Abstract
To create a model in which to study the effects of RAS dysregulation in hematopoietic disease, we developed separate founder lines of transgenic mice, with the tetracycline transactivator (tTA) driven by the Vav hematopoietic promoter in one line and NRASV12 driven by the tetracycline responsive element (TRE2) in the other. When these lines are crossed, doubly transgenic animals uniformly develop a disease similar to human aggressive systemic mastocytosis (ASM) or mast cell leukemia (MCL) when they are between 2 and 4 months of age. Disease is characterized by tissue infiltrates of large, well-differentiated mast cells in the spleen, liver, skin, lung, and thymus. Analysis of bone sections shows small to large foci of similarly well-differentiated mast cells. Results also show that transgene expression and diseases are repressible through the administration of doxycycline in the drinking water of affected animals, indicating that NRASV12 expression is required to initiate and maintain disease in doubly transgenic mice. Our inducible system of transgenes, developed as a model of mutant NRASV12 oncogene–driven myeloid disease, will be useful for studying the role of RAS dysregulation in hematopoietic disease in general and in discrete human diseases, specifically ASM and MCL.
Introduction
NRAS is mutated in a variety of hematologic disorders, including acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and various myeloproliferative diseases (MPDs).1-5 NRAS mutations are also associated with the progression of myelodysplastic and myeloproliferative diseases to overt leukemia.6-9 As many as 30% of AMLs harbor mutations resulting in NRAS dysregulation, and the Ras family members (H-, N-, and K-Ras) have long been the subjects of intense investigation regarding their relation to tumorigenesis and cancer.10-12 Ras becomes dysregulated in hematopoietic disease through a number of mechanisms. Constitutively active tyrosine kinases, such as breakpoint-cluster region/Abelson leukemia (BCR-ABL) and mutant FMS-like tyrosine kinase 3 (FLT3), cause increased RAS activation by upstream signal amplification.13,14 Point mutations in Ras itself (eg, G12V) cause longer association of RAS with guanosine triphosphate (GTP) by reducing GTPase activating protein (GAP) sensitivity of Ras, maintaining an active conformation.15-18 Loss of GAPs, such as NF1, result in slower hydrolysis of the GTP gamma phosphate, resulting in prolonged RAS signaling and increased Ras activation.19 Each of these mechanisms of dysregulation has been implicated in human diseases, including AML.
Despite intense scrutiny, the specific contribution Ras dysregulation makes in the development of these diseases has not been determined. Specific interactions between the leukemic target cell and the genetic and epigenetic contexts in which NRAS mutations occur likely explain how the same mutation can result in related, but distinct, myeloid diseases. Finally, whether RAS oncogene products are appropriate therapeutic targets in various myeloid diseases is unclear. A major impediment to elucidating the contribution NRAS mutation makes to the development and maintenance of hematopoietic disease and its suitability as a target is a lack of an appropriate mouse model.
Here we report the development of a transgenic mouse model of activated NRAS-driven myeloid disease. Mice develop disease with 100% penetrance in a period of time amenable to pharmacologic intervention and cooperating mutagenesis studies. In addition, the model offers the opportunity to evaluate changes in the hematopoietic system in response to transgene repression and derepression by virtue of the tetracycline transactivator system.
Materials and methods
Vector construction and generation of FVB/n transgenic mice
The pVav vector was obtained from Dr Jerry Adams at the Walter and Eliza Hall Institute of Medical Research, Royal Melbourne Hospital (Victoria, Australia). The tetracycline transactivator (tTA) was cut from the pRev-Tet-Off plasmid (Becton Dickinson Clontech, San Jose, CA) and was subcloned into the TOPOII-blunt cloning vector (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The tTA fragment was then cut out with EagI and subcloned into pVav cut with EagI. pTRE2-NRASV12-IRES-hCD2 was generated by inserting the BamHI fragment containing the human NRASV12 coding sequence, with a C-terminal EE epitope tag, from pV12NRAS (a kind gift from Dr Brian Van Ness, University of Minnesota Cancer Center) into the single BamHI site of pTRE2 (Becton Dickinson Clontech).20 The internal ribosomal entry site–human CD2 (IRES-hCD2) was then cut from pMI2 with NheI and subcloned into a single XbaI site of pTRE2-NRASV12.
Plasmid DNA for pronuclear injection into FVB/n strain embryos from pVav-tTA was prepared by linearizing with HindIII. Plasmid DNA for pTRE2-NRASV12-IRES-hCD2 was cut with AatII and SapI. Digested DNA was isolated by agarose electrophoresis and visualized using crystal violet. Linear DNA was extracted from the gel using NA45 DEAE cellulose membrane (S&S Membrane Filters, Dassel, Germany). Fragments were eluted from the membranes in a solution containing 1 M NaCl and 0.05 M arginine-free base. Eluted DNA was phenol/chloroform extracted, ethanol precipitated, and resuspended in a solution of 5 mM Tris, pH 7.4, and 0.1 mM EDTA (ethylenediaminetetraacetic acid). Extracted DNA was quantified using an ultraviolet (UV) spectrophotometer and was visualized on an agarose gel. DNA was adjusted to a concentration of 4 ng/mL for pronuclear injection.
Southern blotting
For detection of the Vav-tTA transgene by Southern blot, 5 μg genomic DNA was digested with BglII, run out on a 1% agarose gel, and transferred to a membrane. The tTA probe used was a 405-bp PstI/BglII fragment from the pVav-tTA plasmid. For detection of the TRE2-NRASV12-IRES-hCD2 transgene, genomic DNA was digested with KpnI, run out on a 1% agarose gel, and transferred to a membrane. The NRASV12 probe used was a 926-bp EcoRV fragment from the pTRE2-NRASV12-IRES-hCD2 plasmid. All restriction enzymes were obtained from New England Biolabs (Beverly, MA).
PCR and RT-PCR
Approximately 15 to 30 ng genomic DNA from offspring was used for polymerase chain reaction (PCR) genotyping. Primers specific for the Vav-tTA transgene were used in combination with primers designed to amplify the endogenous Tsh1 locus as a reaction control. PCR entailed 30 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 1 minute with a hot-start at 94°C for 10 minutes and a final extension at 72°C for 7 minutes. Primers used were tTA-1 (5′-CTCTCTTGCCTGCCTGTG-3′) and tTA-2 (5′-GTAAACTTCTGACCCACTGGAAT-3′) (Figure 1A). Reaction control primers for TshI were 5′-AACGGAGAGTGGGTCATCAC-3′ and 5′-CATTGGGTTAAGCACACAGG-3′. The TRE2-NRASV12-hCD2 transgene was amplified using primers with a 58°C annealing temperature. PCR entailed 35 cycles of denaturation at 95°C for 30 seconds, annealing at 61.5°C for 30 seconds, and extension at 72°C for 45 seconds with a hot-start at 95°C for 10 minutes and a final extension at 72°C for 5 minutes. Primers used were NRAS-1 (5′-AGTGAACCGTCAGATCGCCTGG-3′) and NRAS-2 (5′-CACCCTGTCTGGTCTTGGCTGA-3′) (Figure 1B).
The primer RT NRAS-1 (5′-GTTATAGATGGTGAAACCTGTT-3′) for NRASV12 reverse transcription–PCR (RT-PCR) was designed to have a 2-bp mismatch between human and mouse NRAS at the 3′ end of the annealing site (Figure 1B). The primer RT NRAS-2 (5′-ATCTTGTTACATCACCACACAT-3′) anneals to vector backbone sequence between NRASV12 and the IRES. RNA was prepared by Trizol (Invitrogen) extraction of total bone marrow according to the manufacturer's instructions. RNA was treated with DNaseI (Invitrogen) per manufacturer instructions. RT-PCR was carried out using the Robust-RT kit (Finnzymes; MJ Research, Waltham, MA). RT-PCR entailed incubation at 48°C for 35 minutes followed by 2 minutes at 94°C, then 35 cycles at 94°C for 30 seconds, 48°C for 30 seconds, and 72°C for 45 seconds. These cycles were followed by a final extension at 72°C for 10 minutes.
Diagram of transgenes. The Vav-tTA transgene consists of the 2.3-kb Vav promoter region, the tTA sequence, and 2 enhancer regions of 0.7 and 3.7 kb. The tetracycline-responsive transgene is composed of TRE2, the human NRAS coding sequence containing a G>V mutation at codon 12, then the EMCV IRES and a truncated human CD2 lacking the intracellular signaling portion of the molecule. Arrows indicate primer annealing sites for PCR and RT-PCR.
Diagram of transgenes. The Vav-tTA transgene consists of the 2.3-kb Vav promoter region, the tTA sequence, and 2 enhancer regions of 0.7 and 3.7 kb. The tetracycline-responsive transgene is composed of TRE2, the human NRAS coding sequence containing a G>V mutation at codon 12, then the EMCV IRES and a truncated human CD2 lacking the intracellular signaling portion of the molecule. Arrows indicate primer annealing sites for PCR and RT-PCR.
Western blotting
Cells were washed once in ice-cold phosphate-buffered saline (PBS) and were lysed in ice-cold buffer containing 50 mM Tris, pH 7.4, 2 mM EDTA, pH 8.0, 0.1% NP-40, 250 mM NaCl, 5 mM NaF, Complete Mini protease inhibitor pellet (Boehringer-Mannheim/Roche, Indianapolis, IN), 1 mM NaVO4, and 1 mM Na2PO4. Lysates were electrophoresed on 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membrane, and blocked in PBS-T (PBS, 0.1% Tween-20) with 1% nonfat dry milk and 1% bovine serum albumin or 5% nonfat dry milk. Membranes were incubated overnight in the same solution containing anti-pERK antibody (Cell Signaling, Beverly, MA) or for 90 minutes at room temperature with anti-ERK1 (Santa Cruz Biotechnology, Santa Cruz, CA), respectively. Blots were developed using the appropriate secondary antibody (Amersham Biosciences, Piscataway, NJ) and chemiluminescent reagents (SuperSignal; Pierce, Rockford, IL). Membranes were stripped in a solution of 7 M guanidine hydrochloride for 20 minutes before reprobing.
Flow cytometry
Nucleated cells from total bone marrow were flushed from the long bones of the leg or single-cell suspensions made from spleen tissue. Red cells in the spleen were lysed by suspension in red cell lysis buffer (0.15 M NH4Cl, 1.0 M NaHCO3, 0.1 M Na2 EDTA) for 3 minutes, and then an equal volume of PBS was added to stabilize the remaining cells. Cells were pelleted, and nucleated cells were washed once in fluorescence-activated cell sorter (FACS) buffer (PBS, 2% fetal bovine serum, 0.1% sodium azide), counted and resuspended in FACS buffer. Blocking antibody (anti-CD16/CD32) was added to the cell suspensions and incubated 15 minutes. Specific antibodies were added, and cells were incubated for 25 minutes and then washed. All incubations were performed on ice protected from light to prevent capping. Stained cells were resuspended and stored in 1% formaldehyde in PBS until analysis. All phenotyping antibodies were obtained from Becton Dickinson. All flow cytometry was performed on a FACScalibur (Becton Dickinson Clontech). Acquisition and data analysis were performed using CellQuest Pro software (Becton Dickinson Clontech).
Treatment of mice
Transgenic animals were housed under specific pathogen-free conditions. Animals treated with doxycycline were provided drug in their drinking water at 2 or 5 mg/mL (as indicated) in a solution of 5% sucrose ad libitum. The solution was changed every 3 to 4 days and was protected from light by wrapping with autoclaved paper towels and aluminum foil. Peripheral blood was obtained by retro-orbital puncture. The eye was anesthetized with a 0.5% ophthalmic solution of proparacaine hydrochloride (Falcon Pharmaceuticals, Fort Worth, TX). Blood was collected in EDTA-coated, 50-μL capillary tubes (Drummond Scientific, Broomall, PA) and was transferred to a mini–capillary blood collection tube (Ram Scientific, Needham, MA) for later analysis. On completion of the collection, Neocidin ophthalmic ointment (Major Pharmaceuticals, Livonia, MI) was applied to the eye to prevent infection. Affected animals were humanely killed by cervical dislocation for the harvest of the required tissues. The Institutional Animal Care and Use Committee at the University of Minnesota approved all procedures.
Tissue culture
Cells from spleen and bone marrow were cultured in supportive media (Dulbecco modified Eagle medium [DMEM], 10% fetal bovine serum [FBS], 10% NCTC-109, 1% penicillin/streptomycin, 1% HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 1% nonessential amino acids, 1% sodium pyruvate, β-mercaptoethanol, 0.2 U/mL insulin, 1 mM oxaloacetic acid) with the addition of 10% WEHI3-conditioned media and 50 ng/mL recombinant murine stem cell factor (SCF; Peprotech, Rocky Hill, NJ). All additives were obtained from Mediatech (Herndon, VA). Cultured cells were not used beyond 10 passages.
Peripheral-blood analysis
Peripheral blood was analyzed using a HemaVet Mascot 800 hematology analyzer (CDC Technologies, Oxford, CT). Commercial controls were run each day the instrument was used, and the instrument was calibrated semiannually or as control results indicated. Peripheral blood smears obtained during retro-orbital puncture were fixed within 2 hours of collection in absolute methanol. For staining, fixed slides were immersed in modified Wright-Giemsa stain (Sigma-Aldrich, St Louis, MO) for 30 minutes. Slides were then transferred to a solution of 20% vol/vol stain in 1 × Wright-Giemsa buffer (from a 5 × solution containing 0.25 M KH2PO4 and 0.3 M Na2HPO4. 7 H2O, pH 6.4). Slides were rinsed 3 times in distilled water and allowed to air dry before mounting.
Microscopy
All microscopy was performed on a Zeiss Atto Arc HBO 110W Upright microscope equipped with 4×/0.1, 10×/0.25, 40×/0.95; and 63×/1.40 Zeiss objective lenses (Carl Zeiss, Göttingen, Germany), and using a SPOT camera and software package (Diagnostic Instruments, Sterling Heights, MI). Minor editing of images was done using Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA).
Results
Evaluation of transgene expression
Because the Vav promoter construct is hematopoiesis specific, a transgene driven by the tetracycline responsive element (TRE) will only be expressed in the hematopoietic compartment.21-23 Transgenic mice were created with tTA driven by the Vav hematopoietic promoter (Vav-tTA) (Figure 1A). The tetracycline-responsive transgenic mice harbored NRASV12 under the control of the TRE, followed by the encephalomyocarditis virus (EMCV) IRES, with a truncated hCD2 in the second cistron (Figure 1B). The hCD2 cDNA lacking the intracellular signaling portion of the molecule was included to provide a surrogate marker for transgene expression in response to doxycycline therapy.24 Three independent founder lines of NRASV12 transgenic mice were established, and a single Vav-tTA founder line was used in all experiments. The phenotype described was present in all NRASV12 transgenic lines when crossed to Vav-tTA mice (data not shown).
Before transgenic animals were generated, the constructs in Figure 1 were tested in vitro for function. pVav-tTA was cotransfected with pTRE2-Luc (Becton Dickinson Clontech) into Jurkat cells. Luciferase expression was 3 orders of magnitude higher in cells cultured without doxycycline, and cells cultured with 2 mg/mL doxycycline had expression levels similar to those of controls transfected with pVav-hCD4 and pTRE2-Luc (data not shown). To test pTRE2-NRASV12-IRES-hCD2, HEK293 cells, stably transfected with pRev-Tet-Off, were transiently transfected with pTRE2-NRASV12-IRES-hCD2. A unique 23-kDa band was present on Western blot using an anti–NRAS antibody (sc-519; Santa Cruz Biotechnology) in the lysate from nontreated cells, and the band was markedly diminished in the lysate from cells cultured with doxycycline (data not shown). These results demonstrated that both constructs were functioning as expected in vitro.
On establishing NRASV12 founder lines, we were concerned about the possibility of transcriptional activity from the NRASV12 transgene, independent of the presence of the Vav-tTA transgene. To address this issue, transgenic (tg) animals were assessed for expression of the NRASV12 transgene with and without the tTA transgene. Littermates were identified as tTA tg, NRASV12 tg, or tTA+NRASV12 tg. Animals were killed, and RT-PCR was performed on RNA extracted from total bone marrow (Figure 2). NRASV12 transcript was only detectable by RT-PCR in doubly transgenic animals. These results suggest that the NRASV12 transgene is not actively transcribed in the absence of tTA. Furthermore, no animals genotyped as singly transgenic for the NRASV12 transgene have developed disease of any kind. These animals are fertile and have apparently normal lifespans. Similarly, no Vav-tTA transgenic animals have developed disease of any kind in the absence of the NRASV12 transgene.
In addition to RT-PCR, the TRE2-NRASV12-hCD2 transgene offers the opportunity to evaluate transgene expression by flow cytometry (Figure 3). Total bone marrow from singly and doubly transgenic animals was harvested and stained with anti-hCD2 or isotype control. As seen in Figure 3, a small but detectable population of hCD2+ cells is present in the bone marrow of doubly transgenic animals. Bone marrow from singly transgenic animals stained with anti-hCD2, either from Vav-tTA or NRASV12 mice, was not different from isotype control (data not shown). The low relative number of cells expressing hCD2 on their surfaces may be a result of poor translational efficiency following the IRES.25-27 Interactions between the NRASV12 sequence and the IRES-hCD2 may also alter translation of the second cistron. Variegated expression of the Vav-tTA transgene within the hematopoietic compartment could contribute to fewer cells expressing the tetracycline-responsive transgene in doubly transgenic animals.21
Phenotype
Although these young animals had no overt signs of illness, doubly transgenic animals as young as 2 months of age were found to have enlarged spleens. All doubly transgenic animals became pruritic and had red, swollen eyes and cutaneous lesions from the head to mid-torso by 4 to 5 months of age, at which time they were killed. Examination at necropsy uniformly revealed splenomegaly and cutaneous and subcutaneous lesions, combined with widespread mast cell infiltrates into all organs examined, including bone marrow, lungs, liver, thymus, lymph nodes, and kidney (Table 1; Figures 4, 5).
Peripheral blood was collected from affected animals before their deaths, and blood smears were examined. Myeloid and mast cells were seen in various stages of maturation in the peripheral blood of all doubly transgenic animals (Figure 6). The presence of immature myeloid forms in the peripheral blood suggests either mobilization of this cell type from the marrow as a result of transgene expression or replacement of normal marrow with mast cell infiltrates, causing extramedullary hematopoiesis and inappropriate release of these cells from the intramedullary space. Examination of the marrow reveals diffuse and focal mast cell infiltrates accompanying normal hematopoiesis. Extensive findings of mast cell proliferation and infiltration, along with circulating mast cells, immature myeloid forms, or blasts, is characteristic of a myeloproliferative disorder, specifically aggressive systemic mastocytosis (ASM) or mast cell leukemia (MCL).28-30
Expression of NRASV12 in bone marrow of line 6468 mice by RT-PCR. Total bone marrow was harvested from singly and doubly transgenic littermates, as indicated, and total RNA was extracted. RNA was DNase treated, and NRAS RT-PCR was performed with (+RT) and without (-RT) reverse transcriptase. Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) RT-PCR was performed on the same samples to verify integrity and loading.
Expression of NRASV12 in bone marrow of line 6468 mice by RT-PCR. Total bone marrow was harvested from singly and doubly transgenic littermates, as indicated, and total RNA was extracted. RNA was DNase treated, and NRAS RT-PCR was performed with (+RT) and without (-RT) reverse transcriptase. Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) RT-PCR was performed on the same samples to verify integrity and loading.
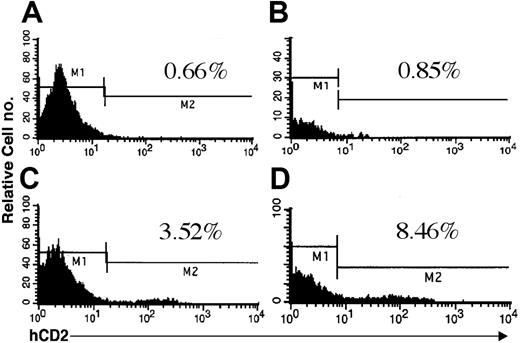
Expression of hCD2 surrogate marker in bone marrow of mice. Doubly transgenic animals were killed, and cells were flushed from the long bones of the hind legs. Flow cytometry was performed as described using either an IgGk1 isotype control (A-B) or an anti-hCD2 antibody (C-D). Jurkat cells were used as a positive control for anti-hCD2 in all experiments (data not shown). Mice from both founder lines 6468 (A,C) and 6543 (B,D) were included in the analysis. The percentage of positive cells is indicated in each panel.
Expression of hCD2 surrogate marker in bone marrow of mice. Doubly transgenic animals were killed, and cells were flushed from the long bones of the hind legs. Flow cytometry was performed as described using either an IgGk1 isotype control (A-B) or an anti-hCD2 antibody (C-D). Jurkat cells were used as a positive control for anti-hCD2 in all experiments (data not shown). Mice from both founder lines 6468 (A,C) and 6543 (B,D) were included in the analysis. The percentage of positive cells is indicated in each panel.
Repression with doxycycline
A unique feature of this disease model is the capacity to repress transgene expression by administering doxycycline (dox). To test this feature of the model system, viably frozen cells from the spleens of affected animals were briefly cultured as described in “Materials and methods” (defined media with 10% WEHI3-conditioned media and 50 ng SCF/mL). The cells were split and treated with increasing concentrations of doxycycline. RT-PCR revealed that the level of NRASV12 transcript was absent at both concentrations of doxycycline tested (Figure 7A). Repression of the transgene in vitro with doxycycline resulted in a reduction in proliferation at low concentrations of SCF (less than 5 ng/mL) or a percentage of WEHI3-conditioned media (less than 2.5%) (data not shown). In serum-starved cells treated with or without doxycycline, stimulation with 5 ng/mL SCF resulted in increased phosphorylation of extracellular regulated kinase (ERK), at longer duration, in the absence of repression (Figure 7B).
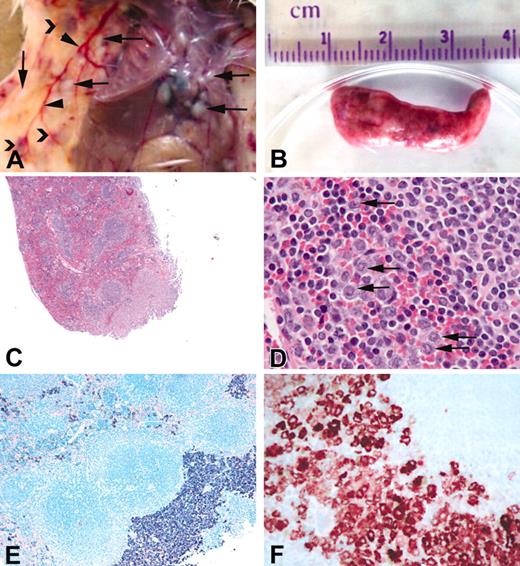
Gross appearance and histology of spleen from affected mice. A portion of the spleen from affected animals was fixed in 10% phosphate-buffered formalin, and histologic examination was performed. (A) Gross appearance of a typical animal including focal tumors in the skin and pleura/peritoneum (arrows), dilated blood vessels (arrowheads), and subcutaneous hematomas (chevrons). (B) Typical spleen from an affected animal. (C) Hematoxylin and eosin stain of spleen from line 6468. (D) Higher magnification of panel C showing infiltration of granulocytes (arrows) in the spleen. (E) Giemsa stain of spleen in panel C showing diffuse and focal infiltrates of cells containing basophilic granules consistent with mast cells. (F) Mast cell tryptase immunohistochemistry of the spleen from an affected animal. Original magnifications: (C) × 4, (D) × 40, (E) × 10, (F) × 40.
Gross appearance and histology of spleen from affected mice. A portion of the spleen from affected animals was fixed in 10% phosphate-buffered formalin, and histologic examination was performed. (A) Gross appearance of a typical animal including focal tumors in the skin and pleura/peritoneum (arrows), dilated blood vessels (arrowheads), and subcutaneous hematomas (chevrons). (B) Typical spleen from an affected animal. (C) Hematoxylin and eosin stain of spleen from line 6468. (D) Higher magnification of panel C showing infiltration of granulocytes (arrows) in the spleen. (E) Giemsa stain of spleen in panel C showing diffuse and focal infiltrates of cells containing basophilic granules consistent with mast cells. (F) Mast cell tryptase immunohistochemistry of the spleen from an affected animal. Original magnifications: (C) × 4, (D) × 40, (E) × 10, (F) × 40.
To test the repressibility of the system in vivo, a cohort of animals was identified with the appropriate genotypes, including singly transgenic animals from the transactivator (n = 2) and responsive lines (n = 1). These littermates were monitored for the appearance of mast cells in the peripheral blood. When mast cells were identified in the peripheral blood of all 4 doubly transgenic animals, doxycycline was added to the drinking water (2 mg/mL) of 2 of them. Treatment continued for 5 weeks, at which time peripheral blood was collected from all the animals before they were killed. Spleen, femurs, and sternum were collected from each animal, along with any other tissues of grossly abnormal appearance. Flow cytometry was performed on spleen cells and bone marrow.
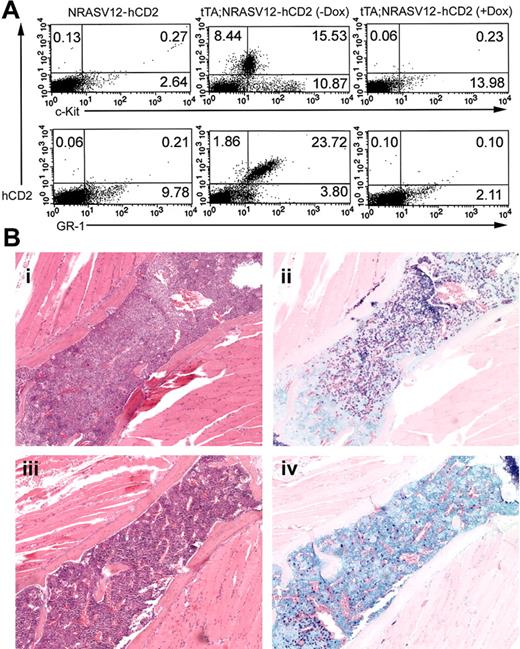
Among the spleens of doubly transgenic animals not treated with doxycycline was a distinct population of hCD2, GR-1, c-Kit–positive cells, as determined by flow cytometry (Figure 8A). This cell population is absent in singly transgenic NRASV12 mice. Spleen cells from animals treated with doxycycline are similar to those of controls, with no expression of hCD2 in the spleen. A concordant population of GR1+, c-Kit+ cells in the spleens of treated animals did not appear, and these markers were reduced to the level of the control. Curiously, examination of sections from spleen and sternum of these animals confirms the presence of mast cells, though the numbers were lower than in animals left untreated (Figure 8B).
Histologic examination of other tissues in affected animals. Hematoxylin (A,C,E,G) and Giemsa (B,D,F,H) stains of various tissues from affected animals. (A-B) Subcutaneous tumor in the skin. (C-D) Widespread infiltration of the lung. (E-F) Mast cell tumor in thymus. (G-H) Small, widespread infiltrates in the liver. Infiltrates were also seen in the kidney and lymph nodes of some animals (data not shown). Original magnification, × 10 in all panels.
Histologic examination of other tissues in affected animals. Hematoxylin (A,C,E,G) and Giemsa (B,D,F,H) stains of various tissues from affected animals. (A-B) Subcutaneous tumor in the skin. (C-D) Widespread infiltration of the lung. (E-F) Mast cell tumor in thymus. (G-H) Small, widespread infiltrates in the liver. Infiltrates were also seen in the kidney and lymph nodes of some animals (data not shown). Original magnification, × 10 in all panels.
Treated animals had no evidence of cutaneous lesions at necropsy, in contrast to the widespread lesions present on untreated animals. Gross examination of other tissues from treated animals did not reveal any abnormalities. Despite the normal appearance and size of the spleens in doxycycline-treated animals, sections of them showed persistence of mast cells. Histologic analysis demonstrated a reduced number of mast cells in the bone marrow of treated animals (Figure 8B). The persistence of mast cells in these animals is perhaps not surprising given estimates of mast cell lifespan of up to 80 days,31 considerably longer than the 35 days of treatment these animals experienced.
Mast cells in the peripheral blood of doubly transgenic animals. Peripheral blood from affected animals was examined for the presence of mast cells (modified Wright-Giemsa stain). Granulocytes in varying stages of development are shown, including immature myeloid forms, each with a few metachromatic granules (top row), immature or atypical mast cells (middle row), and mature mast cells (bottom row). Original magnification, × 63 in all panels.
Mast cells in the peripheral blood of doubly transgenic animals. Peripheral blood from affected animals was examined for the presence of mast cells (modified Wright-Giemsa stain). Granulocytes in varying stages of development are shown, including immature myeloid forms, each with a few metachromatic granules (top row), immature or atypical mast cells (middle row), and mature mast cells (bottom row). Original magnification, × 63 in all panels.
Discussion
In this report we have successfully developed a 2-transgene system of tTA-driven gene expression in the hematopoietic system, which is repressible using doxycycline and is used to drive expression of the NRASV12 oncogene. Our results confirm that NRASV12 expression by itself can induce a profound MPD phenotype, namely ASM. Most features of human ASM are recapitulated in this model, including dramatic skin and lung pathology. What, if any, additional genetic or epigenetic changes might contribute to the ASM phenotype are unclear. However, it is clear from our results that NRASV12 expression is not only sufficient to initiate, but is required to maintain, the ASM phenotype because disease in doxycycline-treated mice was arrested.
Repression of transgene expression in vitro with doxycycline. (A) Cryopreserved spleen cells from affected animals were incubated 80 hours with doxycycline (Dox) at the indicated concentrations. Total RNA was extracted, DNase-treated and NRAS RT-PCR was performed with (+RT) and without (-RT) reverse transcriptase. GAPDH RT-PCR was performed on the same samples to verify integrity and loading. (B) Cultured cells were split, and half were treated with doxycycline for 24 hours. Treatment was continued while cells were serum starved overnight and then stimulated with 5 ng/mL SCF for the times indicated. Lysates were harvested on ice, and Western blot analysis was performed for p-ERK. The blot was stripped and reprobed for total ERK1 to verify equivalent loading.
Repression of transgene expression in vitro with doxycycline. (A) Cryopreserved spleen cells from affected animals were incubated 80 hours with doxycycline (Dox) at the indicated concentrations. Total RNA was extracted, DNase-treated and NRAS RT-PCR was performed with (+RT) and without (-RT) reverse transcriptase. GAPDH RT-PCR was performed on the same samples to verify integrity and loading. (B) Cultured cells were split, and half were treated with doxycycline for 24 hours. Treatment was continued while cells were serum starved overnight and then stimulated with 5 ng/mL SCF for the times indicated. Lysates were harvested on ice, and Western blot analysis was performed for p-ERK. The blot was stripped and reprobed for total ERK1 to verify equivalent loading.
Repressibility of disease in vivo with doxycycline. Five weeks after the start of doxycycline treatment, mice were killed, and spleens and bones were taken for histologic analysis, flow cytometry, and tissue culture. (A) Flow cytometry was performed on single-cell suspensions of spleen tissue. Cells were stained with anti-hCD2 and either anti–c-Kit (top row) or anti–GR-1 (bottom row). Singly transgenic (left column) and doubly transgenic animals were analyzed. A substantial population of c-Kit+, GR-1+ cells, characteristic of mast cells, is present in doubly transgenic mice not treated with doxycycline (middle column), whereas a similar population is absent in mice treated with doxycycline (right column). Representative results are shown, with percentages of positive cells indicated in each quadrant. (B) Histologic analysis showing (i) granulocyte and (ii) mast cell infiltrates in the bone marrow of untreated mice and (iii-iv) reduced infiltration in the bone marrow of treated animals. A single sternal vertebra is shown in each panel. Original magnification, × 10.
Repressibility of disease in vivo with doxycycline. Five weeks after the start of doxycycline treatment, mice were killed, and spleens and bones were taken for histologic analysis, flow cytometry, and tissue culture. (A) Flow cytometry was performed on single-cell suspensions of spleen tissue. Cells were stained with anti-hCD2 and either anti–c-Kit (top row) or anti–GR-1 (bottom row). Singly transgenic (left column) and doubly transgenic animals were analyzed. A substantial population of c-Kit+, GR-1+ cells, characteristic of mast cells, is present in doubly transgenic mice not treated with doxycycline (middle column), whereas a similar population is absent in mice treated with doxycycline (right column). Representative results are shown, with percentages of positive cells indicated in each quadrant. (B) Histologic analysis showing (i) granulocyte and (ii) mast cell infiltrates in the bone marrow of untreated mice and (iii-iv) reduced infiltration in the bone marrow of treated animals. A single sternal vertebra is shown in each panel. Original magnification, × 10.
Previous attempts have been made to recapitulate RAS oncogene-driven hematopoietic disease in the mouse. One model uses a conditional “flox-stop” regulated knock-in of mutant KRASG12D into the endogenous locus.32,33 The KRASG12D allele was turned on in hematopoietic tissue using the inducible MX1-Cre transgene. This work demonstrates that mutant KRAS expressed from its endogenous locus leads to the development of rapidly fatal myeloproliferative disease but does not result in the development of leukemia. Other attempts have been made using RAS oncogenes in retroviral vectors.34,35 Transduction of hematopoietic cells with retroviruses containing the HRAS oncogene resulted in T-cell thymic lymphoma, pre–T-cell thymic lymphomas, and pre–B-cell lymphoblastic lymphomas in recipient mice. There was no evidence of myeloid proliferation in these experiments. Patients with neurofibromatosis type 1 (NF1) develop peripheral nerve sheath tumors that show infiltration with mast cells. Mast cells from NF1 heterozygous mice have been shown to be defective in degranulation and cytokine response.36,37 NF1 patients are also at increased risk for the MPD/MDS syndrome juvenile myelomonocytic leukemia (JMML).38-40 Retroviral transduction with BCR-ABL has been shown to cause mastocytosis in mice.41-43 In addition to retroviral transduction of murine bone marrow with BCR-ABL, one attempt to recapitulate human mutant NRAS-driven myeloid disease in mice has been made. Murine bone marrow was transduced with retrovirus containing activated NRAS, but disease occurred in only a subset of mice, with MPDs predominating (3 of 17).44 Monocytic leukemia did develop in 1 mouse. Interestingly, 3 of 17 mice in this study also had a mast cell disorder, 1 of which occurred concomitantly with the monocytic leukemia. The other 2 incidences of mast cell disease occurred in conjunction with MPD. Notably, our model of NRASV12-driven disease supports previous work in which Ras pathway activation results in mast cell disease or hyperproliferation.41-44
During the preparation of this manuscript, Guo et al45 reported a retroviral transduction model using activated MRAS in the hematopoietic system of mice. In this model, the transplantation of transduced bone marrow resulted in ASM/MCL, further supporting the findings presented in this paper. The authors present evidence that activation phosphatidylinositol 3 kinase (PI3K) is required for transformation in their model, and levels of activated ERK in cells transduced with mutant MRAS were able to drive the proliferation of mast cells. Furthermore, in cultures from HRAS-transduced cells, lower levels of HRAS expression resulted in a slight increase in HRAS-expressing mast cells cultured in and dependent on interleukin-3 (IL-3). This finding is similar to our observations in that we were able to detect a mutant NRAS transcript (Figures 2, 7), and the phosphorylation of ERK (Figure 7), but were unable to demonstrate NRASV12 protein expression in primary or cultured cells (data not shown), suggesting low expression levels or rapid degradation of the protein. Guo et al45 did attempt transduction with activated NRAS retrovirus but, like others, they were unable to generate a consistent phenotype.44 In contrast, our data clearly indicate that NRASV12 is able to drive the development of ASM/MCL in mice. The reasons for disparate phenotypes derived from oncogenic M-, H-, and N- RAS are unclear and may not be attributed solely to differential levels of mitogen-activated protein kinase (MAPK) pathway activation. It has become clear that different RAS isoforms traffic differently in the cell and undergo different posttranslational modifications and that specific mutations of the same isoform can result in different phenotypes.46-52 Furthermore, the effect of oncogenic RAS expression may largely be affected by the cell in which the mutant protein is expressed, regardless of isotype or mutation status, as seen in models of conditionally expressed KRAS.32,33,53,54 Retroviral transduction may not efficiently target cells susceptible to oncogenic NRAS transformation. Subtle differences in retroviral construct and titer may alter target cell specificity and expression levels. To address the question of target cell permissiveness, studies under way in our laboratory are aimed at identifying which cells in the hematopoietic system of mice express tTA and mutant NRAS.
The 2-transgene system used in this model has been used by a number of other investigators and has been the subject of several excellent reviews.55-62 The response to transgene repression in these models has been varied, including transgene-independent growth, regression by apoptosis, differentiation, and growth arrest.58,62,63 In one example, lymphomagenesis has been reported using the tet system, wherein c-Myc is under the control of the transactivator in B-lymphoid cells driven by the μE promoter.64 Lymphoma regressed, through rapidly induced apoptosis, on c-Myc repression in these animals. In a mouse model of doxycycline-induced melanoma, oncogenic HRAS was required for the development of tumors in an INK4a-/- background, and tumors regressed dramatically on removal of the inducer, similarly through apoptosis.65 In another system, BCR-ABL fusion was driven by the TRE, resulting in B-cell leukemia that rapidly regressed on transgene repression.66 Other studies investigating the role of oncogenes in the maintenance of tumors involve the induction of lung adenocarcinomas by tet-regulated KRAS.53 Clearly, in this model, KRAS was required for the development and maintenance of tumors, as was demonstrated in the previously mentioned tumor models. Using a conditionally expressed activated KRAS allele, similar results were obtained by somatic activation of this oncogene and subsequent development of lung carcinoma.54 This same conditional KRAS allele was also used in the hematopoietic system resulting in MPD, described earlier in this section. Of note, where activated KRAS drives the development of lung carcinoma in 2 separate models, it was unable to induce acute leukemia, suggesting that the hematopoietic system is more resistant to oncogenic RAS transformation.
Importantly, in each of these regulatable models of tumorigenesis in which rapid apoptosis occurred on transgene repression, some secondary genetic lesion was either intentionally introduced or was a consequence of expression of the transgene. The melanoma model was derived on an INK4a-/- background. A prominent feature of tumor cells derived from the c-Myc model was genomic instability, primarily a gain of material corresponding to chromosome 15. Last, BCR-ABL itself has been shown to directly induce genomic instability and a higher frequency of mutation in mice and humans.67 Indeed, the authors of this study note that some tumors that developed with longer latency were resistant to repression of the BCR-ABL transgene by doxycycline, suggesting secondary mutations had occurred.66 In our work, as with the hematopoietic activation of KRAS, chronic MPD developed, but acute malignant disease did not. Despite extensive effort, we have been unable to demonstrate the induction of apoptosis on NRASV12 repression in this model (data not shown). This, combined with evidence of residual disease in animals treated with doxycycline (Figure 8), suggests either proliferative arrest or a return to homeostatic proliferation of mast cells on oncogene repression. The level of disease detected at the termination of the experiment may be the extent of disease present at the time of treatment initiation. Alternatively, residual mast cells seen in treated animals that appear to be c-Kit, GR1–negative may represent a pool of progenitor cells present at the time of treatment initiation that have undergone differentiation in the absence of the NRASV12 oncogenic stimulus, similar to other reports of differentiation or cell cycle arrest on oncogene repression.62,63 Whether NRASV12 expression directly alters surface expression of c-Kit and GR1 or these changes are caused by altered differentiation and response to the microenvironment on oncogene repression remains to be determined. Perhaps the introduction of additional mutations would allow an outgrowth of leukemic clones within the hematopoietic system that would be more sensitive to removal of the stimulus oncogenic NRAS provides.
In patients with ASM, codon 816 in the stem cell factor receptor (KIT) is commonly mutated, resulting in constitutive activation.68 Studies of the mutant KIT receptor tyrosine kinase suggest that Janus kinase 2–signal transducer and activator of transcription 3 (JAK2-STAT3) is one of the major signal transduction pathways altered when compared with the wild-type receptor, and it may be informative to examine Jak2-Stat3 activation in our model.69 However, some investigators have implicated PI3K as a major downstream effector of KIT. Activation of KIT causes signal transduction through RAS-mediated signaling pathways.70,71 KIT can interact with PI3K directly or indirectly through interactions with RAS and subsequent RAS-PI3K association. Our data implicate Ras signaling in ASM and may mimic signaling by mutant c-Kit, though we have not yet determined whether activated Kit mutations are present in ASM cells in our model.71
GR1 and c-Kit expression on the surface of mast cells in our model appeared to depend on oncogenic NRAS expression (Figure 8). Alternatively, mast cells observed by histologic examination in treated animals may be quiescent or apoptotic in the absence of NRASV12 expression, causing the loss of c-Kit and GR-1 on their surfaces. One possibility that is difficult to rule out is an unknown effect of doxycycline on the expression of these cell surface markers. Nonetheless, results from this model, in particular the loss of Kit expression on doxycycline treatment, combined with results from previous studies, strongly suggest that signaling through the RAS-MAPK pathway, rather than Jak-Stat3, is important for some of the phenotypic characteristics of mast cells.
The relationship between the NRAS oncogene and Kit expression may be more complicated, however. Our results suggest the NRASV12 might be driving high levels of Kit expression on the malignant mast cells, potentially helping to establish paracrine or autocrine Scf/Kit signaling that itself is driving proliferation. Indeed, increased transcription from KIT in patients with more aggressive forms of mast cell disease associated with MPD has been documented.72 If this is the case, KIT inhibitors may inhibit ASM, even when driven by an RAS oncogene.
Expression of CD2 (LFA-2) on mast cells in human systemic mastocytosis is well documented.73-76 The surrogate hCD2 marker introduced with the NRASV12 transgene may contribute to the development of the ASM phenotype described here. Although the intracellular portion of CD2 has been deleted, the extracellular domain may engage antigen-presenting cells in the mouse. Even if this specific interaction does not result in an effective signal within the mast cell, engagement may induce the release of cytokines and chemokines from antigen-presenting cells, such as IL-3, IL-4, or IL-10, contributing to mast cell proliferation.77 However, the contribution of hCD2 in this model is unlikely. First, control transgenic animals were derived in this experiment using wild-type NRAS in the first cistron of the tetracycline-responsive transgene followed by the IRES and hCD2. To date, none of the control animals has developed a phenotype similar to that in experimental animals. Second, several investigators cited here have reported findings in which Ras-MAPK pathway activation has resulted in mast cell disease or other mast cell abnormalities.
The model described in this report has the advantage of providing a conditionally expressed and repressed NRAS oncogene, a uniform disease picture, and a model that closely parallels a form of human MPD, namely ASM. Disadvantages include the potential that doxycycline itself may affect the ASM phenotype, independently of its effect upon NRASV12 transgene expression. In addition, variegated NRASV12 expression is likely to be occurring. Although this might be an advantage because leukemic and nonleukemic clones coexist in patients with MPD, it may also mean that some aspects of RAS oncogene–driven myeloid diseases may not be easily recapitulated in this model. Because of the potential limitations using the hCD2 surrogate marker in our system, future work with a more sensitive reporter will be directed toward determining precisely which cells express the tTA within the hematopoietic system in Vav-tTA transgenic. Another potential disadvantage is that the level of NRASV12 expression achieved may not be equivalent to that seen in human myeloid diseases. A more physiologic level of NRAS oncogene expression may result in other myeloid proliferative diseases (MPD, AML) or may yield MDS phenotypes. If this is true, varying levels of doxycycline treatment and partial-repression NRASV12 transgene expression may produce varying phenotypes. Nevertheless, the model has the potential to answer questions about the nature of RAS gene activation in myeloid malignancies.
Further investigation using this model may help to elucidate which characteristics of RAS pathway activation may contribute to hematologic abnormalities, including patients with ASM. The possibility of transgene repression in this model will lend itself to the study of oncogenic NRAS-specific phenotypes in hematopoietic disease. Finally, this model may provide a tool with which to investigate the efficacy of targeted molecular therapies and the development of new pharmacologic agents that can be used to treat ASM and related myeloid diseases. In particular, it will be important to compare the effects of genetic suppression of RAS oncogene expression with pharmacologic inhibition of RAS itself or elements of downstream pathways. Such experiments might uncover biologically meaningful molecular end points to be used in clinical trails aimed at the suppression of RAS oncogene activity.
Prepublished online as Blood First Edition Paper, April 14, 2005; DOI 10.1182/blood-2004-08-3306.
Supported by research grants from the Leukemia and Lymphoma Society of America (LLSA) and the Leukemia Research Fund (7019-04). Partially funded by the University of Minnesota Institute for Human Genetics Fellowship Award (S.M.W. and D.A.L.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Sandra Horn in the Mouse Genetics Laboratory, Nicole Kirchoff in the Histopathology Core Facility, and Greg Veltri in the Flow Cytometry Core Facility at the University of Minnesota Cancer Center for their help generating transgenic animals and assisting with the extensive analyses.