL and CML. In addition, in each of these samples and for MYC/IGH for t(8;14), FISH was applied using standard procedures.34
Before therapy, all patients gave their informed consent for participation after having been advised of the purpose and investigational nature of the study and of the potential risks. The study design adhered to the tenets of the Declaration of Helsinki and was approved by the ethics committees of the participating institutions before its initiation.
Leukemia entities selected for identification by gene expression profiling
The focus of the present study was to identify all leukemia subgroups that were clinically relevant with regard to specific treatment and prognostication. Along with the distinctions among the 4 main categories of leukemia (AML, ALL, CML, CLL), these relevant groups also comprised specifically defined subentities. In addition to the group designated “nonleukemia”—those with healthy bone marrow (n = 11), elevated white blood cell (WBC) counts during infection (n = 8), slightly elevated WBC counts of unknown origin (n = 6), staging non-Hodgkin lymphoma without bone marrow involvement (n = 4), staging Hodgkin disease without bone marrow involvement (n = 3), staging cutaneous mastocytosis without bone marrow involvement (n = 3), drug-induced bone marrow failure in early regeneration (n = 3), liver cirrhosis (n = 2), iron deficiency (n = 2), osteoporosis (n = 1), vitamin B12 deficiency (n = 1), or idiopathic thrombocytopenic purpura (n = 1)—the following 12 clinically relevant subgroups were analyzed: AML with t(15;17), AML with t(8;21), AML with inv(16), AML with normal karyotype or so-called “other” cytogenetic abnormalities, AML with 11q23/MLL rearrangement, AML with complex aberrant karyotype, pro–B-ALL with t(11q23), mature B-ALL with t(8;14), c-ALL/pre–B-ALL with or without t(9;22), T-cell ALL (T-ALL), CML, and CLL.
Gene expression profiling, data analysis, and real-time PCR
Additional information on methods with regard to gene expression profiling, data analysis, and real-time PCR are given in Supplemental Document S1 (see the Supplemental Materials link at the top of the online article, at the Blood website).
Results
Prediction of 13 subgroups
Predicting the respective leukemia type or subtype based on differential gene expression signatures was approached using SVMs. The complete data set was randomly, but equally, split into training and independent test cohorts for the 13 subgroups. Then differentially expressed genes were identified in the training set calculated by means of t test statistic, and an SVM model was built based on the genes that demonstrated differential expression among the respective subclasses in the training set.
This SVM model was used to predict samples in the test cohort. Application of the top 100 genes per group resulted in the best prediction accuracy (superior to top 20, 50, 150, 200, 250, and 300 genes, respectively; data not shown) and were used for all subsequent analyses (for a description of the genes, see the supplemental material). Table 3 represents a confusion matrix of subgroup predictions based on their gene expression signatures using a 10-fold cross-validation approach (9 of 10 for training and 1 of 10 for testing, with 10 iterations so that each sample is classified once). Overall, a 95.1% accuracy of subgroup prediction was achieved analyzing 13 subgroups. Specifically, the highest accuracy was achieved for 7 of the 13 subgroups—AML with t(15;17), 100% accurate predictions; AML with inv(16), 98%; CLL, 97.8%; CML, 97.3%; AML normal/other, 97.3%; pro–B-ALL with t(11q23), 96.2%; and AML with t(8;21), 94.7%. The presence of a cryptic rearrangement (inv(16), n = 1; t(16;16), n = 3; AML with t(11q23)/MLL, n = 1; ALL with t(9;22), n = 2; CML with t(9;22), n = 2; and CML with variant t(9;22), n = 3)35 had no impact on the accuracy of classification. These patients, therefore, were grouped together with patients carrying the respective overt cytogenetic abnormality. For the other 6 subgroups, the percentages of accurate predictions ranged between 83.3% and 93.3%.
Most of the misclassifications occurred in subgroups that either had relatively low sample numbers or were characterized by high intra-subgroup biologic heterogeneity. The first aspect clearly applies to mature B-ALL with t(8;14) with a sample number of 12 and 83.3% accurate predictions. The latter aspect is reflected in AML with t(11q23) (89.4% accurate predictions) with balanced translocations involving the MLL gene and 6 different fusion partner genes (AF4, AF6, AF9, AF10, ELL, ENL). Another example of biologic heterogeneity is AML with complex aberrant karyotype (88% accurate predictions) composed of a wide range of 3 to 30 chromosomal abnormalities (median, 9). As anticipated, most of the misclassifications of these groups (4 of 5 for AML with t(11q23) and 8 of 9 for AML with complex aberrant karyotype) were attributed to a prediction of the samples as AML normal/other. A third aspect to consider is the relative similarity of distinct subgroups with regard to specific characteristics, such as flow cytometrically detected expression of myeloid antigens on immature T-ALL samples.36 Probably because of these complexities, 4 of 32 patients with T-ALL in the present series were classified as having AML normal/other. Importantly, the inclusion of “AML normal” and “AML other” into the analyses as 2 separate groups did not result in superior classification accuracy (Supplemental Tables S3-S6).
To assess the robustness of class prediction, a resampling approach was applied, which is to say the complete SVM classification procedure was repeated 100 times. For each of the 100 runs, all samples were randomly divided into a training set (two thirds of all samples; n = 625) and a test set (one third of all samples; n = 312). Thus, the test set for each run consisted of c-ALL/pre–B-ALL (n = 28), pro–B-ALL with t(11q23) (n = 9), mature B-ALL with t(8;14) (n = 4), T-ALL (n = 10), AML with t(15;17) (n = 14), AML with t(8;21) (n = 12), AML with inv(16) (n = 16), AML with t(11q23) (n = 16), AML with complex karyotype (n = 25), AML with normal karyotype or other aberrations (n = 123), CLL (n = 15), CML (n = 25), and nonleukemia samples (n = 15). The matrix in Table 4 gives the average number of class predictions as determined after 100 runs of SVM-based classifications. For example, 9 pro–B-ALL with t(11q23) samples were predicted by the algorithm 900 times (each sample 100 times). Of the 900 predictions, the class label pro–B-ALL with t(11q23) was assigned correctly 854 times (ie, on average 8.54 per run). In 2 individual predictions, pro–B-ALL with t(11q23) samples were predicted as c-ALL/pre–B-ALL, and in 44 predictions they were predicted as AML with normal karyotype or other aberrations.
Confirming the data obtained by 10-fold cross-validation, the overall median accuracy amounts to 93.8% (95% CI, 91.4%-95.8%). In particular, and similar to the 10-fold cross-validation approach, a very high degree of accurate predictions was achieved in 7 of the 13 subgroups: AML with t(15;17), 100% median accuracy; AML with inv(16), 98.1%; CLL, 97.5%; CML, 95.3%; AML normal/other, 96.8%; pro–B-ALL with t(11q23), 94.9%; and AML with t(8;21), 95.3%. For the other 6 subgroups, the median prediction accuracy rates ranged between 64.3% and 92.3%. Thus, the results obtained for the subgroups by applying the resampling approach are highly consistent with those obtained by 10-fold cross-validation and strongly confirm the capability of gene expression profiling to predict leukemia subtypes. The reasons for the misclassifications are most likely the same as those described, in particular the relatively low sample number of patients with mature B-ALL with t(8;14).
Sensitivities and specificities of the predictions for each of the 13 subclasses are given in Table 5. According to the accuracy data given, the specificity overall is very high—more than 99% for all but one subgroup. Because most misclassified samples were classified as AML normal/other, the specificity of this subgroup was slightly lower than it was for other subgroups and amounted to 93.65%. The median sensitivity ranged between 75% and 100% for all subgroups because of the reasons discussed for the results of the 10-fold cross-validation.
Cluster analysis of 13 subgroups
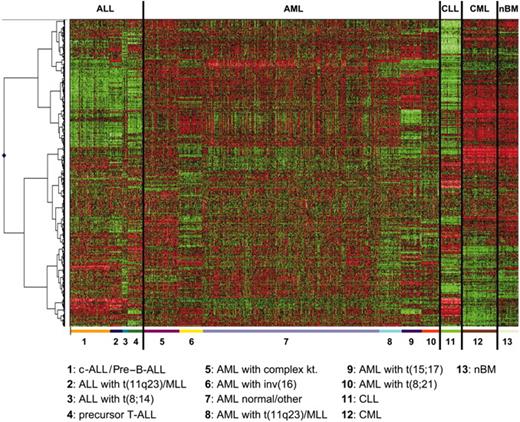
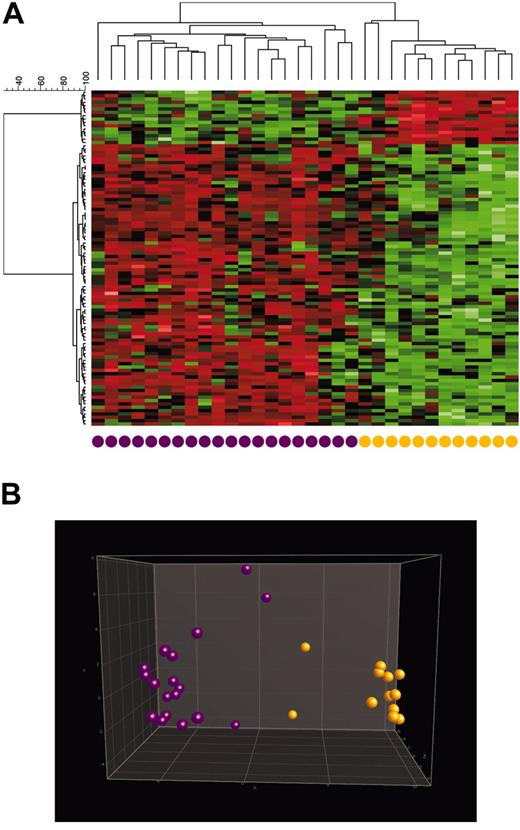
To further validate the findings described, supervised cluster analyses (CAs) and principal component analyses (PCAs) using the top 100 differentially expressed genes for each subgroup were performed for the 13 groups analyzed and for the paired comparison of selected groups. CAs of all of the analyzed samples reflect the clearly differing gene expression patterns of the 13 groups, resulting in a highly accurate separation of this large and comprehensive series of 937 samples (Figure 1). Applying 3-dimensional PCA (Figure 2), the power of the gene expression profile–based leukemia classification is demonstrated by the clear separation of T-precursor ALL from c-ALL/pre–B-ALL (with or without t(9;22)/BCR/ABL). Similarly, 3-dimensional PCAs provide a clear distinction between both t(9;22)–positive entities, CML and c-ALL/pre–B-ALL (Figure 3). Interestingly, the one sample of t(9;22)–positive c-ALL/pre–B-ALL shown in proximity to the CML samples carries the BCR-ABL type M-bcr (p210), which is more frequently present in CML than in ALL. Furthermore, this sample is characterized by only 50% leukemic bone marrow infiltration; thus, the normal hematopoiesis present in this sample, which is largely myelomonocytic, and the forced assignment to either of the 2 groups are the likely reasons for this result.
Hierarchical cluster analysis of 937 samples. Analysis of 937 samples (columns) using a set of 1019 differentially expressed genes (rows). The normalized expression value for each gene is coded by color (SD from mean). Red cells indicate high expression, and green cells indicate low expression. Bars separate the major leukemia types. For each of the 13 classes, the top 100 differentially expressed genes, according to t test statistic, were used. Of the 1300 genes, 281 were repeatedly identified as important diagnostic markers and overlapped among the lists of the top 100 genes, resulting in 1019 nonoverlapping genes.
Hierarchical cluster analysis of 937 samples. Analysis of 937 samples (columns) using a set of 1019 differentially expressed genes (rows). The normalized expression value for each gene is coded by color (SD from mean). Red cells indicate high expression, and green cells indicate low expression. Bars separate the major leukemia types. For each of the 13 classes, the top 100 differentially expressed genes, according to t test statistic, were used. Of the 1300 genes, 281 were repeatedly identified as important diagnostic markers and overlapped among the lists of the top 100 genes, resulting in 1019 nonoverlapping genes.
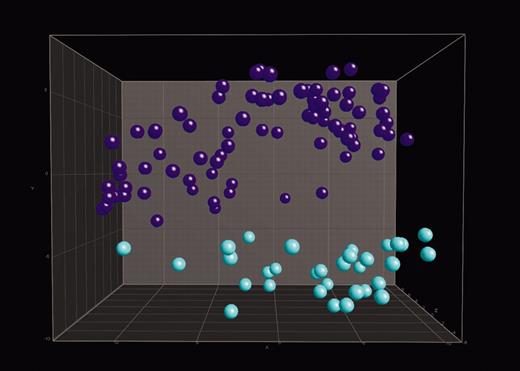
Distinction between precursor B-ALL and T-ALL. In 3-dimensional PCA, 114 ALL samples were projected into the feature space consisting of a combination of the top 100 differentially expressed genes when comparing precursor B-ALL with the other 12 classes or T-ALL with the other 12 classes. Data points with similar characteristics will cluster together. Here, a single color-coded sphere represents each patient's expression pattern. The respective label (ie, precursor B-ALL or T-ALL) was unknown to the algorithm. Labels and coloring of the classes were added after the analysis of means for better visualization. Pre–B-ALL samples (n = 82) are blue and include 42 c-ALL/pre–B-ALL with t(9;22) and 40 c-ALL/pre–B-ALL without t(9;22). T-ALL samples (n = 32) are turquoise.
Distinction between precursor B-ALL and T-ALL. In 3-dimensional PCA, 114 ALL samples were projected into the feature space consisting of a combination of the top 100 differentially expressed genes when comparing precursor B-ALL with the other 12 classes or T-ALL with the other 12 classes. Data points with similar characteristics will cluster together. Here, a single color-coded sphere represents each patient's expression pattern. The respective label (ie, precursor B-ALL or T-ALL) was unknown to the algorithm. Labels and coloring of the classes were added after the analysis of means for better visualization. Pre–B-ALL samples (n = 82) are blue and include 42 c-ALL/pre–B-ALL with t(9;22) and 40 c-ALL/pre–B-ALL without t(9;22). T-ALL samples (n = 32) are turquoise.
Identification of cortical T-ALL and pre–B-ALL with t(9;22)
To further define the classification capabilities of our approach, we aimed at identifying the clinically distinct entities, c-ALL/pre–B-ALL with t(9;22) and cortical T-ALL, from among the groups classified as c-ALL/pre–B-ALL and T-ALL, respectively.
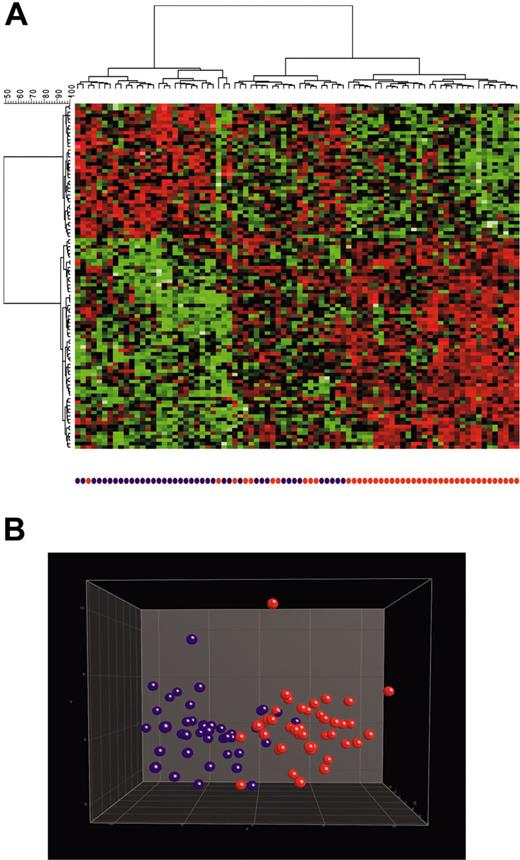
With regard to c-ALL/pre–B-ALL, cluster analysis (Figure 4) showed that most (61 of 82; 74%) of the patients had c-ALL/pre–B-ALL without t(9;22) or with t(9;22). The remaining 21 (26%) patients had a third branch characterized by a gene expression profile clearly different from that of the other 2 groups. Accordingly, the 10-fold cross-validation analysis (allowing separation into 2 groups only) revealed an accuracy of 82.9%. Misclassifications occurred in both directions. Patients with t(9;22) were classified as without it and vice versa. Resampling of the training and test sets, respectively, applying 100 runs of SVM-based classification (median accuracy, 77.8%; range, 61%-90.8%) indicated that these misclassifications were not limited to distinct samples. Percentages of misclassifications per sample ranged from 3.1% to 88.1%, probably reflecting a significant overlap of gene expression signatures between both groups or resulting from the presence of a clinically unidentified third group of c-ALL/pre–B-ALL.
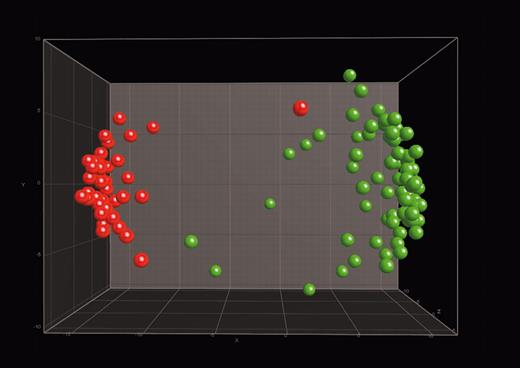
Distinction between c-ALL/pre–B-ALL with t(9;22) and CML. In 3-dimensional PCA, 117 samples were projected into the feature space consisting of a combination of the top 100 differentially expressed genes when comparing c-ALL/pre–B-ALL with t(9;22) samples with the other 12 classes and CML with the other 12 classes. Data points with similar characteristics will cluster together. A single color-coded sphere represents each patient's expression pattern. The respective label (pre–B-ALL or CML) was unknown to the algorithm. Labels and coloring of the classes were added after the analysis of means for better visualization. c-ALL/pre–B-ALL with t(9;22) samples (n = 42) are red, and CML samples (n = 75) are green.
Distinction between c-ALL/pre–B-ALL with t(9;22) and CML. In 3-dimensional PCA, 117 samples were projected into the feature space consisting of a combination of the top 100 differentially expressed genes when comparing c-ALL/pre–B-ALL with t(9;22) samples with the other 12 classes and CML with the other 12 classes. Data points with similar characteristics will cluster together. A single color-coded sphere represents each patient's expression pattern. The respective label (pre–B-ALL or CML) was unknown to the algorithm. Labels and coloring of the classes were added after the analysis of means for better visualization. c-ALL/pre–B-ALL with t(9;22) samples (n = 42) are red, and CML samples (n = 75) are green.
Identification of c-ALL/pre–B-ALL samples with or without t(9;22). Analysis of 82 c-ALL/pre–B-ALL samples based on a supervised identification of differentially expressed genes among 42 patients demonstrating a t(9;22)/BCR-ABL and 40 patients without t(9;22). The labels and coloring of the classes were added after the analysis of means for better visualization. (A) In the hierarchical cluster analysis, the normalized expression value for each gene (given in rows) is coded by color (SD from mean). Red cells indicate high expression, and green cells indicate low expression. Most (61 of 82; 74%) patients were in the branch of c-ALL/pre–B-ALL without t(9;22) (left branch) or in the branch of c-ALL/pre–B-ALL with t(9;22) (right branch). The remaining 21 (26%) patients were in a third branch characterized by a gene expression profile clearly different from the 2 other groups (middle branch). (B) In 3-dimensional PCA, the c-ALL/pre–B-ALL samples were projected into the feature space consisting of the top 100 differentially expressed genes when comparing t(9;22)–positive (red) with t(9;22)–negative (purple) patients. Data points with similar characteristics cluster together. A single color-coded sphere represents each patient's expression pattern.
Identification of c-ALL/pre–B-ALL samples with or without t(9;22). Analysis of 82 c-ALL/pre–B-ALL samples based on a supervised identification of differentially expressed genes among 42 patients demonstrating a t(9;22)/BCR-ABL and 40 patients without t(9;22). The labels and coloring of the classes were added after the analysis of means for better visualization. (A) In the hierarchical cluster analysis, the normalized expression value for each gene (given in rows) is coded by color (SD from mean). Red cells indicate high expression, and green cells indicate low expression. Most (61 of 82; 74%) patients were in the branch of c-ALL/pre–B-ALL without t(9;22) (left branch) or in the branch of c-ALL/pre–B-ALL with t(9;22) (right branch). The remaining 21 (26%) patients were in a third branch characterized by a gene expression profile clearly different from the 2 other groups (middle branch). (B) In 3-dimensional PCA, the c-ALL/pre–B-ALL samples were projected into the feature space consisting of the top 100 differentially expressed genes when comparing t(9;22)–positive (red) with t(9;22)–negative (purple) patients. Data points with similar characteristics cluster together. A single color-coded sphere represents each patient's expression pattern.
With regard to patients with T-ALL, the separation of cortical T-ALL samples from immature T-ALL samples is clear, as shown in the cluster analysis (Figure 5). Interestingly, 2 samples of immature T-ALL show a gene expression profile slightly different from that of the other immature T-ALL samples, as visualized by cluster analysis (Figure 5). By standard diagnostics, these 2 samples indicated pre–T-ALL without any specific feature for cortical T-ALL. They are negative for CD1a in immunophenotyping and show no specific cytogenetic abnormality: 46,XX and 46,XX, t(8;14)(q24;q11),+10, t(11;14)(p12;q11). In fact, these 2 samples are those lying nearest to the cortical T-ALL samples in the PCA. According to the relative vicinity of these 2 samples to samples of cortical T-ALL, the accuracy of the 10-fold cross-validation is 84.38%, and resampling applying 100 runs of SVM-based classification results in a median accuracy of 80% (range, 60%-100%).
The separation of samples with AML and normal karyotypes from those with AML and “other” cytogenetic aberrations was not approached in the present study because the prognosis for each subgroup was identical when applying a standardized treatment approach30,37,38 (Figure S1). We intended to approach the data from a clinical point of view, but there was no clear-cut relevance for the distinction between these 2 groups. Additional data with regard to technical aspects of sample target preparation, scan quality, and validation of significant genes by real-time PCR are given in the supplemental material (Supplemental Figure S2).
Discussion
Diagnosing and classifying leukemias is clinically a highly relevant task that requires a comprehensive and well-structured approach in the laboratory to guarantee the appropriateness of the results. Significant resources with regard to time, well-trained and skilled personnel, and laboratory space and equipment are needed to cover this approach. Furthermore, the interlaboratory reproducibility of currently applied diagnostic methods (cytomorphology, cytochemistry, immunophenotyping, cytogenetics, and molecular genetics) ranges between only 56% and 90% in experienced hands; clearly, then, improvement is needed.9,39-43 Gene expression profiling using microarray technology may optimize leukemia diagnostics and overcome the shortcomings of current methods.
Distinction between immature and cortical T-ALL samples. Analysis of 32 T-ALL samples based on a supervised identification of differentially expressed genes between 12 immature T-ALL samples and 20 cortical T-ALL samples. Labels and coloring of the classes were added after analysis for better visualization. (A) In hierarchical cluster analysis, the normalized expression value for each gene (given in rows) is coded by color (SD from mean). Red cells indicate high expression, and green cells indicate low expression. (B) In 3-dimensional PCA, T-ALL samples were projected into the feature space consisting of the top 100 differentially expressed genes when comparing patients with immature (orange) and cortical (purple) T-ALL. Data points with similar characteristics cluster together. A single color-coded sphere represents each patient's expression pattern.
Distinction between immature and cortical T-ALL samples. Analysis of 32 T-ALL samples based on a supervised identification of differentially expressed genes between 12 immature T-ALL samples and 20 cortical T-ALL samples. Labels and coloring of the classes were added after analysis for better visualization. (A) In hierarchical cluster analysis, the normalized expression value for each gene (given in rows) is coded by color (SD from mean). Red cells indicate high expression, and green cells indicate low expression. (B) In 3-dimensional PCA, T-ALL samples were projected into the feature space consisting of the top 100 differentially expressed genes when comparing patients with immature (orange) and cortical (purple) T-ALL. Data points with similar characteristics cluster together. A single color-coded sphere represents each patient's expression pattern.
The present study focused on the identification of all clinically relevant subtypes of leukemia by gene expression profiling and found significant differences for intragroup separation within the 4 main leukemia groups (AML, ALL, CML, and CLL). With regard to AML, more than 50 recurrent cytogenetic abnormalities have been described. However, reliable data about their prognostic impact are available for only the most frequent ones. These include t(15;17), t(8;21), and inv(16), which are associated with a favorable outcome, and complex aberrant karyotypes and t(11q23), which are associated with an unfavorable outcome.8-10,32,44 The remaining abnormalities, normal karyotypes, and so-called other cytogenetic abnormalities, are associated with an intermediate prognosis. Supplemental Figure S1 demonstrates that the separation between these subgroups results in highly different prognoses, supporting the clinical relevance of the selection of AML subgroups in the present study. The same applies for the cohort selected for the present microarray analysis. In addition, the age distribution of the analyzed cohort is similar to the true age distribution of patients with AML and the other diseases analyzed. With regard to clinical relevance, similar characteristics applied to the different entities of ALL. Besides the separation of T-precursor ALL from B-precursor ALL, clinically relevant because of different treatment strategies, it is important to identify patients with pro–B-ALL and t(11q23) c-ALL or pre–B-ALL and t(9;22) and with mature B-ALL and t(8;14). These subentities differ highly with respect to prognostic impact and require substantially different therapies, which is true for mature B-ALL in particular.11,45 The overall smaller numbers of participants with CLL, CML, and nonleukemia were chosen because these entities are biologically and clinically more homogeneous than the acute leukemia cases discussed.
The present study demonstrates a very high degree of accuracy for the correct assignment of bone marrow samples to all clinically relevant subgroups of leukemia and to normal bone marrow, respectively. An essential basis for the achievement of this accuracy was the careful and comprehensive use of standard methods to characterize all the samples before they were subjected to microarray analysis. In addition to the use of cytomorphology and cytochemistry, the samples were processed through immunophenotyping, cytogenetics, and molecular genetics to allow the identification of subtype-specific gene expression patterns and to exclude any misclassification of samples or overlaps among the subcategories in the microarray analyses.
Six different AML subgroups were detected in the present study. For the classification of AML with t(15;17), t(8;21), and inv(16), the highest degree of accuracy was achieved, respectively, with 42 of 42, 36 of 38, and 48 of 49 correct assignments by 10-fold cross-validation and an average number of correct predictions of 14 of 14, 11.43 of 12, and 15.7 of 16, respectively, by resampling. Accordingly, all the median sensitivities and specificities were 100%. This is in line with previous reports describing a unique biologic background for these subentities,46-48 which is reflected in distinct gene expression profiles.49-52 However, because the latter have not yet been assessed by microarray analysis in the context of the full spectrum of AML and the other leukemias, the present study adds important information by clearly demonstrating that, based on their distinct features, these subentities can be accurately predicted even in the context of the heterogeneous background of other leukemias. The other 3 AML subgroups—AML with t(11q23)/MLL, AML with complex karyotype, and AML normal/other—are biologically more heterogeneous, as reflected by different partner genes of the MLL gene and overall heterogeneity with regard to cytogenetic and molecular genetic aberrations. With these complexities in mind, it was anticipated that misclassifications would occur. Of the 24 misclassifications (total, 491 classifications) in these subgroups during 10-fold cross-validation, only 4 were misclassifications into the non-AML subgroups. As a consequence, though the median specificities for AML with t(11q23) and for AML with complex aberrant karyotypes were very high (99.66% and 99.65%, respectively), the median specificity for AML normal/other of 93.65% points to the need for further improvement in the applied method or for the use of supplemental analyses, particularly because a small number of patients with ALL (n = 11), CLL (n = 1), CML (n = 2), and nonleukemia (n = 2) were classified into this subgroup by 10-fold cross-validation.
With the exception of these 3 samples, there were no misclassifications in CLL and CML by 10-fold cross-validation. Accordingly, there were 14.62 of 15 and 23.82 of 25 correct assignments, respectively, by resampling in these entities. As a result, the median sensitivities (100% and 96%) and the clinically most important median specificities (100% and 99.65%) were very high for these distinct disease entities.
All 4 subgroups of ALL analyzed in the present study could be classified with a high median accuracy (99.65% for c-ALL/pre–B-ALL; 100% for the other subgroups). As discussed, most (11 of 13) misclassifications occurred in the AML normal/other group. Interestingly, these samples did not feature the immunophenotype of an aberrant expression of myeloid antigens, which is often observed in patients with ALL.
Given that previous studies reported on the difficulties in distinguishing c-ALL/pre–B-ALL with t(9;22) from other types of B-precursor ALL, resulting in a prediction accuracy rate of 80%,26 the approach in the present study was to include patients with c-ALL/pre–B-ALL combined as one subgroup, irrespective of the presence of t(9;22), in the analysis and to separate patients positive for t(9;22) from those without it. Although the separation of c-ALL/pre–B-ALL from the other entities has been straightforward, we also observed difficulties in separating t(9;22)–positive patients from t(9;22)–negative patients and achieved only 82.9% accuracy. Interestingly, cluster analysis demonstrated that most patients were accurately classified in 1 of the 2 categories; however, a third branch became evident, revealing a gene expression pattern distinct from that of the other 2 groups. The hypothesis that a further and not yet identified genetic lesion could be responsible for this third branch has been discarded because cluster analysis and SVM did not reveal a reproducible gene expression pattern different from that of the other 2 groups (data not shown). Furthermore, the use of SVM with differentially expressed genes selected based on the comparison of only the first 2 more homogeneous groups did not result in a more accurate assignment of samples of the third group either (data not shown). Taken together, these points support the concept that BCR/ABL represents a type 1 mutation53 and that downstream pathways are shared by many other master genes. Thus, the gene expression profile of patients with BCR/ABL-positive ALL is not highly reproducible (Figure 4), and future microarray-based diagnostic tools should include oligonucleotides targeting the bcr/abl fusion transcript to accurately predict BCR/ABL-positive ALL with greater accuracy. It is anticipated this would increase the sensitivities and specificities for the classification of the other subgroups.
Another clinically relevant subgroup has been approached in a second step. After T-ALL was distinguished from all other entities, immature T-ALL was distinguished from cortical T-ALL, which, in the clinical setting, is characterized by a favorable prognosis.36 Again, the separation of both entities has been highly accurate, with the exception of 2 samples that originally were classified by immunophenotyping as immature T-ALL. It is important to note that the definition of cortical T-ALL in this context is based only on positivity for CD1a,54 whereas other T-cell markers, such as CD7, CD2, CD5, CD4, and CD8, may be positive in either subgroup. Intriguingly, though the use of CD1a is a diagnostic standard, the present analysis suggests that in the 2 misclassified patients, the overall gene expression profile is similar to that of the cortical T-ALL signature. Thus, these 2 patients may have cortical T-ALL featuring an aberrant lack of CD1a expression rather than truly immature T-ALL. As a consequence of our studies, the classification of cortical T-ALL may be based not only on positivity for CD1a but also on other markers, such as PAWR.55
Further implications may be gained from analyzing the cellular function of differentially expressed genes. It is known that dexamethasone leads to the down-regulation of CARD4,56 which encodes a proapoptotically acting protein.57,58 Because CARD4 is highly expressed in cortical T-ALL, corticoid therapy may be less effective in this entity than in immature T-ALL. However, clinical studies are needed to prove this hypothesis.
A particularly important issue that has not yet been substantially addressed in other microarray studies59-61 is the identification of nonleukemic bone marrow and its distinction from all leukemia subtypes. In the present study, 42 of 45 nonleukemia samples have been predicted accurately, whereas 1 sample was classified as AML with t(11q23) and 2 samples were classified as AML normal/other by 10-fold cross-validation. Accordingly, the median accuracy applying resampling is 13.2 of 15. Importantly, the median specificity for nonleukemia is 99.7%, and the sensitivity is 90%. Thus, until improvements of the applied methods are achieved that better characterize the heterogeneous subgroup of AML normal/other, it seems appropriate to add conventional methods if the microarray analysis result assigns a sample to the latter subgroup. In contrast, because of its high specificity, the nonleukemia classification can be the basis to exclude the presence of leukemia in a given sample analyzed.
In general, there are 2 strategies to handle the occurrence of misclassifications obtained by microarray analysis. The first is to identify the most frequent false-positive result—the subgroup with the lowest specificity—and to add conventional diagnostics to confirm or revise a diagnosis of malignancy. Clearly, this applies for AML normal/other with a median specificity of 93.7% (95% CI, 90.2%-96.6%). Through the use of cytochemistry, immunophenotyping, and cytogenetics, distinguishing this subgroup from c-ALL/pre–B-ALL, AML with t(11q23), and AML with complex aberrant karyotype is straightforward though resource consuming. Another possible application for additional methods is the use of PCR to identify or exclude the presence of the BCR/ABL fusion gene once c-ALL/pre–B-ALL is diagnosed.
The second and more promising strategy would be an improvement in the capabilities of microarray technology by taking advantage of the additional representation of fusion gene–specific oligonucleotides. By this approach, many of the misclassifications should be avoidable; for example, c-ALL/pre–B-ALL with t(9;22) should be identifiable by the detection of BCR/ABL, as should AML with t(11q23) by the detection of fusion genes involving MLL and various partners.62 Following this approach would potentially result in even higher rates of accuracy in the subgroups discussed and in improving accuracy in the other subgroups.
Even more subgroups, particularly of AML, have been suggested to feature a homogeneous biologic background with potential impact on the clinical course of patients affected by these entities.63 Examples are mutations of CEBPA,64 length mutations of FLT3,33 and partial tandem duplications of MLL.65 However, because this evidence is still under evaluation in clinical trials, these subgroups are not within the focus of the present study.
A growing body of published microarray studies address the identification of specific gene-expression profiles in distinct subentities of leukemia. Along this line, the respective groups of AML with recurrent balanced translocations and of AML with trisomy 8 have been described to carry a typical genetic signature, which, in some cases, is highly specific.28,50,66,67 The present analysis followed these important studies and provided the opportunity, by focusing on all clinically relevant subtypes of chronic and acute leukemias in a single comprehensive approach, to build on these signatures for a highly accurate diagnostic tool capable of predicting leukemia subtypes. In addition, the separation of leukemia samples from samples with nonmalignant diseases and healthy samples has been accomplished. In accordance with these analyses of leukemia, it is anticipated that similar approaches can be taken to diagnose and classify myelodysplastic syndromes and lymphomas.61,68,69
One possible result of this study could be the widespread use of microarray technology entailing a carefully designed, comprehensive approach to the diagnosis of leukemia and representing a significant improvement over current diagnostic procedures through greater accuracy and efficiency. Thus, the 1-day (cytomorphology, immunophenotyping) to 1-week (metaphase cytogenetics) turn-around time for current procedures may be reduced to 1 to 2 days, or even less, with microarray protocols. This technology should also provide significant insight into the specific genetic alterations of distinct entities, allowing the detection of novel markers that can be targeted by PCR-based methods and multiparameter flow cytometry to quantify MRD during the course of antileukemia treatment.70 Identifying prognostic markers or marker constellations that will predict the response to antileukemia treatment is another clinically relevant topic and will be covered by future microarray trials.52,71 Clearly, large, well-designed, multicenter-driven prospective validation trials assessing microarray-based and current standard diagnostics in parallel are needed.
Prepublished online as Blood First Edition Paper, May 5, 2005; DOI 10.1182/blood-2004-12-4938.
Supported in part by a grant from the German José Carreras Foundation (DJCS-R00/13). Gene expression studies in the Laboratory for Leukemia Diagnostics are further supported in part by Roche Diagnostics, Integrated Cancer Care Unit (ICCU), Basel, Germany.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank our technicians for their excellent assistance.