Abstract
Multiple myeloma (MM) plasma cells (PCs) express receptor for hyaluronan-mediated motility (RHAMM), a hyaluronan-binding, cytoskeleton and centrosome protein. The most abundant RHAMM isoforms in MM are full-length RHAMM (RHAMMFL) and the splice variant RHAMM-exon4. We separately examined the significance of RHAMM expression, and isoform balance, in 2 groups of MM patients. In oligonucleotide microarray experiments (n = 210, Arkansas), increasing RHAMM mRNA expression in MM PCs is strongly associated with osteolytic bone lesions (P < .001), and event-free (P = .05) and overall (P = .04) survival. Semiquantitative determination of RHAMM isoform expression (Alberta, Canada) used capillary electrophoretic detection and measurement of RHAMM-exon4/RHAMMFL reverse-transcriptase-polymerase chain reaction (RT-PCR) products. RHAMM isoforms are rarely expressed concurrently in single MM PCs; the pattern of isoform expression, at the single-cell level, is approximated in larger numbers of cells by the RHAMM-exon4/RHAMMFL ratio. Absolute RHAMM expression and the RHAMM-exon4/RHAMMFL ratio are only partially correlated in MM PCs; in cell lines, absolute RHAMM expression is elevated in mitosis, while RHAMM ratios remain stable. Temporal examination of MM patients' peripheral blood reveals that the RHAMM-exon4/RHAMMFL ratio increases with disease burden. The RHAMM-exon4/RHAMMFL ratio in diagnostic bone marrow samples (n = 101, Alberta) is an independent prognostic factor. Thus, expression and splicing of RHAMM are important molecular determinants of disease severity in MM. (Blood. 2004;104:1151-1158)
Introduction
Multiple myeloma (MM) is a B-cell neoplasm characterized by accumulation of clonal plasma cells (PCs) in the bone marrow (BM), secretion of a monoclonal protein (M-protein), and osteolytic lesions accompanied by bone pain and anemia.1 MM is further characterized by chromosomal instability (CIN) and a defining VDJ rearrangement of the immunoglobulin heavy chain (IgH) gene locus, termed clonotypic.2,3 Recurrent genetic translocations into switch recombination regions of the IgH locus occur in up to 75% of myelomas4,5 ; one of the most common of these is t(4;14)(p16.3;q32).4,6 MM PCs contain extensive chromosomal abnormalities, both numeric and structural.5 The presence of specific genetic lesions is prognostic.1,4,6
An additional ubiquitous feature of MM is the expression of the receptor for hyaluronan-mediated motility (RHAMM).7 RHAMM was cloned in 1992 and described as a hyaluronan-binding protein.8,9 A variant of RHAMM, which is predominantly cytoplasmic and involved in multiple signaling pathways,10,11 is transforming and essential for ras transformation.12 Human full-length RHAMM (RHAMMFL) was cloned in 1996 as a predicted 85- to 95-kDa protein encoded by 18 exons.13 RHAMMFL has been described as an actin- and microtubule-associated protein that localizes to the centrosome and mitotic spindle pole and is essential for mitotic stability.14,15 The NH2-terminus of RHAMM directly interacts with microtubules, via exon 4, while the COOH-terminus targets the centrosome, presumably through an indirect interaction with dynein.14,15 Loss of exon 4 inhibits the ability of RHAMM to interact with interphase microtubules14 ; disruption of the COOH-terminus, by microinjection of polyclonal antiserum, affects mitotic integrity and induces tripolar and tetrapolar spindles.15
While RHAMM is ubiquitously expressed in murine tissues (with elevated expression in testis, thymus, and spleen),16 consistent detection of RHAMM expression within healthy human tissues appears to be limited to the testis.17,18 Reports of RHAMM expression have identified the testis, spleen, colon, and stomach17 ; the testis, placenta, and thymus18 ; and the testis, placenta, lung, and pancreas19 as RHAMM-positive tissues (in decreasing orders). RHAMM expression is rare in normal peripheral blood7,17,18 and purified CD19+7 or CD34+ peripheral blood mononuclear cells (PBMCs).19 RHAMM expression influences tumor progression and metastasis in melanoma, pancreatic, breast, and endometrial carcinomas,20-23 and RHAMM is detected as a tumor-associated antigen in acute myeloid leukemia (AML), chronic myeloid leukemia (CML),18 and colon cancer.17 A splice variant of exon 4, RHAMM-exon4 (RHAMM-48), has been described in myeloma patients and in diverse tumors and cancer cell lines.7,14,17 RHAMM-exon4 is also found in healthy tissues, and the relative ratio of these 2 isoforms differs between paired cancerous and adjacent healthy tissue in colon cancer patients.17 While RHAMMFL and RHAMM-exon4 expression is associated with malignancy, until now it has been unclear whether RHAMM isoforms are expressed concurrently in the same cell and what, if any, is the clinical significance of their expression.
In this study we report MM PC gene expression array data showing that elevated RHAMM mRNA levels correlate with the presence of osteolytic bone lesions, and that high RHAMM expression negatively correlates with event-free and overall survival. We identify a partial correlation in MM PCs between elevated RHAMM expression and increased splicing of RHAMM-exon4. In cell lines, while absolute RHAMM expression increases through mitosis, exon 4 splicing remains consistently low. We further demonstrate that individual myeloma plasma cells rarely express both isoforms concurrently and that patients differ in their RHAMM isoform expression profiles. RHAMM isoform ratios in the blood vary within individual patients through the course of disease, decreasing after treatment and increasing with disease relapse. High RHAMM-exon4/RHAMMFL ratios in the bone marrow at the time of diagnosis correlate with poor survival, independent of standard prognostic factors.
Patients, materials, and methods
Patients and clinical data
In Alberta, Canada, total RNA extracts from BM biopsies of 101 newly diagnosed MM patients were used to assess the prognostic impact of RHAMM exon 4 splicing. Sorted BM PCs from an additional 22 patients were used to examine RHAMM expression and splicing at the single-cell level (n = 8) and absolute RHAMM expression at the population level (n = 14). Peripheral blood cellular RNA samples from 5 t(4;14)+ MM patients were obtained at various time points in the course of disease, as described previously,6 and were used to examine temporal changes in RHAMM expression patterns.
In Arkansas, samples for microarray experiments included purified PCs from a separate cohort of 210 newly diagnosed cases of MM. Isolation from mononuclear cell fraction was performed and purity was confirmed as previously described.24 Skeletal roentgenogram surveys (n = 199) and skeletal magnetic resonance imaging (MRI; n = 193) were performed on the Arkansas cases to subclassify patients as having no focal bone lesions, at least 1 focal lesion, and 3 or more focal lesions. Arkansas patients were treated with Total Therapy II25,26 and were followed for event-free and overall survival. Indications for treatment in patients lacking bone lesions included cytopenias, hypercalcemia, and renal dysfunction.
Total RNA isolation, cDNA synthesis, preparation of labeled cRNA, hybridization to microarrays, and analysis of GeneChip data (Arkansas patients)
Total RNA was isolated from PCs using the RNeasy Mini kits (QIAGEN, Valencia, CA). Detailed protocols for cDNA synthesis, cRNA preparation, hybridization to the U95Av2 microarray (Affymetrix, Santa Clara, CA), and analysis of raw data have been described.24 All microarray data used in the analyses were derived from Affymetrix 5.0 software. GeneChip 5.0 output files provide a signal that represents the difference between the intensities of the sequence-specific perfect match probe set and mismatch probe set, or as a detection of present, marginal, or absent as determined by the GeneChip 5.0 algorithm. Gene arrays were scaled to a target intensity of 1500 and then analyzed independently. All signal calls were transformed by the log base 2, and each sample was normalized to give a mean of 0 and variance of 1. The oligonucleotide probes target the carboxy terminal sequences of RHAMM mRNA and therefore detect both RHAMM-exon4 and RHAMMFL without distinguishing the 2 variants. High and low RHAMM gene expression is defined as expression above and below the median.
RNA extraction, cDNA synthesis, and RT-PCR (Alberta patients)
Patient bone marrow mononuclear cells (BMMCs) and peripheral blood mononuclear cells (PBMCs) were purified on Ficoll-Hypaque Plus (Amersham Pharmacia Biotech, Uppsala, Sweden) density gradients. For single-cell analysis, BMMCs were stained with CD138 or CD38 to mark PCs. Individual PCs, or 1000 cells/well, were sorted into polymerase chain reaction (PCR) tubes using a direct lysis protocol, followed by single-cell reverse-transcriptase (RT)-PCR as previously described.27 For aggregate populations (PBMCs, BMMCs, or sorted PCs), cells were suspended in TRIzol Reagent (Invitrogen, Carlsbad, CA) at 2 to 10 × 106 cells/mL and total RNA was extracted according to the manufacturer's instructions. Poly-A-tailed RNA was reverse transcribed for 1 hour at 42°C followed by enzyme inactivation at 99°C for 3 minutes from 1 μg total RNA with 500 μM dT15, 500 μM each deoxynucleoside triphosphate (dNTP), 10 mM dithiothreitol (DTT), 50 mM Tris (tris[hydroxymethyl]aminomethane)-HCl, 75 mM KCl, 3 mM MgCl2, and 200 U Superscript RNase H Reverse Transcriptase (Invitrogen) in a 20-μL reaction.
All RT-PCRs contained 20 mM Tris-HCl, 50 mM KCl, 2.0 mM MgCl2, 200 μM each dNTP, 1.0 U Platinum Taq DNA polymerase (Invitrogen), and 0.2 μM each PCR primer. Reactions were carried out in 25-μL reactions using 4% of the cDNA as template. Reactions consisted of an initial 5-minute denaturation at 94°C, followed by 30 cycles of amplification at an annealing temperature of 64°C and 30-second extensions at 72°C. RHAMM primers were as follows: 5′ primer, TGACAAAGATACTACCTTGCCTGCT; 3′ primer, CAGCATTTAGCCTTGCTTCCATC.
The primers were designed to flank exon 4, so that 2 RT-PCR products of different sizes could be detected in a single reaction, representing full-length RHAMM and the splice variant lacking exon 4. A 6-carboxy fluorescein (6-FAM) fluoresceinated label was added to the 3′ primer for detection using fragment capillary electrophoresis (FCE, ABI3100; Applied Biosystems, Foster City, CA). RT-PCR products were visualized by ethidium bromide (EtBr) staining on 2% agarose (Invitrogen) gels or by capillary electrophoresis as described.28,29 Single-cell RT-PCR products were precipitated prior to capillary electrophoresis, to increase sensitivity of detection. Sequencing of clonotypic VDJ was performed as described.27
The relative amount of exon 4 splicing in a given sample was estimated by calculating the ratio of fluorescence intensity of the RHAMM-exon4 and RHAMMFL products following RT-PCR and FCE (ie, the RHAMM-exon4/RHAMMFL ratio, hereafter “RHAMM ratio”). Each sample was analyzed in triplicate. Fluorescence intensity was analyzed using GeneScan software (Applied Biosystems).
Quantitative RT-PCR (Alberta)
BMMCs were stained with anti-CD138 microbeads (Miltenyi Biotec, Auburn, CA) as suggested by the manufacturer, and single-column isolations were performed on an autoMACS Magnetic Cell Sorter (Miltenyi Biotec). Purity of the selected CD138+ PCs was verified to be more than 90% by cytospin and morphology examination. RNA isolation, sample quality examination, and reverse transcription were as suggested by the manufacturer (Applied Biosystems). Each quantitative RT-PCR reaction was performed in a 50-μL volume consisting of 1 × Universal PCR Master Mix No AmpErase UNG (Applied Biosystems), 2 μL of the respective Taqman Assays-on-Demand Gene Expression Products primer and probe mix for glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Hs99999905_m1) or RHAMM (Hs00234864_m1) (Applied Biosystems), and 5 ng RNA converted to cDNA as template. Reactions were run on an ABI PRISM 7700 Sequence Detection System (Applied Biosystems). Quantitation using the Relative Standard Curve Method was performed using a 2-log dilution range of Raji cDNA to generate a standard curve for each reaction.
Synchronization, immunoprecipitations, and quantitation (Alberta)
HeLa and Raji cells were synchronized by double thymidine block. Briefly, cells were plated at subconfluency and incubated with, or without, 2 mM thymidine for 14 to 16 hours. Cells were washed, released from thymidine block, and incubated in fresh media for 8 hours. Cells were washed and the synchronized populations were incubated with 2 mM thymidine for 14 to 16 hours. Cells were released into fresh media for 3 to 4 hours, and synchronized populations were incubated in 300 ng/mL nocodazole (Sigma, St Louis, MO) for 10 to 12 hours, washed with phosphate-buffered saline (PBS), and released by shake-off. Unsynchronized HeLa populations were released from plates with 1 × trypsin. Both mitotic and unsynchronized populations were then washed 3 times with PBS and lysed at 5 × 106 to 107 cells/mL in 1% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propane-sulfonic acid) plus 10 μg/mL leupeptin, 10 μg/mL antipain, and 1 mM phenylmethylsulfonyl fluoride (all from Sigma). Protein quantitation used the Odyssey v1.1 Infrared imaging system (LI-COR Biosciences, Lincoln, NE) with detection of polyclonal sera using IRDye 800 conjugated anti-rabbit IgG (Rockland, Gilbertsville, PA).
Statistical methods
Statistical analyses were performed with the software packages SPSS (SPSS, Chicago, IL) and SAS v8 for Windows (SAS, Cary, NC). Differences in proportions were compared using Fisher exact test. Between-group comparisons of continuous variables were with Student t test or the Wilcoxon rank sum test. Linear relationships were determined by Pearson or Spearman correlation coefficient. Survival curves were plotted using the Kaplan-Meier method. Comparison of survival distributions was with the log-rank test. Logistic regression was used to assess the independent associations of RHAMM and DKK1 with osteolytic lesions. Cox proportional hazards analysis was used to assess the independent prognostic significance of the RHAMM ratio. Statistical significance was set at a level of P < .05 using 2-sided analysis.
Results
Increased RHAMM expression levels correlate with bone lesions, but not with DKK1 expression (Arkansas patients)
The characteristics of the 210 Arkansas patients are outlined in Table 1, for the total population as well as for the 2 subgroups of patients defined based on the median level of RHAMM expression. Patients expressing RHAMM at levels below the median are classified as “low” RHAMM expressers; patients above the median are classified as “high” RHAMM expressers. Among the conventional measures of disease severity listed in Table 1, only lactate dehydrogenase (LDH) is correlated with RHAMM at a P value of .05 or less.
Preliminary microarray experiments had identified RHAMM as one of the genes most highly correlated with the presence of bone lesions in myeloma,24 prompting us to investigate further the association between RHAMM and bone lesions. The proportion of patients with 1+ MRI-detected focal bone lesions was significantly higher in the “high” than in the “low” RHAMM expressers (Table 1). The association between bone lesions and RHAMM expression was also found to be significant when using the less sensitive technique of skeletal x-rays as the basis for assessment of bone disease, or when comparing only those patients with multiple bone lesions to the group lacking bone lesions (Table 1).
A recent publication by the Arkansas group has demonstrated a significant correlation between expression of the DKK1 gene and the presence of osteolytic lesions.30 We examined the correlation between RHAMM and DKK1 expression in the microarray experiments. Remarkably, there was no statistically significant correlation between the expression of RHAMM and DKK1 (Spearman r = 0.077, P = .26). Logistic regression modeling of MRI-detectable bone lesions revealed that RHAMM and DKK1 are both independently and significantly associated with the presence of lytic bone disease (odds ratio [OR] for bone lesions in 2-variable regression model: for DKK1, OR = 1.37 [95% CI, 1.18-1.60], P < .0001; for RHAMM, OR = 1.72 [95% CI, 1.14-2.59], P < .001).
RHAMM expression correlates with poor disease-free and overall survival (Arkansas patients)
Figure 1A-B shows overall and event-free survival for 210 Arkansas patients stratified by high versus low RHAMM expression (median cutoff) as assessed by microarray. High RHAMM expression is significantly associated with poor outcome. The 2-year overall survival estimate for patients with high RHAMM expression is 76% and with low RHAMM is 88% (P = .04). The 2-year event-free survival in high RHAMM expressers is 76%, versus 88% in the low RHAMM expressers (P = .05).
RHAMM expression predicts event-free and overall survival. (A-B) Kaplan-Meier overall and event-free survival distributions for 210 Arkansas patients stratified by high versus low expression of RHAMM (median cutoff) according to Affymetrix U95Av2 microarray experiments. High RHAMM expression is significantly and negatively associated with both event-free and overall survival.
RHAMM expression predicts event-free and overall survival. (A-B) Kaplan-Meier overall and event-free survival distributions for 210 Arkansas patients stratified by high versus low expression of RHAMM (median cutoff) according to Affymetrix U95Av2 microarray experiments. High RHAMM expression is significantly and negatively associated with both event-free and overall survival.
RHAMM isoforms are rarely coexpressed in individual MM plasma cells
Previous examination of RHAMM expression within myeloma revealed the presence of splice variants; RHAMM-exon4 was qualitatively detected at a level approximating RHAMMFL, while detection of RHAMM-exon13 transcripts required more sensitive methodologies.7 Within colon cancer patients, the ratio of RHAMM-exon4/RHAMMFL increases qualitatively in tumor tissue compared with autologous normal tissue.17 Moreover, in AML, CML, and renal cell carcinoma patients, RHAMMFL expression was accompanied with RHAMM-exon4 expression, while RHAMM-exon13 was undetectable.18 We thus investigated the differential expression of RHAMM-exon4 and RHAMMFL within single MM PCs.
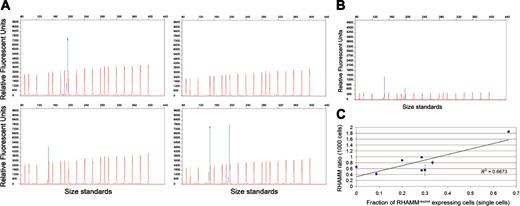
To investigate the differential expression of RHAMM-exon4 and RHAMMFL, we used RT-PCR, with fluoresceinated RHAMM primers, and FCE to detect RHAMM isoform transcripts at the single-cell level (Figure 2A). Optimal PCR conditions were determined to provide reproducible RHAMM ratios from purified PC, PBMC, and BM samples (Figure 2B).
Single-cell analysis of RHAMM isoform expression patterns in MM plasma cells. (A) Electropherogram analysis of sorted, single, clonotypic MM (Patient 8) plasma cell (PC) expressing (clockwise from top left) RHAMMFL alone, neither RHAMMFL nor RHAMM-exon4, both RHAMMFL and RHAMM-exon4, and RHAMM-exon4 alone. All samples were RT-PCR positive, by ethidium bromide detection, for β2-microglobulin and clonotypic message. RT-PCR products were EtOH precipitated prior to GeneScan analysis. To maximize the sensitivity of the single-cell assay, large amounts of RT-PCR product were analyzed, as seen by the large product peaks. (B) Electropherogram analysis of RHAMM-specific RT-PCR amplification of cDNA from 1000 sorted MM (patient 8) plasma cells. For ratio determination, product peaks were kept below 3500 relative fluorescent units (RFU), and size standards were within the manufacturer's suggested levels (ie, 150-600 RFU). (C) Plot of RHAMM ratio (1000 cells) versus the fraction of RHAMM-exon4 single cells. A Pearson R2 correlation coefficient of 0.667 indicates a significant linear relationship (P < .02) between the 2 parameters. Error bars indicate ± 2 standard deviations.
Single-cell analysis of RHAMM isoform expression patterns in MM plasma cells. (A) Electropherogram analysis of sorted, single, clonotypic MM (Patient 8) plasma cell (PC) expressing (clockwise from top left) RHAMMFL alone, neither RHAMMFL nor RHAMM-exon4, both RHAMMFL and RHAMM-exon4, and RHAMM-exon4 alone. All samples were RT-PCR positive, by ethidium bromide detection, for β2-microglobulin and clonotypic message. RT-PCR products were EtOH precipitated prior to GeneScan analysis. To maximize the sensitivity of the single-cell assay, large amounts of RT-PCR product were analyzed, as seen by the large product peaks. (B) Electropherogram analysis of RHAMM-specific RT-PCR amplification of cDNA from 1000 sorted MM (patient 8) plasma cells. For ratio determination, product peaks were kept below 3500 relative fluorescent units (RFU), and size standards were within the manufacturer's suggested levels (ie, 150-600 RFU). (C) Plot of RHAMM ratio (1000 cells) versus the fraction of RHAMM-exon4 single cells. A Pearson R2 correlation coefficient of 0.667 indicates a significant linear relationship (P < .02) between the 2 parameters. Error bars indicate ± 2 standard deviations.
FCE was approximately 5000 times more sensitive than agarose detection, detecting PCR products resulting from amplification of attogram (10-18 g) levels of template. Moreover, FCE could discriminate the RHAMM-exon4 and/or RHAMMFL products resulting from amplification of attogram amounts of template, while agarose detection required 100 femtograms of template for such discrimination. Sequencing of randomly selected RT-PCR products confirmed the identity of the 2 fragments as RHAMMFL and RHAMM-exon4. Consistent with previous reports17 the RHAMM clones (n = 12), sequenced from 3 patients, all exhibited an additional triplet (AAG), compared with the sequence reported by Assmann et al14 (1998) at the 5′ end of exon 4.
CD38+ or CD138+ BM PCs from 8 Alberta MM patients, for whom patient-specific clonotypic IgH primers had been derived, were sorted into PCR tubes at 1 or 1000 cells per well and frozen until analyzed. For each patient, 24 or 48 single-cell RT-PCR reactions were performed depending on available samples (mean, 31.5 cells). For each single-cell sample, cDNA was produced and divided to detect β2-microglobulin (β2M; to control for RNA integrity), clonotypic VDJ (to identify clonal PC), and RHAMM. Table 2 illustrates the findings for single PC cDNA preparations from these 8 patients that were positive for clonal transcripts (mean, 26.8 cells per patient). Interestingly, RHAMMFL and RHAMM-exon4 transcripts are rarely detected within the same clonal MM PCs (mean, 6.1% of PCs; range, 0%-17.4%). Rather, a population of PCs within individual myeloma patients predominantly expresses RHAMMFL, RHAMM-exon4, or neither transcript (Figure 2A). Single and 1000 PC samples were isolated from 3 control patients and evaluated for RHAMM expression. Interestingly, in control samples RHAMM-expressing PCs predominantly express both isoforms. A greater fraction of single PCs (63% vs 43%) lack detectable RHAMM transcripts in the control populations examined; due to low expression levels we were unable to determine RHAMM ratios within purified 1000 PC control samples. We expanded our analysis to include total RNA from unpurified BM of additional control patient (n = 4) samples. Again, expression levels were too low to determine RHAMM ratios from these samples. These results strongly suggest that within populations of control PCs the absolute levels of RHAMM transcripts, although detectable by extremely sensitive FCE at the single-cell level, are so insignificant as to be undetectable at the bulk cell population level.
RHAMM ratios measured in populations of MM PCs reflect the distribution of RHAMM isoforms seen in individual clonal cells
RHAMM ratios were determined for 1000 sorted PCs from the same 8 patients who were examined at the single PC level. As expected from previous data,7,17 RT-PCR of aggregate populations of PCs using exon 4 spanning primers resulted in 2 fluoresceinated peaks (198 and 150 bp) representing both isoforms (Figure 2B). To determine the RHAMM ratios for individual patients, RT-PCR was performed, in triplicate, and the area under the peak for the 150-bp fragment (RHAMM-exon4) was divided by the area under the peak for the 198-bp fragment (RHAMMFL). Of 4 patients whose individual PCs predominantly expressed RHAMMFL, 3 patients (1, 2, and 4) had RHAMM ratios lower than 0.7, while the patient whose individual PCs predominantly expressed RHAMM-exon4 (patient 8) had a dramatically higher RHAMM ratio in the 1000-cell samples (1.8 ± 0.01). There were 2 patients whose individual PCs tended not to express detectable RHAMM (patients 5, 7), as well as 1 RHAMMFL expresser (patient 6), who had moderate RHAMM isoform ratios in 1000-cell samples ranging from 0.8 to 1.0. Thus, there is a strong relationship between the single-cell results and the 1000-cell results in these patients. Regression analysis revealed a significant linear relationship (P < .02) between single PC isoform expression and 1000 PC RHAMM ratio (Figure 2C). From these results we conclude that the RHAMM ratio in aggregate populations of cells, detected semiquantitatively by RT-PCR, provides a valid indicator of the RHAMM isoform expression profile at the single-cell level.
Elevated RHAMM expression, in MM patient PCs, correlates modestly with a relative increase in RHAMM-exon4 expression
We investigated whether elevated RHAMM expression was associated with preferential expression of one isoform (high or low RHAMM ratio). RNA was isolated from purified CD138+ PCs from 14 MM patients, quantitative RT-PCR was performed as described, and ratios were determined. RHAMM expression was normalized to the lowest RHAMM expresser and plotted against the corresponding RHAMM ratio for individual patients. As shown in Figure 3, the patients clustered into 3 groups based upon their RHAMM ratios; 3 patients exhibited ratios less than 0.9, 6 patients exhibited ratios of between 0.9 and 1.2, and 5 patients exhibited ratios more than 1.2. Of 4 of the lowest RHAMM expressers, 3 were clustered in the less than 0.9 group, while 4 of 6 of the highest RHAMM expressers were clustered in the more than 1.2 group. While these 2 parameters demonstrated a statistically significant linear relationship, the degree of correlation was modest (r2 = 0.337, P < .03), and one patient exhibited an elevated RHAMM ratio (1:33) in the absence of elevated absolute RHAMM expression (1.2-fold). The partial correlation of RHAMM expression and splicing results suggests that RHAMM expression and splicing are 2 phenomena that are linked but are also, in part, independently regulated.
Plot of absolute RHAMM expression versus RHAMM ratios for 14 MM patients. Elevated expression of RHAMM is significantly related to an increase in RHAMM-exon4 expression within MM patients. RHAMM expression within purified MM CD138+ PCs was determined by quantitative RT-PCR analysis. Relative RHAMM expression was normalized to the lowest patient expresser. RHAMM ratio (± standard error) was determined as described. Statistical comparison used the Pearson correlation coefficient.
Plot of absolute RHAMM expression versus RHAMM ratios for 14 MM patients. Elevated expression of RHAMM is significantly related to an increase in RHAMM-exon4 expression within MM patients. RHAMM expression within purified MM CD138+ PCs was determined by quantitative RT-PCR analysis. Relative RHAMM expression was normalized to the lowest patient expresser. RHAMM ratio (± standard error) was determined as described. Statistical comparison used the Pearson correlation coefficient.
RHAMM expression, but not exon 4 deletion, is up-regulated during mitosis
Previous experimentation demonstrated that RHAMM expression is up-regulated during mitosis in adherent (HeLa) and suspension (Raji) cell lines (C.A.M., J.J.K., A.R.B., L.M.P., and T.R., submitted April 2004). We investigated whether RHAMM ratios were also altered during mitosis (Table 3).
Interestingly, while both RHAMM message and protein levels were elevated during mitosis, the RHAMM ratio did not change significantly. Within both mitotic and unsynchronized populations of cycling cell lines, the level of RHAMMFL was much greater than RHAMM-exon4. This fits with our previous work suggesting that RHAMM may function in the cross-linking and stabilization of the mitotic spindle, through microtubule contacts at its NH2- and COOH-termini15 ; as RHAMM-exon4 is inhibited in its NH2-terminal interphase microtubule interactions and may not associate/stabilize spindle poles as efficiently as RHAMMFL, amplified expression of this variant during mitosis may have debilitating effects on spindle integrity. Thus, while increases in RHAMM expression levels could reflect in part the mitotic rate in MM PCs, increases in RHAMM exon 4 splicing are not expected to be associated with an increased mitotic rate.
RHAMM ratios in the peripheral blood reflect disease burden over time
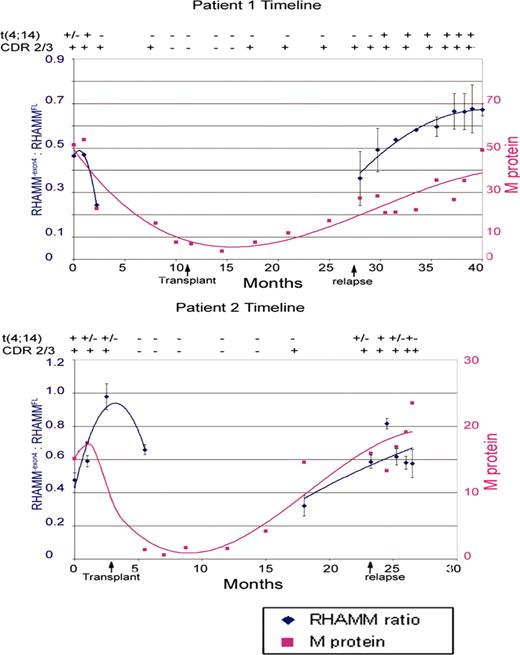
Our lab has previously published the temporal examination of fibroblast growth factor receptor 3 (FGFR3) expression within t(4;14)-positive myeloma patients.6 t(4;14)-positive patients can be identified by RT-PCR amplification of IgH-MMSET fusion transcripts.31 This provides an extremely specific, sensitive assay for the detection of the myeloma cells. To investigate whether the expression of RHAMM-exon4 relative to RHAMMFL changes over the disease course, PBMC samples from sequential clinic visits were examined for 5 MM patients. The use of PBMCs allows examination of multiple time points throughout the course of disease but necessitates detection and differentiation between samples with, or without, transformed cells. PBMC samples from patients, previously identified as t(4;14)+, with sequenced clonotypic transcripts were examined. The advantage of using blood samples from these particular patients is that 3 RT-PCR reactions can detect malignant clones within the blood: amplification of clonal transcript (CDR2/CDR3), Iμ1-MMSET, and JH-MMSET hybrid transcripts. The sensitivity of the single-stage Iμ1-MMSET and the single-stage JH-MMSET assay is 2% positive cells.6 Thus, for all IgH-MMSET+ blood samples, at least 2% of the total PBMC population is malignant. The integrity of each cDNA sample (n = 88) was confirmed with β2-microglobulin. Only the t(4;14) status of the patient was known to the experimenter; the analysis was blind for all other parameters. For each patient, the RHAMM ratio was plotted against time and compared with a standard clinical measure of BM PC burden, the serum M protein level (g/L). RHAMM ratios from samples that were positive for clonotypic transcript and for at least one t(4;14) RT-PCR reaction, as well as the immediate neighboring negative samples, are plotted, and trendlines, generated with Microsoft Excel software (Microsoft, Seattle, WA), are given. Figure 4 illustrates results typical of the patients examined. For samples in which t(4;14) message was detected, an increase in the relative RHAMM-exon4 message was associated with increasing serum M protein levels. For samples lacking detectable IgH-MMSET transcript (not shown), the RHAMM ratio fluctuated from approximately 0.6 to 0.9. As shown in Figure 4, the fluctuations of the RHAMM ratio closely approximate that of the M protein prior to transplantation and following relapse. In 4 of 5 patients examined, clinical relapse coincided with a dramatic increase in RHAMM ratios. These observations illustrate a significant association between RHAMM-exon4 mRNA expression in PBMCs and tumor burden.
Timeline analysis of RHAMM-exon4/RHAMMFL ratios within t(4;14)+ MM patient blood samples. PBMC samples were acquired and archived between 1998 and 2002 as described previously.6 Detection of t(4;14) fusion transcript used 2 RT-PCR reactions targeting first JH/MMSET and then Iμ/MMSET.6 Samples are classified as MMSET negative (-), MMSET positive for both reactions (+), or MMSET positive for one reaction (±). Samples are also classified as containing clonotypic CDR2/3 message (+) or lacking clonotypic CDR2/3 message (-). Only samples that were MMSET positive in at least one reaction are shown along with the immediately neighboring MMSET-negative samples. Polynomial trendlines for RHAMM-exon4/RHAMMFL ratios (Microsoft Excel) are indicated for samples with detectable t(4;14) message. Serum M protein levels at the time of each PBMC sample are also indicated. Arrows indicate time of transplantation and disease relapse.
Timeline analysis of RHAMM-exon4/RHAMMFL ratios within t(4;14)+ MM patient blood samples. PBMC samples were acquired and archived between 1998 and 2002 as described previously.6 Detection of t(4;14) fusion transcript used 2 RT-PCR reactions targeting first JH/MMSET and then Iμ/MMSET.6 Samples are classified as MMSET negative (-), MMSET positive for both reactions (+), or MMSET positive for one reaction (±). Samples are also classified as containing clonotypic CDR2/3 message (+) or lacking clonotypic CDR2/3 message (-). Only samples that were MMSET positive in at least one reaction are shown along with the immediately neighboring MMSET-negative samples. Polynomial trendlines for RHAMM-exon4/RHAMMFL ratios (Microsoft Excel) are indicated for samples with detectable t(4;14) message. Serum M protein levels at the time of each PBMC sample are also indicated. Arrows indicate time of transplantation and disease relapse.
High RHAMM ratios in the diagnostic bone marrow correlate with poor survival in MM
We investigated the prognostic significance of relative RHAMM-exon4 expression in diagnostic bone marrow samples. The RHAMM ratio was measured in total RNA extracts from the bone marrow of 101 MM patients from Alberta, Canada. The fact that RHAMM ratios in control BM were too low to determine RHAMM ratios indicates that the RHAMM ratio in total BM from MM patients is largely contributed by the malignant cells, justifying the use of the available total BM samples for this experiment. These patients are a subset of a previously described cohort.6 The characteristics of the 101 Alberta patients are listed in Table 4.
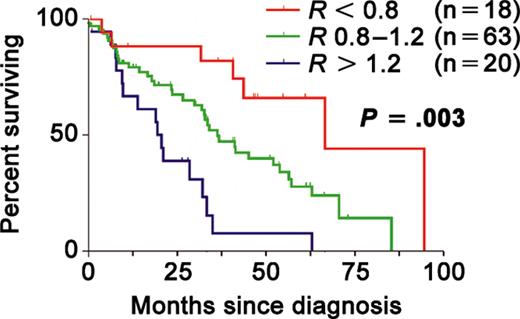
There was no statistically significant correlation between the RHAMM ratio and any baseline characteristic examined (age, sex, clinical isotype, M-protein level, creatinine, calcium, LDH, or β-2 microglobulin). Although patients in this portion of the study were treated heterogeneously, there was no association between RHAMM ratio and the type of therapy received. There was no correlation between the RHAMM ratio and the presence of x-ray-detectable lytic bone lesions; these patients did not undergo MRI scanning. In spite of the correlation between disease burden and the RHAMM ratio seen in our studies of circulating cells, there was no correlation between the BM RHAMM ratio and the level of BM plasmacytosis. Of the 101 patients, 18 had RHAMM ratios less than 0.8. There were 20 patients within the cohort who had ratios more than 1.2. Thus, patients were classified as RHAMMFL expressers (n = 18, R < 0.8), RHAMM-exon4 expressers (n = 20, R > 1.2), and intermediate expressers (n = 63, 0.8 > R < 1.2), analogous to what was seen in the sorted PC experiments. These 3 groups exhibited significantly differing overall survival (OS) distributions (Figure 5), with increasingly poorer outcomes associated with increasingly higher RHAMM ratios (P = .003).
RHAMM isoform balance in BM samples is prognostic in multiple myeloma. Kaplan-Meier survival curves for myeloma patients with RHAMM ratios of less than 0.8 (red), 0.8 to 1.2 (green), and more than 1.2 (blue) in the bone marrow at diagnosis. Survival is increasingly poor with increasing RHAMM ratio (P = .003 by the log-rank test). RHAMM ratios were determined using RT-PCR and capillary fragment analysis.
RHAMM isoform balance in BM samples is prognostic in multiple myeloma. Kaplan-Meier survival curves for myeloma patients with RHAMM ratios of less than 0.8 (red), 0.8 to 1.2 (green), and more than 1.2 (blue) in the bone marrow at diagnosis. Survival is increasingly poor with increasing RHAMM ratio (P = .003 by the log-rank test). RHAMM ratios were determined using RT-PCR and capillary fragment analysis.
In a single-variable Cox model, an increasing RHAMM ratio significantly correlated with poor survival (hazard ratio [HR] = 2.18; 95% CI, 1.11-4.26; P = .024). The RHAMM ratio was an independent predictor of survival when included in a multivariable Cox regression model with albumin and β2M as covariates (HR = 3.34; 95% CI, 1.14-9.79; P = .028). Albumin and β2M are the factors used in the Southwest Oncology Group (SWOG) model,32 a currently accepted standard prognostication method in MM; thus, the RHAMM ratio adds independent information to standard prognostic factors.
Discussion
The clinical impact of RHAMM-exon4 was evaluated in 101 newly diagnosed MM patients by correlating the RHAMM-exon4/RHAMMFL ratio in the diagnostic BM sample to survival. A high RHAMM ratio strongly correlates with poor survival and provides information on prognosis that is independent of standard factors such as β2-microglobulin and albumin. In 5 t(4;14)+ MM patients, we performed time-course analysis of blood samples; we found that in samples containing malignant cells, as assessed with both t(4;14) and clonotypic mRNA assays, the RHAMM ratio decreased after autologous transplantation and increased at the time of relapse. While the RHAMM ratio in circulating malignant cells correlates with tumor burden, a high RHAMM ratio in the BM does not appear to be simply a measure of disease burden, since it does not correlate with the degree of marrow plasmacytosis nor with LDH or β2-microglobulin levels.
Also interesting is the finding that, in 8 patients, RHAMM-exon4 and RHAMMFL are rarely coexpressed within individual malignant MM PCs, despite the fact that both isoforms are present in all patients examined. RHAMM exon 4 splicing appears to be largely an “all or none” phenomenon in individual plasma cells. There is clearly heterogeneity of RHAMM splicing among MM PCs from individual patients, suggesting the presence of aggressive myeloma subclones, characterized by preferential RHAMM-exon4 expression, which may mediate disease progression.
RHAMM expression levels, irrespective of exon 4 splicing, correlate with the presence of bone disease in microarray studies, as well as event-free and overall survival. Osteolytic lesions in myeloma presumably result from a disruption of the bone deposition/resorption balance toward resorption. DKK1 is an inhibitor of osteoblast differentiation that is secreted by myeloma cells in patients with lytic bone disease.30 However, the impact of RHAMM expression in MM cells on this balance of bone growth and bone loss is unclear. Given the lack of correlation between RHAMM and DKK1 expression, and given the lack of any currently known biologic link between the 2 genes, the mechanism underlying the association between RHAMM and osteolytic lesions is in all probability quite distinct from that for DKK1. The simplest explanation for the association between RHAMM levels and disease severity would be that RHAMM expression is intimately linked to myeloma proliferation; elevated RHAMM expression may be analogous to an elevated bromodeoxyuridine plasma cell labeling index (PCLI), a measure of proliferative activity that is an important prognostic factor in newly diagnosed MM.33 However, actively cycling cell lines predominantly express RHAMMFL, as indicated by their reduced RHAMM ratios. Further, within MM patients, elevated RHAMM expression did not correlate with a high PCLI (Table 1). A parallel hypothesis is that altered RHAMM expression may dramatically affect chromosomal segregation and chromosomal instability (CIN) within the malignant clone, analogous to the mitotic errors seen previously in our RHAMM immunoblocking experiments.15 We have previously postulated that RHAMM mitotic function depends upon microtubule cross-linking resulting from microtubule interactions at the NH2- and COOH-termini.15 Antibody targeting of the COOH-terminus of RHAMM disrupts spindle integrity leading to fragmentation and multipolar spindle architecture.15 Elevated expression of RHAMM-exon4, a variant incapable of interacting with interphase microtubules,14,15 may have similar deleterious effects on spindle integrity. Increasing CIN within the malignant clone may have substantial effects on gene expression profiles in malignant “subclones” with dramatic consequential effects on the bone marrow microenvironment.
The association of RHAMM expression and isoform balance with disease relapse and survival is intriguing, particularly the clinical impact of clonal expansion by individual MM plasma cells preferentially expressing RHAMM-exon4. Standard markers of poor prognosis may not have any direct bearing on disease pathogenesis, but are simply markers of tumor burden and/or poor physiologic functioning (eg, serum β2-microglobulin and albumin levels).32 At present, standard prognostic factors in MM are not particularly useful for determining the best treatment for each patient. Newer, molecular prognostic factors, such as RHAMM, t(4;14)6 , and other IgH translocations,34,35 are exciting not just for their prognostic value but because their associations with survival are also linked to a biologically plausible reasoning behind the observed association. It is hoped that such molecular epidemiologic studies as this one will be more clinically useful than older studies of prognosis by helping us to understand the biology of myeloma, and more particularly by identifying novel targets for therapy.
In summary, we speculate that abnormal RHAMM-exon4 splice variant expression in MM promotes mitotic abnormalities, genetic instability, and possibly spread of disease, providing mechanistic insight into the adverse clinical impact of RHAMM-exon4 on survival. We show that coexpression of both isoforms is rare in individual MM plasma cells, and, at the cell population level, elevated expression of RHAMM-exon4 relative to RHAMMFL coincides with disease relapse and is prognostic of poor outcome when assessed at the time of diagnosis. The association of RHAMM expression with lytic bone disease, disease-related events, and survival in microarray experiments provides further evidence of the importance of this gene in myeloma. Further study of the role of RHAMM in myeloma pathogenesis is warranted. There may be value in the development of therapeutic strategies to target RHAMM, with the goal of slowing myeloma disease progression.
Prepublished online as Blood First Edition Paper, April 22, 2004; DOI 10.1182/blood-2003-11-4079.
Supported by National Cancer Institute grants CA80963 (L.M.P., A.R.B.), CA55819 (J.S.), and CA97513 (J.S.); by a grant from the Canadian Institutes of Health Research (CIHR) (L.M.P., T.R., A.R.B.); and through private funding to J.S. L.M.P. is a Canada Research Chair in Biomedical Nanotechnology. C.A.M. is supported by a Natural Sciences and Engineering Research Council of Canada, Alberta Heritage Foundation for Medical Research (AHFMR) and the Department of Oncology PhD Endowed studentships. J.J.K. is supported by CIHR and AHFMR studentships. S.A. is supported by AHFMR, Alberta Cancer Board, and Department of Oncology Multiple Myeloma studentships.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.