Abstract
Chromosomal rearrangements of the 11p15 locus have been identified in hematopoietic malignancies, resulting in translocations involving the N-terminal portion of the nucleoporin gene NUP98. Fifteen different fusion partner genes have been identified for NUP98, and more than one half of these are homeobox transcription factors. By contrast, the NUP98 fusion partner in t(11;20) is Topoisomerase I (TOP1), a catalytic enzyme recognized for its key role in relaxing supercoiled DNA. We now show that retrovirally engineered expression of NUP98-TOP1 in murine bone marrow confers a potent in vitro growth advantage and a block in differentiation in hematopoietic precursors, evidenced by a competitive growth advantage in liquid culture, increased replating efficient of colony-forming cells (CFCs), and a marked increase in spleen colony-forming cell output. Moreover, in a murine bone marrow transplantation model, NUP98-TOP1 expression led to a lethal, transplantable leukemia characterized by extremely high white cell counts, splenomegaly, and mild anemia. Strikingly, a mutation to a TOP1 site to inactivate the isomerase activity essentially left unaltered the growth-promoting and leukemogenic effects of NUP98-TOP1. These findings, together with similar biologic effects reported for NUP98-HOX fusions, suggest unexpected, overlapping functions of NUP98 fusion genes, perhaps related to common DNA binding properties. (Blood. 2004;104:1127-1136)
Introduction
A heterogenous group of hematopoietic malignancies has been described that are characterized by chromosomal translocations that create fusion genes involving the N-terminal portion of the nucleoporin gene, NUP98, on chromosome 11p15.1 To date, 15 distinct fusion partners have been identified in NUP98 translocations. The most frequently observed fusion partners for NUP98 belong to the homeobox family of transcription factors and include HOXA9,2,3 HOXD13,4 HOXD11,5 HOXA11,6 HOXA13,7,8 HOXC11,9 and HOXC1310 as well as the nonclustered homeobox gene PMX1.11 Recent studies in murine model systems have clearly demonstrated that both NUP98-HOXA9 and NUP98-HOXD13 play a causal and overt role in the pathogenesis of leukemia.12,13 Furthermore, the disease onset can be greatly accelerated in these models by coexpression of the NUP98-HOX fusion gene and the HOX co-factor Meis1. These reports are consistent with the observations that HOX genes play key roles in the regulation of hematopoiesis and that overexpression of select HOX genes (eg, HOXA10,14 HOXB315 ) lead to leukemia.
In addition to the HOX genes, 7 “variant” partner genes that are associated with a wide range of biologic functions have been identified in fusion with NUP98 in hematopoietic malignancies. These include DDX10,16,17 RAP1GDS1,18,19 Topoisomerase I,20 LEDGF,21 NSD1,22 NSD3,23 and Adducin 3.24 These NUP98 fusion partner genes are diverse in function and, in contrast to the HOX fusion partners, are not known to have a specific or unique function in hematopoiesis.
An intriguing example of one such non-HOX partner is Topoisomerase I (TOP1), a ubiquitous enzyme identified as the NUP98 fusion partner in t(11;20)(p15;q11).20 The t(11;20) translocation has been described in patients with polycythemia vera, therapy-related myelodysplastic syndrome (t-MDS), and acute myeloid leukemia.25-33 TOP1 is a ubiquitously expressed protein initially recognized for its role in the unwinding or relaxing of supercoiled DNA (reviewed in Pommier et al34 and Wang35,36 ). This activity is dependent on an active site tyrosine located in the C-terminus,37 which forms a phosphodiester bond with the 3′ strand of DNA and generates transient, single-stranded DNA breaks. Through the generation of DNA topologic transformations, TOP1 has been implicated in cellular processes, including DNA replication, transcription, recombination, and chromosome condensation, and has further been targeted clinically by the anticancer agent camptothecin and its derivatives.34 Interestingly, TOP1 has also been reported to function independent of the active site tyrosine and catalytic unwinding activity as a splicing factor kinase,38,39 a transcriptional cofactor,40-42 and a p53 interacting protein.43,44
The contribution of NUP98 to leukemic transformation is largely unknown. Although many genes recurrently rearranged in leukemia encode transcription factors, NUP98 is a ubiquitously expressed member of the nuclear pore complex (NPC) and functions in the nuclear-cytosolic transport of RNA and protein complexes.45 Intriguingly, all NUP98 fusions reported to date retain the FG repeat motif in the N-terminus, known for their role in the docking of import substrates.
In the NUP98-TOP1 fusion protein generated from t(11;20), the N-terminal portion of NUP98 (amino acid 1-514) is fused to most of TOP1 (amino acids 170-765), including the core, linker, and catalytic domains (Figure 1A). Given the seemingly different function of TOP1 compared with HOX genes, we wanted to ascertain whether NUP98-TOP1 as a single agent is sufficient to perturb hematopoietic growth and differentiation. Moreover, by abolishing the active site tyrosine of TOP1 in the fusion protein, we sought to determine whether the topoisomerase activity was required for transformation.
NUP98-TOP1 fusion protein demonstrates nuclear localization. (A) Schematic representation of NUP98, TOP1, and NUP98-TOP1 proteins. Fusion breakpoints are indicated with vertical arrows. Functional domains of NUP98 include FXFG repeats (▨), GLEBS domain (□), ribonucleoprotein (RNP)-binding domain (▥), and nuclear localization signals (NLSs) (▦). TOP1 functional domains comprise N-terminal NLS, core domain (▪), linker (□), and C-terminus ( ). (B) Western blot analysis of total cell lysates from calcium-phosphate-transfected 293T cells detected by anti-GFP antibody. Cells were transfected with GFP-fusion constructs as described in “Materials and methods.” Arrows indicate the size of the expected full-length NUP98-TOP1 protein (150 kDA) and a smaller processed fragment. (C) Fluorescent microscopy images of 293T cells transfected as in panel B. Panels i, iv, vii, and x depict cells stained with 4′, 6-diamidino-2-phenylindole, dilactate (DAPI) for visualization of nuclei; panels ii, v, viii, and xi show visualization of GFP expression; panels iii, vi, ix, and xii show the overlay of DAPI and GFP. Top row (i-iii) results with empty pEGFP-C1 vector, demonstrating pan-cellular GFP expression in nucleus and cytosol. Second row (iv-vi) results with GFP-NUP98-TOP1 fusion, showing that NUP98-TOP1 directs nuclear expression. Third row (vii-ix) GFP-NT-Y723F also exhibits nuclear localization. Bottom row (x-xii) results with GFP-SH2-containing inositol-5-phosphatase (SHIP) used as positive control for cytosolic localization. (D) Retroviral vectors used to express NUP98-TOP1 and NT-Y723F in murine bone marrow. The expected sizes of full-length proviral transcripts are indicated. LTR indicates long terminal repeats; GFP, green fluorescent protein; IRES, internal ribosomal entry site.
). (B) Western blot analysis of total cell lysates from calcium-phosphate-transfected 293T cells detected by anti-GFP antibody. Cells were transfected with GFP-fusion constructs as described in “Materials and methods.” Arrows indicate the size of the expected full-length NUP98-TOP1 protein (150 kDA) and a smaller processed fragment. (C) Fluorescent microscopy images of 293T cells transfected as in panel B. Panels i, iv, vii, and x depict cells stained with 4′, 6-diamidino-2-phenylindole, dilactate (DAPI) for visualization of nuclei; panels ii, v, viii, and xi show visualization of GFP expression; panels iii, vi, ix, and xii show the overlay of DAPI and GFP. Top row (i-iii) results with empty pEGFP-C1 vector, demonstrating pan-cellular GFP expression in nucleus and cytosol. Second row (iv-vi) results with GFP-NUP98-TOP1 fusion, showing that NUP98-TOP1 directs nuclear expression. Third row (vii-ix) GFP-NT-Y723F also exhibits nuclear localization. Bottom row (x-xii) results with GFP-SH2-containing inositol-5-phosphatase (SHIP) used as positive control for cytosolic localization. (D) Retroviral vectors used to express NUP98-TOP1 and NT-Y723F in murine bone marrow. The expected sizes of full-length proviral transcripts are indicated. LTR indicates long terminal repeats; GFP, green fluorescent protein; IRES, internal ribosomal entry site.
NUP98-TOP1 fusion protein demonstrates nuclear localization. (A) Schematic representation of NUP98, TOP1, and NUP98-TOP1 proteins. Fusion breakpoints are indicated with vertical arrows. Functional domains of NUP98 include FXFG repeats (▨), GLEBS domain (□), ribonucleoprotein (RNP)-binding domain (▥), and nuclear localization signals (NLSs) (▦). TOP1 functional domains comprise N-terminal NLS, core domain (▪), linker (□), and C-terminus ( ). (B) Western blot analysis of total cell lysates from calcium-phosphate-transfected 293T cells detected by anti-GFP antibody. Cells were transfected with GFP-fusion constructs as described in “Materials and methods.” Arrows indicate the size of the expected full-length NUP98-TOP1 protein (150 kDA) and a smaller processed fragment. (C) Fluorescent microscopy images of 293T cells transfected as in panel B. Panels i, iv, vii, and x depict cells stained with 4′, 6-diamidino-2-phenylindole, dilactate (DAPI) for visualization of nuclei; panels ii, v, viii, and xi show visualization of GFP expression; panels iii, vi, ix, and xii show the overlay of DAPI and GFP. Top row (i-iii) results with empty pEGFP-C1 vector, demonstrating pan-cellular GFP expression in nucleus and cytosol. Second row (iv-vi) results with GFP-NUP98-TOP1 fusion, showing that NUP98-TOP1 directs nuclear expression. Third row (vii-ix) GFP-NT-Y723F also exhibits nuclear localization. Bottom row (x-xii) results with GFP-SH2-containing inositol-5-phosphatase (SHIP) used as positive control for cytosolic localization. (D) Retroviral vectors used to express NUP98-TOP1 and NT-Y723F in murine bone marrow. The expected sizes of full-length proviral transcripts are indicated. LTR indicates long terminal repeats; GFP, green fluorescent protein; IRES, internal ribosomal entry site.
). (B) Western blot analysis of total cell lysates from calcium-phosphate-transfected 293T cells detected by anti-GFP antibody. Cells were transfected with GFP-fusion constructs as described in “Materials and methods.” Arrows indicate the size of the expected full-length NUP98-TOP1 protein (150 kDA) and a smaller processed fragment. (C) Fluorescent microscopy images of 293T cells transfected as in panel B. Panels i, iv, vii, and x depict cells stained with 4′, 6-diamidino-2-phenylindole, dilactate (DAPI) for visualization of nuclei; panels ii, v, viii, and xi show visualization of GFP expression; panels iii, vi, ix, and xii show the overlay of DAPI and GFP. Top row (i-iii) results with empty pEGFP-C1 vector, demonstrating pan-cellular GFP expression in nucleus and cytosol. Second row (iv-vi) results with GFP-NUP98-TOP1 fusion, showing that NUP98-TOP1 directs nuclear expression. Third row (vii-ix) GFP-NT-Y723F also exhibits nuclear localization. Bottom row (x-xii) results with GFP-SH2-containing inositol-5-phosphatase (SHIP) used as positive control for cytosolic localization. (D) Retroviral vectors used to express NUP98-TOP1 and NT-Y723F in murine bone marrow. The expected sizes of full-length proviral transcripts are indicated. LTR indicates long terminal repeats; GFP, green fluorescent protein; IRES, internal ribosomal entry site.
Materials and methods
Mice
Mice were bred and maintained in micro-isolator cages in the animal care facility of the British Columbia Cancer Research Centre. Recipients of bone marrow cells were (C57Bl/6J × C3H/HeJ)F1 ((B6C3)F1) and donor mice were (C57Bl/6Ly-Pep3b × C3H/HeJ)F1 ((PepC3)F1). For bone marrow transplantation experiments, recipient mice were lethally irradiated with 950 cGy from a 137Cs source prior to intravenous injection with 5 × 105 transduced donor bone marrow cells.
Retroviral constructs and GFP expression vectors
The NUP98-TOP1 cDNA was composed of nucleotides 33 to 1686 of the NUP98 reference sequence (NM 139132.1) fused in frame to nucleotides 828 to 2580 of the TOP1 reference sequence (NM 003286.2). This cDNA was cloned as an 3.3-kb EcoRI fragment upstream of an IRES-GFP-linked cassette in an murine stem cell virus (MSCV)-derived retroviral vector46 using standard procedures. An MSCV vector (MIG) carrying the IRES-GFP cassette only was used as a control (GFP CTL) as previously described.13 To generate a flag-tagged version of NUP98-TOP1, the flag sequence within the pSuperCatch plasmid (Clontech, Palo Alto, CA) was excised as a HindIII-XhoI fragment and cloned into the HindIII site of pBlueScript vector (Stratagene, La Jolla, CA) in which the BstXI-EcoRV fragment was previously removed (pBS-flag). With the use of primers to NUP98-TOP1 (forward (Fwd), 5′-AGA CTC ATT TTG GGA TCC TT AAC AAA-3′; and reverse (Rev), 5′-AGA ACT CTG CCT CTC GAG ACT-3′) the NUP98-TOP1 cDNA was inserted as a BamHI-XhoI fragment into pBS-flag (herein referred to as pBS-flag-NT), excised by SstI-XhoI and ligated into the MIG vector. The catalytic mutant of NUP98-TOP1 engineered to abolish the TOP1 active site (NT-Y723F), was constructed by using the QuikChange Site Directed Mutagenesis Kit (Stratagene), and primers designed to convert amino acid 723 of TOP1 from tyrosine to phenylalanine (Fwd, 5′-ACC TCC AAA CTC AAT TTT CTG GAC CCT AGG ATC-3′; Rev, GA GGT-3′). The NUP98 5′ and TOP1 3′ portions of the fusion and NT ΔC-term mutant were amplified by polymerase chain reaction (PCR) from pBS-flag-NT as BamHI-XhoI fragments using the primers (NUP98 5′; Fwd, M13 fwd; and Rev, 5′-GGG TTT TTT CAA CTC GAG TTA CTC TTC CTT-3′; TOP1 3′; Fwd, 5′-CCT AAG AAG AAG GGA TCC GAT GGT-3′; and Rev, M13 rev, and NT ΔC-term; Fwd, M13 fwd; Rev, 5′-TGT CAG CTC GAG TAG CTA CTG CTG TAG-3′). The NT-ΔCD mutant was amplified as 2 fragments from pBC-flag-NT (NT CD 5′ Fwd, M13F; Rev, 5′-TAG GAA TTT CCA CTT GGC GCC TTC-3′; and NT CD 3′ Fwd, 5′-TGT AAC CAT CAG AGG GGA GAA CCA-3′; Rev, M13 rvs) digested with BamHI-NarI or Nar1-XhoI, respectively, and ligated into MIG. The MSCV-IRES-YFP-Meis1 construct was as described in Pineault et al.13
For fluorescent microscopy visualization, the 3.3-kb NUP98-TOP1 cDNA insert (or mutant forms as described in this section) was excised as a BamHI-XhoI fragment from Pbs-flag and ligated in-frame (BglII-SalI) to the C-terminus of enhanced GFP (EGFP) in the pEGFP-C1 vector (Clontech).
Generation of retrovirus
Infection of primary murine bone marrow cells
Bone marrow (BM) cells were harvested from mice treated 4 days prior with 150 mg/kg 5-fluorouracil (Pharmacia and Upjohn, Mississauga, ON, Canada) and prestimulated for 48 hours in Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (FBS) in the presence of 6 ng/mL murine interleukin 3 (mIL-3), 10 ng/mL human interleukin 6 (hIL-6), and 100 ng/mL murine stem cell factor (mSCF). BM cells were transduced by cocultivation for an additional 48 hours with irradiated (1500 cGy X-ray) GP + E86 viral producer cells in the presence of 5 μg/mL protamine sulfate (Sigma, Oakville ON, Canada). Loosely adherent and nonadherent cells were recovered and injected immediately into the tail vein of recipient mice, used for in vitro assays as described in the next section, or cultured for an additional 48 hours to allow expression of GFP. All media components, including growth factors, were from StemCell Technologies Inc (STI), Vancouver, BC, Canada.
In vitro liquid cultures and clonogenic progenitor assays
For liquid culture assays, 1 × 105 cells were placed in DMEM supplemented with serum and cytokines as described in “Infection of primary murine bone marrow cells.” Every few days cultures were harvested, total cell numbers were evaluated, the proportion of GFP-expressing cells was measured by flow cytometric analysis (FACSort; Becton Dickinson, Mississauga, ON, Canada), and cells were plated back into culture at a density less than 2 × 105 cells/mL. Colony-forming cells (CFCs) were assayed in methylcellulose (Methocult M3234; STI) supplemented with 10 ng/mL mIL-3, 10 ng/mL hIL-6, 50 ng/mL mSC,F and 3 U/mL erythropoietin (EPO). Colonies were scored microscopically 9 to 11 days after plating by using standard criteria.
Spleen colony-forming cell assay
Transduced (GFP+) donor BM cells were isolated 48 hours after infection by FACS (FACS Vantage; Becton Dickinson), and their ability to form spleen colonies (day 0) was measured by transplanting 5000 GFP+ cells per lethally irradiated recipient mice. Spleen colony-forming cell numbers following 8 days of culture were determined by injecting the progeny of 250 to 2 × 105 input (day 0) cells. Between 4 and 6 mice were injected at each cell dose. Macroscopic spleen colonies were counted 12 days after injection following fixation in Telleyesnickzky solution.50
DNA and RNA analysis
Southern blot analysis was used to assess the presence and number of proviral integrations using standard techniques. Genomic DNA was isolated with DNAzol (Invitrogen, Burlington ON, Canada) and digested with XbaI, which cuts in the long terminal repeats (LTRs) to release the provirus or digested with EcoR1 (GFP ctl) or BglII (NUP98-TOP1 and NT-Y723F) to assess the presence of different clones.
Total cellular RNA was isolated by using Trizol (Invitrogen) and analyzed by Northern blot with use of standard techniques. Membranes were probed with a 731-base pair PCR-generated fragment of pEGFP-C1 vector (Clontech) labeled with p32 deoxycytidine triphosphate (dCTP).
Flow cytometry
Flow cytometric analysis was performed by using a FACsort. The red cells in peripheral blood (PB) and single-cell suspensions of BM or spleen were lysed with ammonium chloride. Recovered nucleated cells were rinsed then incubated on ice with primary phycoerythrin (PE)-labeled antibodies (Gr-1, Mac-1, B220, CD4/CD8, Ly5.1, Ter119) purchased from Pharmingen, San Diego, CA.
Cytospins and PB smears
Morphologic analysis of PB, BM, and spleen cells was carried out on modified Wright-Giemsa-stained smears and cytospin preparations.
Cell transfections and Western blot analysis
NUP98-TOP1 was expressed in 293T cells by calcium-phosphate transfection (CellPhect Transfection Kit; Amersham Biosciences, Buckinghamshire, England). Whole-cell lysates were solubilized 48 hours after transfection with sodium dodecyl sulfate (SDS) sample buffer, electrophoresed on 8% SDS-polyacrylamide gel electrophoresis (PAGE) gel and blotted to polyvinylidene diflouride (PVDF) membranes (Pall Corporation, Ann Arbor MI). Membranes were probed with an anti-GFP monoclonal antibody (Roche Diagnostics, Indianapolis, IN) and donkey horseradish peroxidase (HRP)-conjugated antimouse antibody (Jackson ImmunoResearch Lab, West Grove, PA). Protein expression was detected with Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer, Boston, MA).
To assess the subcellular localization of NUP98-TOP1, 293T cells were grown on poly-L lysine (Sigma) treated coverslips and transfected as described earlier. Cells were fixed with 4% paraformaldehyde and stained with 2 ng/mL DAPI. Cells were visualized with a × 63, 1.4 Plan-Apochromat oil immersion lens using a Zeiss axioplan microscope (Thornwood, NY). Images were captured with a CCD camera and Applied Imaging software (Santa Clara, CA).
Statistical analysis
Data were analyzed statistically using the Student t test (Microsoft Excel; Microsoft, Redman, WA). Differences of P values less than .05 were considered statistically significant.
Results
NUP98-TOP1 enhances the expansion of hematopoietic cells in vitro
The NUP98-TOP1 cDNA used in this study represents the fusion gene found in the t-MDS/AML (acute myeloid leukemia)-associated t(11;20)(p15;q11).20 The integrity of the cDNA was confirmed by DNA sequencing and by transfection of NUP98-TOP1 as a GFP-C-terminal fusion into 293T cells (Figure 1B). Fluorescent microscopy analysis of these cells revealed a primarily nuclear localization for GFP-NUP98-TOP1, in contrast to GFP alone or a fusion between GFP and the signaling molecule SHIP used as a control for pancellular and cytosolic localization, respectively, (Figure 1C).
To study the effect of NUP98-TOP1 expression on hematopoietic growth and differentiation, the fusion gene was introduced into primary murine BM with use of an MSCV-based retrovirus carrying the NUP98-TOP1 cDNA upstream of an IRES-linked GFP selectable marker (Figure 1D). Immediately following retroviral infection and without any preselection, BM cells transduced with GFP control or NUP98-TOP1 were plated in liquid culture suspensions supplemented with mIL-3, hIL-6, and mSCF. As depicted in Figure 2A, the proportion of GFP+ cells in the control cultures remained similar to input levels following extended culture. In contrast, NUP98-TOP1 GFP+ cells comprising ∼1% of the population at the time of infection, strongly out-competed the growth of nontransduced cells, expanding to more than 80% of the culture by 28 days. This was a cell autonomous effect evidenced by a decreased doubling time for NUP98-TOP1 GFP+-transduced cells, whereas the nontransduced (GFP-) cells in the NUP98-TOP1 culture cells had a doubling time similar to the GFP+ and GFP- populations in the control culture (Figure 2B). Following 4 weeks of culture, NUP98-TOP1 cells displayed a blastlike morphology on cytospin preparations (Figure 2C), suggesting that NUP98-TOP1 induces a block in differentiation. Metaphase spreads were prepared from control uninfected, NUP98-HOXD13-, and NUP98-TOP1-infected BM cells cultured for more than 4 weeks. No abnormalities in chromosome number or gross chromosomal rearrangements were observed, suggesting that expression of NUP98-TOP1 does not induce obvious genomic instability (data not shown).
Expression of NUP98-TOP1 in murine bone marrow confers in vitro proliferative advantage. (A) NUP98-TOP1 expression results in expansion of transduced (GFP+) BM cells in liquid cultures. Graph is representative of 3 independent experiments. (B) Expression of NUP98-TOP1 leads to decreased doubling time (Td) calculated from the slope of the curve representing change in cell number over time. (C) Representative Wright-Giemsa-stained cytospin preparation of GFP cytotoxic T lymphocyte (CTL; left) or NUP98-TOP1 (right) transduced bone marrow cultured for 4 weeks (original magnification, × 600).
Expression of NUP98-TOP1 in murine bone marrow confers in vitro proliferative advantage. (A) NUP98-TOP1 expression results in expansion of transduced (GFP+) BM cells in liquid cultures. Graph is representative of 3 independent experiments. (B) Expression of NUP98-TOP1 leads to decreased doubling time (Td) calculated from the slope of the curve representing change in cell number over time. (C) Representative Wright-Giemsa-stained cytospin preparation of GFP cytotoxic T lymphocyte (CTL; left) or NUP98-TOP1 (right) transduced bone marrow cultured for 4 weeks (original magnification, × 600).
The proliferative effect of NUP98-TOP1 was further evident in hematopoietic precursors assayed for colony formation in methylcellulose. No statistically significant differences were observed in CFC numbers or colony type between control and NUP98-TOP1 GFP+-purified BM cells plated immediately following sorting. However, on secondary replating, NUP98-TOP1 cells gave rise to large granulo-monocytic (GM) colonies (1394.5 ± 427 secondary colonies per primary colony; ∼5% of plated cells harvested from the primary culture gave rise to secondary colonies), in contrast to mast cell colonies derived from GFP control cells (data not shown).
NUP98-TOP1 induces a competitive myeloid growth advantage in vivo
To assess the effects of NUP98-TOP1 expression in vivo, mice were transplanted with 5 × 105 unselected BM cells 48 hours after infection. The initial proportion of GFP+ cells in the transplant inoculum ranged from 7% to 55% for GFP control and ∼1% to 4% for NUP98-TOP1 in 4 independent experiments. In mice transplanted with GFP control-transduced BM, the proportion of GFP+ cells in the PB remained relatively constant over time (Figure 3A). In contrast, analysis of mice transplanted with NUP98-TOP1-transduced cells revealed a large increase in the proportion of GFP+ PB cells, expanding from an initial transplant frequency of less than 4% to more than 40% by 4 weeks and rising ultimately to more than 80% at 24 weeks after transplantation.
NUP98-TOP1 expression leads to in vivo myeloproliferation. (A) NUP98-TOP1 leads to in vivo proliferative advantage as assessed by increased peripheral blood GFP+ content at various times after transplantation. Graph is representative of 4 independent experiments with at least 4 mice in each group (mean ± SD; GFP CTL [□]; NUP98-TOP1 [▪]). (B) NUP98-TOP1 leads to a progressive increase in PB white cell counts. Results are shown for individual mice in a representative experiment. Median values are depicted with a horizontal line. Open symbols indicate GFP CTL mice; filled symbols, NUP98-TOP1 mice. (C) Differential counts performed on Wright-Giemsa-stained PB smears of mice at 2 (top) and 6 (bottom) months after transplantation. Counts are the average of 3 or 5 CTL GFP or NUP98-TOP1 mice, respectively. ▧ indicates lymphoid cells; ▪, myeloid cells. (D) Representative experiment depicting PB of 5 mice stained with antibodies to Mac-1 and B220 at 2 months (top panel) and 6 months (bottom panel) after transplantation. ▪ represents transduced (GFP+) cells; ▧, GFP-. Mice 1 to 5 are GFP CTL; mice 6 to 10 are NUP98-TOP1. (E) Representative Wright-Giemsa-stained PB smear of NUP98-TOP1 mouse at 6 months after transplantation. Original magnification, × 1000. See Figure 5B for control.
NUP98-TOP1 expression leads to in vivo myeloproliferation. (A) NUP98-TOP1 leads to in vivo proliferative advantage as assessed by increased peripheral blood GFP+ content at various times after transplantation. Graph is representative of 4 independent experiments with at least 4 mice in each group (mean ± SD; GFP CTL [□]; NUP98-TOP1 [▪]). (B) NUP98-TOP1 leads to a progressive increase in PB white cell counts. Results are shown for individual mice in a representative experiment. Median values are depicted with a horizontal line. Open symbols indicate GFP CTL mice; filled symbols, NUP98-TOP1 mice. (C) Differential counts performed on Wright-Giemsa-stained PB smears of mice at 2 (top) and 6 (bottom) months after transplantation. Counts are the average of 3 or 5 CTL GFP or NUP98-TOP1 mice, respectively. ▧ indicates lymphoid cells; ▪, myeloid cells. (D) Representative experiment depicting PB of 5 mice stained with antibodies to Mac-1 and B220 at 2 months (top panel) and 6 months (bottom panel) after transplantation. ▪ represents transduced (GFP+) cells; ▧, GFP-. Mice 1 to 5 are GFP CTL; mice 6 to 10 are NUP98-TOP1. (E) Representative Wright-Giemsa-stained PB smear of NUP98-TOP1 mouse at 6 months after transplantation. Original magnification, × 1000. See Figure 5B for control.
This increase in the proportion of GFP+ PB cells in NUP98-TOP1 mice was associated with a progressive increase in the number of circulating white blood cells (Figure 3B). As early as 2 months after transplantation, NUP98-TOP1 mice displayed a significant increase (P < .05) in the average number of circulating white blood cells (mean, 15.8 × 106 cells/mL; range, 6.3-30 × 106 cells/mL) compared with control GFP mice (mean, 7.0 × 106 cells/mL; range, 3.1-8.6 × 106 cells/mL). By 6 months after transplantation, the white blood cell (WBC) numbers in NUP98-TOP1 mice increased further, reaching an average of 75.6 × 106 cells/mL (range, 9.1-201 × 106 cells/mL).
Differential counts performed on Wright-Giemsa-stained peripheral blood smears revealed that NUP98-TOP1 induced a significant increase in the proportion of mature myeloid cells by 2 months after transplantation (59.2% ± 15.2% versus 27.8% ± 7.5% in control, P < .005), which was further exacerbated at 6 months (83.5% ± 6.8% versus 21.9% ± 6.1% in control, P < .005) (Figure 3C). Morphologically, the PB cells were predominantly segmented neutrophils consistent with a myeloproliferative-like disorder (Figure 3E). Flow cytometric analysis of PB with lineagespecific markers confirmed that NUP98-TOP1 expression led to an increase in the proportion of transduced (GFP+) myeloid cells (Mac-1+ and Gr-1+), which resulted in an increase in the absolute numbers of circulating myeloid cells (Figure 3D and data not shown). Although the proportion of GFP+-transduced B cells (B220+) and T cells (CD4+/CD8+) in the NUP98-TOP1 mice were significantly reduced compared with control, the absolute numbers of circulating lymphoid cells were comparable to control (P = .06) (Figure 3D). Moreover, although some NUP98-TOP1 mice showed increased numbers of GFP+ B220+ circulating cells, this population of cells exhibited a different profile (B220lo) on FACS analysis and were further determined to be Mac-1+/B220+.
The NUP98-TOP1 mice became progressively anemic with an average red cell count of 2.20 ± 3.4 × 109 cell/mL at 6 months (P < .002) (Table 1). In 4 independent experiments, 13 of 17 mice did not show any transduced (GFP+) red cells, whereas 4 mice had red blood cells (RBCs) that were equal in GFP content to that observed in the white cells. These observations provide evidence that in vivo NUP98-TOP1 induces a strong proliferative advantage to nonerythroid myeloid cells.
NUP98-TOP1 induces a lethal myeloid leukemia
Ultimately, all NUP98-TOP1 mice (n = 17) became moribund and were killed or died with a median survival of 233.5 days (range, 149-321 days) after transplantation (Figure 4A). Southern and Northern blot analyses of diseased mice revealed full-length NUP98-TOP1 proviral integration and expression (Figure 4B-C). The disease was monoclonal or oligoclonal in nature (Figure 4D); however, this may be due, in part, to the initial low gene transfer efficiency.
NUP98-TOP1 expression induces a lethal leukemia. (A) Survival curve for mice that received transplants with CTL GFP-transduced cells (dashed line, n = 10) or NUP98-TOP1-transduced cells (solid line, n = 17, average latency 225 ± 56 days). Secondary recipients transplanted with 102 to 106 primary BM cells succumbed to disease with reduced latency (42-147 days). (B) Southern blot analysis of genomic DNA from primary mice reveals full-length proviral integration. DNA was digested with XbaI, which cuts once in each LTR. (C) Northern blot analysis of total RNA from NUP98-TOP1 mice. (D) Southern blot analysis reveals that NUP98-TOP1 induces monoclonal or oligoclonal disease, which is transplantable to secondary recipients. The labeling indicates the experiment number and the mouse used to transplant secondary recipients. Genomic DNA from moribund mice was digested with BglII (NUP98-TOP1) or EcoRI (NT-Y723F), which cut once in the provirus sequence. Membranes were hybridized with a probe specific to GFP. In panel D, 1° indicates primary; 2°, secondary.
NUP98-TOP1 expression induces a lethal leukemia. (A) Survival curve for mice that received transplants with CTL GFP-transduced cells (dashed line, n = 10) or NUP98-TOP1-transduced cells (solid line, n = 17, average latency 225 ± 56 days). Secondary recipients transplanted with 102 to 106 primary BM cells succumbed to disease with reduced latency (42-147 days). (B) Southern blot analysis of genomic DNA from primary mice reveals full-length proviral integration. DNA was digested with XbaI, which cuts once in each LTR. (C) Northern blot analysis of total RNA from NUP98-TOP1 mice. (D) Southern blot analysis reveals that NUP98-TOP1 induces monoclonal or oligoclonal disease, which is transplantable to secondary recipients. The labeling indicates the experiment number and the mouse used to transplant secondary recipients. Genomic DNA from moribund mice was digested with BglII (NUP98-TOP1) or EcoRI (NT-Y723F), which cut once in the provirus sequence. Membranes were hybridized with a probe specific to GFP. In panel D, 1° indicates primary; 2°, secondary.
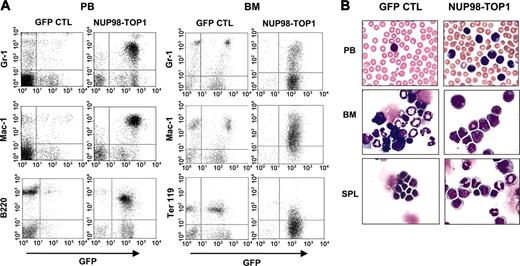
Flow cytometric analysis of moribund NUP98-TOP1 mice demonstrated an increased proportion of myeloid cells (Gr-1+, Mac-1+) in the PB, BM, and spleen and a reduction in Ter119+ BM cells (Figure 5A). Interestingly, several mice (n = 5) had BM cells that were more than 90% for B220; however, these cells exhibited a lower fluorescent intensity than control B220+ cells, suggesting they are of a different population, and we thus describe them as B220lo. Many of the NUP98-TOP1 PB, BM, and spleen cells were morphologically “immature forms/blasts,” consistent with the diagnosis of a myeloid leukemia51 (Figure 5B). The BM from NUP98-TOP1 mice showed greatly increased numbers of both CFC (20-fold; P < .004) and spleen colony-forming cells (5-fold increased; P < .03) compared with controls, and the mice had marked splenomegaly (0.67 ±0.24 g) (Table 1).
Immunophenotype of hematopoietic cells from NUP98-TOP1 mice. (A) Representative FACS profiles of PB (left panels) and BM (right panels) from a GFP control and diseased NUP98-TOP1 mouse. (B) Wright-Giemsa-stained PB smear, BM, and spleen cytospin (original magnification, × 600).
Immunophenotype of hematopoietic cells from NUP98-TOP1 mice. (A) Representative FACS profiles of PB (left panels) and BM (right panels) from a GFP control and diseased NUP98-TOP1 mouse. (B) Wright-Giemsa-stained PB smear, BM, and spleen cytospin (original magnification, × 600).
NUP98-TOP1 leukemia is transplantable and decreases the latency period
The NUP98-TOP1-induced leukemia was transplantable, with all secondary recipients showing greatly elevated numbers of circulating GFP+ nucleated cells having an immature myeloid/monocytic blastlike morphology. The disease course in secondary animals was accelerated compared with primary recipients (Figure 4A), with a maximum survival of less than 77 days and less than 147 days with a transplantation dose of 1 × 105 or 1 × 103 BM cells, respectively (Table 2). Southern blot analysis confirmed full-length proviral transmission in secondary recipients and revealed that the same clone present in primary donors was present and un-rearranged in secondary animals (Figure 4D). Moreover, transplanting as few as 100 BM cells from a diseased primary animal along with 5 × 105 helper cells was sufficient to induce a lethal disease in secondary recipients within 88 days after transplantation, demonstrating that the frequency of the “leukemia-propagating cell” is more than 1 in 100. The aggressiveness of the secondary disease was further evident as transplantation of 1 × 106 BM cells from a leukemic primary mouse into nonmyeloablated secondary recipients was sufficient to induce a lethal disease within 85 days after transplantation. These observations indicate that NUP98-TOP1 is sufficient to induce a lethal, transplantable myeloid leukemia.
NUP98-TOP1 induces a lethal leukemia in the presence of a mutation to the TOP1 catalytic active site
A series of NUP98-TOP1 deletion mutants was tested in vitro in an attempt to functionally dissect the contributing domains of TOP1 to the leukemogenesis. Protein expression of all mutants expressed as GFP fusion constructs was confirmed by Western blot analysis and fluorescent microscopy (data not shown). Neither the NUP98 (NUP98 5′) portion nor the TOP1 (TOP1 3′) portion of the fusion alone was sufficient to induce the growth advantage observed for NUP98-TOP1 in liquid culture assays, demonstrating that each partner contributes an essential function required for leukemogenesis (Figure 6C-D).
NT-Y723F behaves similarly to wild-type NUP98-TOP1 to induce an in vitro and in vivo proliferative advantage. (A) NT-Y723F induces an increase in the proportion of transduced (GFP+) BM cells in liquid cultures similar to that observed for NUP98-TOP1. Graph is representative of 2 independent experiments. Values are mean ± SD of single experiment plated in triplicate. Inset, cytospin preparation taken at day 28. Inset magnification, × 100. (B) Frequency of day 12 spleen colonies obtained immediately after FACS sorting (day 0; left) or following 8 days of liquid culture (day 8; right) (mean ± SD; n > 2). Inset, representative pictures of day 12 spleens following injection of transduced cells cultured for 8 days. GFP CTL spleen was derived from a culture initiated with 2 × 104 cells; in contrast both NUP98-TOP1 and NT-Y723F gave rise to enlarged spleen from only 500 input cells. (C) Schematic representation of NUP98-TOP1 mutants used in this study. (D) Fold increase in the proportion of GFP+ cells in liquid culture measured 4 weeks after culture initiation. Representative graph of n = 2 independent experiments with each sample plated in triplicate (Avg ± SD).
NT-Y723F behaves similarly to wild-type NUP98-TOP1 to induce an in vitro and in vivo proliferative advantage. (A) NT-Y723F induces an increase in the proportion of transduced (GFP+) BM cells in liquid cultures similar to that observed for NUP98-TOP1. Graph is representative of 2 independent experiments. Values are mean ± SD of single experiment plated in triplicate. Inset, cytospin preparation taken at day 28. Inset magnification, × 100. (B) Frequency of day 12 spleen colonies obtained immediately after FACS sorting (day 0; left) or following 8 days of liquid culture (day 8; right) (mean ± SD; n > 2). Inset, representative pictures of day 12 spleens following injection of transduced cells cultured for 8 days. GFP CTL spleen was derived from a culture initiated with 2 × 104 cells; in contrast both NUP98-TOP1 and NT-Y723F gave rise to enlarged spleen from only 500 input cells. (C) Schematic representation of NUP98-TOP1 mutants used in this study. (D) Fold increase in the proportion of GFP+ cells in liquid culture measured 4 weeks after culture initiation. Representative graph of n = 2 independent experiments with each sample plated in triplicate (Avg ± SD).
The most well-characterized function of TOP1 is the unwinding of supercoiled DNA. It has been well documented that mutating Tyr723 to phenylalanine (Y723F) abrogates the catalytic ability of TOP1, rendering it unable to relax supercoiled DNA.37 To test whether the myeloproliferative effects of NUP98-TOP1 were dependent on TOP1 isomerase activity, in vitro mutagenesis was used to change the corresponding tyrosine residue in the NUP98-TOP1 fusion to phenylalanine, herein called NT-Y723F.
Surprisingly, NT-Y723F had the same growth-promoting activity as NUP98-TOP1 in liquid culture assays, out-competing the growth of nontransduced cells and increasing the proportion of GFP+ cells 82.5-± 4.0-fold above input over 38 days (Figure 6A). Both NUP98-TOP1 and NT-Y723F resulted in a dramatic increase in the yield of spleen colony-forming cell output after 8 days in culture, increasing some 210- and 250-fold, respectively, compared with GFP control (Figure 6B). Transplanting the progeny of as few as 500 NUP98-TOP1 or NT-Y723F cells cultured for 8 days gave rise to a massive enlargement in spleen size 12 days after transplantation.
Following transplantation, NT-Y723F-transduced BM behaved essentially identical to NUP98-TOP1-transduced BM, yielding an increased proportion of GFP+ myeloid PB cells, elevated white cells counts (120 ± 77 × 106), and mild anemia (6.0 ± 1.0 × 109 cells/mL) at 237 days after transplantation (Table 1). NT-Y723F mice became moribund with a median survival of 270.5 days (n = 4) (Table 3), and the disease was transplantable to secondary recipients with kinetics essentially identical to that observed for NUP98-TOP1 (Table 2). Together, these data strongly argue that the leukemia resulting from NUP98-TOP1 expression occurs independent of TOP1 catalytic activity.
NUP98-TOP1 fusion exhibits both overlapping and distinct properties compared with NUP98-HOX fusions
It has been hypothesized that NUP98-HOX fusions act as aberrant transcription factors, evidenced by the concomitant loss of in vitro proliferative advantage with mutation to the DNA binding home-odomain.13 TOP1 binds to DNA like a clamp with portions of the core domain and C-terminal domain, forming the upper and lower halves, respectively.52,53 To ascertain whether these DNA binding domains are necessary for NUP98-TOP1 transformation, mutant constructs were engineered that deleted either the TOP1 core domain (NT ΔCD) or the C-terminal domain (NTΔC-term). Both mutants lost their in vitro growth-promoting activity as assayed in competitive liquid culture assays (Figure 6C-D) or spleen colony-forming cell expansion assays (data not shown).
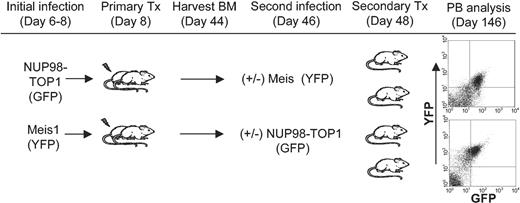
To further ascertain whether NUP98-TOP1 has some overlapping mechanisms with NUP98-HOX fusions, a bone marrow transplantation experiment was performed by using BM coexpressing Meis1 (linked to yellow fluorescent protein [YFP]) and NUP98-TOP1 (linked to GFP). As outlined in Figure 7, a novel 2-stage infection strategy was devised to coexpress both trans-genes, as the transduction efficiency of NUP98-TOP1 is too low to efficiently cotransduce 2 genes. As early as 28 days after transplantation in the secondary mice, there was readily detectable repopulation by doubly transduced cells in most of the mice (GFP+/YFP+; 6.8%-22.7%). Although 5 of 8 mice were reconstituted with both GFP+/YFP+ peripheral blood (Figure 7), the median survival of these mice was 242 days after transplantation, similar to that previously observed for expression of NUP98-TOP1 alone (Table 3). Moreover, in this experiment 2 mice receiving NUP98-TOP1 BM without Meis1 had a comparable survival (175 and 187 days), further suggesting that in this model, Meis1 does not accelerate NUP98-TOP1-induced leukemogenesis.
Outline of strategy used to transplant BM cells coexpressing NUP98-TOP1 and Meis1. Five-fluorouracil (5-FU) BM was harvested, transduced, and transplanted into primary mice as described in “Materials and methods.” Five weeks after transplantation, non-5-FU BM was harvested from the primary mice and transduced, and 1 × 106 cells were transplanted into secondary mice along with 3 × 105 helper cells. FACS profiles are representative of PB at 98 days after secondary transplantation. Top plot indicates NUP98-TOP1 BM infected with Meis1; bottom, Meis1 BM infected with NUP98-TOP1.
Outline of strategy used to transplant BM cells coexpressing NUP98-TOP1 and Meis1. Five-fluorouracil (5-FU) BM was harvested, transduced, and transplanted into primary mice as described in “Materials and methods.” Five weeks after transplantation, non-5-FU BM was harvested from the primary mice and transduced, and 1 × 106 cells were transplanted into secondary mice along with 3 × 105 helper cells. FACS profiles are representative of PB at 98 days after secondary transplantation. Top plot indicates NUP98-TOP1 BM infected with Meis1; bottom, Meis1 BM infected with NUP98-TOP1.
Discussion
Our findings reveal potent and rapid perturbations in hematopoiesis induced by engineered expression of NUP98-TOPI, the fusion recognized in t(11;20)(p15q11). The observed effects included a potent in vitro growth advantage, a multilog increase in generation of spleen colony-forming cells, and induction of a lethal, transplantable myelomonocytic leukemia in mice. Strikingly, these perturbations appear to be independent of TOP1 catalytic activity because a mutation known to abrogate TOP1 DNA-isomerase activity essentially did not alter the NUP98-TOP1 proliferative and oncogenic potentials.
To date, more than 15 different partner genes have been identified in translocations involving the nucleoporin gene, NUP98, on chromosome 11p15.1 More than one half of the partners are homeobox genes, with 7 belonging to paralogs 9, 11, and 13 of the abdominal-B cluster of HOX. Moreover, at least 2 such fusions, NUP98-HOXA9 and NUP98-HOXD13, were shown to have direct, potent effects on hematopoiesis and to induce a lethal MPD/AML in mice.12,13
Leukemia induced by expression of NUP98-HOX fusions is consistent with the mounting evidence that HOX transcription factors play an integral role in hematopoiesis, and their altered expression and/or function are leukemogenic.54-56 The mechanism driving NUP98-TOP1-induced leukemia is more enigmatic given that TOP1 has no known unique role in hematopoiesis. Most provocative is the observation that the active site tyrosine required for TOP1 catalytic activity is dispensable for leukemic transformation, suggesting that NUP98-TOP1 does not act through up-regulated TOP1 DNA-unwinding activity. We cannot yet exclude the possibility that NUP98-TOP1 has a dominant-negative effect on topoisomerase activity. However, Topoisomerase I is necessary for growth and development of Drosophila melanogaster, and homozygous mutant murine embryos are embryonic lethal between the 4- and 16-cell stage, demonstrating that TOP1 is essential for cell growth.57,58 Furthermore, increased levels of TOP1 have been observed in cancer cells,59 and TOP1 inhibitors are widely used clinically in the treatment of leukemia and other cancers.34 These observations strongly argue against a dominant-negative effect on TOP1, as decreased levels of TOP1 are inconsistent with a malignant phenotype. Moreover, no gross chromosomal abnormalities were observed in NUP98-TOP1 BM cells cultured for more than 4 weeks, suggesting that NUP98-TOP expression does not interfere with normal TOP1 activity generating genomic instability.
Our experimental observations suggest that other key domains and/or functions of TOP1 retained in the fusion may play essential roles in the leukemogenic effects of NUP98-TOP1. TOP1 consists of 4 conserved domains (Figure 1): an N-terminal domain containing several nuclear targeting signals, a core domain necessary for DNA binding, a nonconserved linker domain, and a C-terminal domain that is also required for DNA binding and further contains the active site tyrosine in position 723 and a kinase domain implicated in phosphorylation of SF2/ASF splicing factors.
TOP1 has also been identified as a coactivator of transcription, enhancing basal transcription from a TATA promoter in the presence of an activator and is able to directly interact with TATA binding protein in the transcription factor IID (TFIID) transcriptional complex.40-42 Moreover, a TOP1 mutant (Y723F) lacking DNA-relaxing activity was equally active in enhancing transcription. This transcriptional coactivator property of TOP1 provides a possible functional similarity to HOX genes and other nuclear NUP98 fusion partners such as NSD1, NSD3, and LEDGF. We can hypothesis that NUP98 fusions act as aberrant transcription factors with binding to DNA occurring by way of the NUP98 fusion partner gene, and recruitment of the transcriptional machinery occurring by ways of recruiting CBP/p300 or other unknown transcription coactivators through NUP98.60 However, other contributions of NUP98 remain to be ruled out. Support for the hypothesis of NUP98-TOP1 as an aberrant transcription factor is backed by the observation of its nuclear localization. Other NUP98 fusions, including HOXA9,60 PMX1,11 and RAP1GDS1,61 are similarly observed to reside in the nucleus. Intriguingly, many of the non-HOX NUP98 fusion partners also contain domains predicted to adopt a coiled-coil conformation,62 and this domain has been verified for TOP1 by x-ray crystallography.52,53 Whether this domain promotes dimerization or is required for leukemic transformation, as is observed for several mixed-lineage leukemia (MLL) fusions,63 awaits further investigation. Also intriguing is the observation of PB and BM from NUP98-TOP1 leukemic mice that exhibit high proportion of B220lo cells. A bi-phenotypic leukemia exhibiting the phenotype (c-kit+/B220+/Mac-1+/CD19-/Gr-1-) has recently been reported on expression of several MLL fusion genes.64,65 Whether NUP98-TOP1 similarly transforms a novel progenitor capable of bi-phenotypic differentiation or NUP98-TOP1-transformed progenitors simply exhibit aberrant expression of lineage associated genes will be of interest to dissect in future experiments.
It is intriguing that many of the in vitro and in vivo effects induced by NUP98-TOP1 expression are similar to those observed following expression of NUP98-HOXA9 and NUP98-HOXD13.12,13 For example, in vitro, all fusions are associated with an increased proliferative capacity of transduced BM as measured in CFC spleen colony assay and/or liquid culture assays. In vivo, NUP98-TOP1 induced a progressive and ultimately lethal leukemia in all transplant recipients characterized by elevated WBC counts, splenomegaly, and a long latency, behavior similar to that documented for both NUP98-HOXA9 and NUP98-HOXD13. The long survival period observed for diseased NUP98-TOP1 primary mice, coupled with the accelerated onset of secondary disease, suggests the possible acquisition of additional genetic changes. However, the nature of this second hit is unclear. Of note, a rapid and lethal AML could be induced on coexpression of either NUP98-HOXA9 or NUP98-HOXD13 with the HOX-cofactor Meis1.12,13 However, when mice were transplanted with BM coexpressing both NUP98-TOP1 and Meis1, there was no obvious decrease in mouse survival compared with NUP98-TOP1 alone, suggesting distinctive transforming properties compared with NUP98-HOX fusions.
Our study provides a mouse model and an initial characterization of the leukemia-associated fusion NUP98-TOP1. It suggests that the isomerase activity of TOP1 is dispensable for the leukemic effects and may provide unique insights into the molecular pathogenesis of NUP98 fusions.
Prepublished online as Blood First Edition Paper, April 20, 2004; DOI 10.1182/blood-2003-10-3550.
Supported in part by the National Cancer Institute of Canada, with funds from the Terry Fox Foundation and by Genome BC. R.M.G. is a recipient of a scholarship from the Michael Smith Foundation for Health Research and the Natural Sciences and Engineering Research Council of Canada.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Nicolas Pineault for helpful discussions, Patty Rosten for technical assistance, Gloria Shaw for microscopy assistance and metaphase spreads, and Colleen MacKinnon for secretarial support.



![Figure 3. NUP98-TOP1 expression leads to in vivo myeloproliferation. (A) NUP98-TOP1 leads to in vivo proliferative advantage as assessed by increased peripheral blood GFP+ content at various times after transplantation. Graph is representative of 4 independent experiments with at least 4 mice in each group (mean ± SD; GFP CTL [□]; NUP98-TOP1 [▪]). (B) NUP98-TOP1 leads to a progressive increase in PB white cell counts. Results are shown for individual mice in a representative experiment. Median values are depicted with a horizontal line. Open symbols indicate GFP CTL mice; filled symbols, NUP98-TOP1 mice. (C) Differential counts performed on Wright-Giemsa-stained PB smears of mice at 2 (top) and 6 (bottom) months after transplantation. Counts are the average of 3 or 5 CTL GFP or NUP98-TOP1 mice, respectively. ▧ indicates lymphoid cells; ▪, myeloid cells. (D) Representative experiment depicting PB of 5 mice stained with antibodies to Mac-1 and B220 at 2 months (top panel) and 6 months (bottom panel) after transplantation. ▪ represents transduced (GFP+) cells; ▧, GFP-. Mice 1 to 5 are GFP CTL; mice 6 to 10 are NUP98-TOP1. (E) Representative Wright-Giemsa-stained PB smear of NUP98-TOP1 mouse at 6 months after transplantation. Original magnification, × 1000. See Figure 5B for control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/4/10.1182_blood-2003-10-3550/6/m_zh80160465290003.jpeg?Expires=1766274879&Signature=gMHDT2Do1pRMfpZB~WF0m3lWH-q4-URCGOl~MqJEgn4ZCCpOfvKRXN-XPyVilQlTPWZ4t4p6h03JEUlsOrKxjq9Dg~JvLYGslpoa-6P4PDE~DZ-YKQH-txCeqr9ZfEJpJC7tRSa6D-rWb4zWA~oKBCWYpCfEawyRqiiVEkMb1qc2nwv2ySIXw3rp6ug~rfNRCRRu4Jr-zIOdc0-eaNhI0IlrFjyjwR7~dkOLvJpZ7b9lfF8KZpvWrfN0BixGNFADR25bRAxDD9lYSN-NKwVe5PzgdmXYGqV~QaY6TYhqUvI6DPsYWw4NyvuHZBy5wEBfHuFYb3-iq35UKAhrSK9NdA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



 ). (B) Western blot analysis of total cell lysates from calcium-phosphate-transfected 293T cells detected by anti-GFP antibody. Cells were transfected with GFP-fusion constructs as described in “Materials and methods.” Arrows indicate the size of the expected full-length NUP98-TOP1 protein (150 kDA) and a smaller processed fragment. (C) Fluorescent microscopy images of 293T cells transfected as in panel B. Panels i, iv, vii, and x depict cells stained with 4′, 6-diamidino-2-phenylindole, dilactate (DAPI) for visualization of nuclei; panels ii, v, viii, and xi show visualization of GFP expression; panels iii, vi, ix, and xii show the overlay of DAPI and GFP. Top row (i-iii) results with empty pEGFP-C1 vector, demonstrating pan-cellular GFP expression in nucleus and cytosol. Second row (iv-vi) results with GFP-NUP98-TOP1 fusion, showing that NUP98-TOP1 directs nuclear expression. Third row (vii-ix) GFP-NT-Y723F also exhibits nuclear localization. Bottom row (x-xii) results with GFP-SH2-containing inositol-5-phosphatase (SHIP) used as positive control for cytosolic localization. (D) Retroviral vectors used to express NUP98-TOP1 and NT-Y723F in murine bone marrow. The expected sizes of full-length proviral transcripts are indicated. LTR indicates long terminal repeats; GFP, green fluorescent protein; IRES, internal ribosomal entry site.
). (B) Western blot analysis of total cell lysates from calcium-phosphate-transfected 293T cells detected by anti-GFP antibody. Cells were transfected with GFP-fusion constructs as described in “Materials and methods.” Arrows indicate the size of the expected full-length NUP98-TOP1 protein (150 kDA) and a smaller processed fragment. (C) Fluorescent microscopy images of 293T cells transfected as in panel B. Panels i, iv, vii, and x depict cells stained with 4′, 6-diamidino-2-phenylindole, dilactate (DAPI) for visualization of nuclei; panels ii, v, viii, and xi show visualization of GFP expression; panels iii, vi, ix, and xii show the overlay of DAPI and GFP. Top row (i-iii) results with empty pEGFP-C1 vector, demonstrating pan-cellular GFP expression in nucleus and cytosol. Second row (iv-vi) results with GFP-NUP98-TOP1 fusion, showing that NUP98-TOP1 directs nuclear expression. Third row (vii-ix) GFP-NT-Y723F also exhibits nuclear localization. Bottom row (x-xii) results with GFP-SH2-containing inositol-5-phosphatase (SHIP) used as positive control for cytosolic localization. (D) Retroviral vectors used to express NUP98-TOP1 and NT-Y723F in murine bone marrow. The expected sizes of full-length proviral transcripts are indicated. LTR indicates long terminal repeats; GFP, green fluorescent protein; IRES, internal ribosomal entry site.