Abstract
Bryostatin 1 is known to exhibit in vitro and in vivo activity against chronic lymphocytic leukemia (CLL) cells by inducing their further maturation into plasmalike cells. Signal transducer and activator of transcription (STAT) proteins play a central role in B-lymphocyte growth and function and are aberrantly phosphorylated on serine residues in CLL cells. To determine whether STAT transcription factors are important in Bryostatin 1–induced differentiation of CLL cells, primary CLL cells were examined for signaling events following exposure to Bryostatin 1 in vitro. Western analysis and electrophoretic mobility shift assays revealed that Bryostatin 1 induced tyrosine phosphorylation and DNA binding of STAT1, yet there was no effect on constitutive serine phosphorylation of STAT1. Bryostatin 1–induced STAT1 activation occurred in a manner that was dependent on protein kinase C (PKC), mitogen-activated protein kinase (MAPK), and Janus tyrosine kinase (JAK) activation. Evidence indicates that Bryostatin 1 induces STAT1 activation through an interferon γ (IFNγ) autocrine loop. However, STAT1 activation by IFNγ stimulation alone was not sufficient to induce differentiation. This insufficiency is due to the broader effect on gene expression caused by Bryostatin 1 compared with IFNγ, as demonstrated by microarray analysis. Both up-regulation of CD22 expression and immunoglobulin M (IgM) production, markers of CLL differentiation, were inhibited by a decoy oligonucleotide for STAT1, indicating that STAT1 is necessary for Bryostatin 1–induced differentiation of CLL cells. This study implicates STAT transcription factors as important mediators of Bryostatin 1–induced differentiation of CLL cells and could possibly lead to improved therapeutic approaches for the treatment of CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common form of adult leukemia in the United States, accounting for 25% of all leukemias.1 CLL is currently an incurable disease, characterized by the gradual accumulation of mature B lymphocytes arrested in the G0 phase of the cell cycle. CLL cells coexpress mature B-cell antigens, such as CD19, CD20, and CD23, and display a typical B-cell morphology. The underlying pathogenesis of the disease remains unknown, but the slow-growing CLL cells are thought to accumulate from a defect in apoptosis.2 Current drug therapy has some activity against CLL but has minimal effect on survival. Improvement in the treatment of CLL depends on identifying new therapeutic approaches.
One promising new drug for treating CLL is the macrocyclic lactone, Bryostatin 1, a naturally derived compound from a marine bryozoan. The antineoplastic effects of Bryostatin 1 are attributed to its ability to interact with protein kinase C (PKC). Like phorbol esters, Bryostatin 1 modulates PKC activity with a distinct pattern of activation of PKC isoforms in a highly tissue-specific manner.3,4 Unlike phorbol esters, Bryostatin 1 does not have tumor-promoting properties, which makes it an attractive therapeutic agent. Bryostatin 1 is considered both an agonist and an antagonist of PKC activity in that it binds to the diacylglycerol (DAG) binding site of PKC, resulting in the activation of PKC with acute exposure and down-regulation and loss of PKC activity with chronic exposure.
Among the potent biologic effects of Bryostatin 1 is the ability to induce differentiation of both myeloid and lymphoid leukemic cells.5-7 Bryostatin 1 induces differentiation of CLL cells8 and has antileukemic activity in vivo.9,10 Treatment of CLL cells with Bryostatin 1 induces the cells to undergo phenotypic changes consistent with further B-cell maturation, including morphologic changes, increased RNA synthesis, up-regulation of the CD22 cell surface marker, and immunoglobulin secretion. Bryostatin 1 can also sensitize hematologic tumor cells to conventional cytotoxic agents by increasing their susceptibility to apoptosis and/or further leukemic cell maturation.11 The observation that Bryostatin 1 has antineoplastic activity, causes differentiation of CLL cells, and can potentiate the cytotoxic activity of other drugs led to clinical trials testing Bryostatin 1 in patients with CLL. Phase 1 and 2 clinical trials have demonstrated that Bryostatin 1 has the potential for the treatment of refractory acute leukemias and indolent hematologic malignancies.9,11-13 Bryostatin 1 is currently undergoing phase 1 and 2 analysis in combination with other chemotherapeutic agents, including fludarabine, cladribine, and vincristine, for indolent hematologic malignancies, including CLL.
The exact mechanism of action of Bryostatin 1 remains unclear and likely involves signaling pathways in addition to PKC to achieve the full spectrum of its biologic effects. In considering signaling pathways that may be modulated by Bryostatin 1, we have focused on signal transducer and activator of transcription (STAT) proteins. STATs are a family of transcription factors that are key mediators of extracellular stimuli in lymphocytes.14 Following stimulation by ligands such as cytokines, STATs are phosphorylated on tyrosine residues by Janus kinases (JAKs) and on serine residues by a variety of serine/threonine kinases. STATs then dimerize, translocate to the nucleus, and bind DNA where they initiate the transcription of target genes. Tyrosine phosphorylation is necessary for STAT dimerization, nuclear translocation, and DNA binding, whereas serine phosphorylation is associated with enhanced transcriptional activity. Previous studies demonstrated that, in contrast to healthy B cells, STAT1 and STAT3 are constitutively phosphorylated on serine residues in CLL cells.15 The nature of this constitutive serine phosphorylation in CLL cells is not well understood, but it is suggestive of a role for STAT1 and STAT3 in the pathogenesis of CLL. Fludarabine, a nucleoside analog used in the treatment of CLL, was found to cause depletion of STAT1 protein and mRNA, demonstrating that STATs can be targets of antineoplastic agents.16 Given the importance of STAT signaling in lymphocyte biology and the aberrant serine phosphorylation of STATs in CLL cells, we examined the role of STAT signal transduction in Bryostatin 1–induced differentiation of CLL cells. The present study demonstrates that Bryostatin 1 activates STAT1 in a PKC-dependent manner by inducing an interferon γ (IFNγ) autocrine loop. These data further demonstrate that STAT1 activation is critical for Bryostatin 1–induced differentiation of CLL cells.
Materials and methods
Cells and culture conditions
After informed consent, CLL cells were isolated from heparinized blood of untreated patients diagnosed with CLL, according to clinical and immunophenotypic criteria. Mononuclear cells were isolated by Ficoll density gradient centrifugation. More than 95% of Ficoll-separated cells coexpressed CD5 and CD19 as determined by flow cytometry. In some cases, purified B cells were isolated by negative selection of the separated cells by using magnetic bead depletion (MACS B cell isolation kit; Miltenyi Biotech, Auburn, CA). The enriched cell population was more than 95% CD5/CD19+. Fresh and cryopreserved CLL cells were studied. For studies in which frozen cells were used, CLL cells were frozen in fetal bovine serum (FBS) containing 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen. CLL cells were cultured in RPMI-1640 containing 10% FBS.
Reagents
Bryostatin 1 (NSC 339555) was obtained from the Cancer Therapy Evaluation Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute (Rockville, MD), resuspended at a concentration of 500 μM in DMSO, and stored at –20°C. Just prior to use, Bryostatin 1 was diluted to 5 μM in DMSO and used at a final concentration of 10 nM. AG490, Gö6850, and Gö6976 were purchased from Calbiochem (San Diego, CA). PD98059 and U0126 were purchased from Cell Signaling (Beverly, MA). Cycloheximide was purchased from Sigma-Aldrich (St Louis, MO). Anti-PKC antibodies were purchased from BD Transduction Labs (San Diego, CA). Recombinant human IFNγ (285-IF) and antihuman IFNγ antibody (MAB285) were purchased from R & D Systems (Minneapolis, MN).
Western blot analysis
CLL cells were lysed in buffer containing 50 mM Tris (tris(hydroxymethyl) aminomethane)–Cl, pH 8.0, 250 mM NaCl, 0.5% NP40 (Nonidet P40), 2 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 2 μg/mL pepstatin. For Western analysis, 80 μg protein were resolved on 7% sodium dodecyl sulfate (SDS)–polyacrylamide gels and transferred to nitrocellulose membranes. Blots were incubated with an antibody specific for the tyrosine or serine phosphorylated form of STAT115,17 by using a 1:10 000 dilution of the antibody in Tris-buffered saline/0.05% Tween 20 (TBST) for 45 minutes at room temperature. The blots were stripped and reprobed with an antibody that recognizes total STAT1 (SC-346; Santa Cruz Biotechnology, Santa Cruz, CA) by using a 1:20 000 dilution of the antibody. Blots were incubated with goat antirabbit horseradish peroxidase–conjugated secondary antibodies (Calbiochem, La Jolla, CA), and detection was performed by using the Renaissance chemiluminescent ECL (enhanced chemiluminescence) kit (NEN Dupont, Boston, MA).
Nuclear extract preparation and electrophoretic mobility shift assay
Nuclear extracts were prepared by resuspending cells in hypotonic buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 6 mM MgCl2, 1 mM β-mercaptoethanol [βME], 1 mM sodium orthovanadate, 10 μg/mL PMSF, 2 μg/mL pepstatin, 2 μg/mL leupeptin, and 10 mM aprotinin) followed by incubation on ice for 5 minutes. Cells were centrifuged for 10 seconds at 12 000g, resuspended in hypotonic buffer, and disrupted using a Dounce homogenizer (Type B pestle, 30 strokes; Wheaton, Millville, NJ). The nuclei were collected by centrifugation for 10 seconds at 12 000g and washed once with hypotonic buffer. The nuclear pellet was resuspended in 1 pellet volume of high salt buffer (20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol, 0.2 mM EDTA (ethylenediaminetetraacetic acid), 10 μg/mL PMSF, 1 mM sodium orthovanadate, and 1 mM βME) and incubated for 30 minutes at 4°C followed by centrifugation for 3 minutes at 12 000g. Two microliters nuclear extract was incubated with a double-stranded 32P-labeled oligonucleotide (1 ng) gamma activation sequence (GAS), derived from the ciliary neurotropin factor (CNTF) response element (5′-CAGCCTGATTTCCCCGAAATGACGGCG-3′ and its complement) in 10 μL binding buffer (25 mM HEPES, pH 7.9, 100 μM EGTA (ethylene glycol tetraacetic acid), 200 μM MgCl2, 500 μM dithiothreitol, 1 μg/mL bovine serum albumin [BSA], 0.2 μg/μL poly dI:dC, 1% Ficoll, and 0.1 μg/μL salmon sperm DNA) for 15 minutes at room temperature. For supershift analysis, the nuclear extracts were incubated with 1 μg anti-STAT1 antibody (Santa Cruz Biotechnology) for 20 minutes on ice prior to adding the binding buffer. For competition experiments, 50 to 100 molar excess of double-stranded unlabeled oligonucleotide was added to the nuclear extract just prior to incubation with the binding buffer. Protein-DNA complexes were resolved by nondenaturing gel electrophoresis and detected by autoradiography.
Immunoprecipitation and protein kinase C activity
CLL cells were treated with 10 nM Bryostatin 1 for 30 minutes then washed with phosphate-buffered saline (PBS) and lysed with radioimmune precipitation assay lysis buffer (20 mM HEPES, pH 7.4, 10 mM EDTA, 125 mM NaCl, 1 mM DTT (dithiothreitol), 1% NP40, 1 mM sodium orthovanadate, 10 μg/mL PMSF, and 2 μg/mL pepstatin for PKCα, PKCβ, and PKCγ assays; 50 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% Tween 20, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 1 mM sodium orthovanadate, 10 μg/mL PMSF, and 2 μg/mL pepstatin for PKCδ and PKCϵ assays). Lysates were clarified by centrifugation at 14 000 rpm for 30 minutes at 4°C. Between 300 μg and 500 μg protein lysate was combined with 2.5 μg isoform-specific PKC antibody (BD Transduction Laboratories) and incubated overnight at 4°C. Immunocomplexes were collected following incubation with 40 μL Protein A/G agarose beads (Santa Cruz Biotechnology) for 1 hour at 4°C. PKC activity was assayed by measuring the incorporation of γ-[32P]ATP (adenosine triphosphate; NEN Dupont) into a specific PKC substrate using a PKC assay kit (Upstate Biotechnologies, Lake Placid, NY) according to the manufacturer's instructions. The peptide substrate QKRPSQRSKYL was used for PKCα, PKCβ, and PKCγ assays, and the peptide substrate ERMRPRKRQGSVRRRV (Upstate Biotechnologies) was used for PKCδ and PKCϵ assays. Total PKC levels present in the immunoprecipitation were measured by immunoblotting with anti-isoform–specific PKC antibodies.
Reverse transcriptase (RT)–PCR analysis
RNA was isolated using the RNEasy purification method (Qiagen, Valencia, CA). Five micrograms total RNA was reverse transcribed using random primers and polymerase chain reaction (PCR) amplified using IFNγ-specific primers (5′-ATGAAATATACAAGTTATATCTTGGCTTT-3′ and 3′-GATGCTCTTCGACCTCGAAACAGCAT-5′) or glyceraldehyde phosphate dehydrogenase (GAPDH)–specific pirmers (5′-GAAGGTGAAGGTCGGAGTC-3′ and 3′-GAAGATGGTGATGGGATTC-5′). Amplification of IFNγ and GAPDH was optimized in an MJ Research PTC 200 thermal cycler (MJ Research, Waltham, MA) for the following PCR parameters: 94°C for 45 seconds, 57°C for 30 seconds, and 72°C for 30 seconds for 30 cycles (IFNγ) or 35 cycles (GAPDH) followed by a final elongation step at 72°C for 2 minutes. PCR products were resolved on 1.5% agarose gels.
Immunostaining
Cells (1 × 106) were washed in PBS and then analyzed for surface expression of CD5, CD19, and CD22 by the addition of anti-CD5–fluorescein isothiocyanate ([FITC] IM0468; Beckman Coulter, Miami, FL), anti-CD19–phycoerythrin-cyanin 5.1 ([PC5] IM2643; Beckman Coulter), and anti-CD22–phycoerythrin ([PE] MHC2204; Caltag Laboratories, Burlingame, CA) conjugated monoclonal antibodies. Anti-IgG1 (6603482; Beckman Coulter) isotype control was used for each staining. Following incubation on ice for 20 minutes, the cells were washed with PBS, and a minimum of 10 000 CD5/CD19 positive cells were analyzed for CD22 expression by using a Beckman-Coulter EPICS-XL-MCL flow cytometer.
Measurement of immunoglobulin and cytokine production
Culture supernatants (200 μL) were analyzed for the presence of IgM or IFNγ by using enzyme-linked immunosorbent assay (ELISA; IgM ELISA, Bethyl Laboratories, Montgomery, TX; IFNγ ELISA, Cell Sciences, Norwood, MA) according to the manufacturers' instructions. The ELISA systems had detection limits of 10 ng/mL for IgM and less than 5 pg/mL for IFNγ.
Decoy oligonucleotides and transfection of CLL cells
Double-stranded decoy oligonucleotides (ODNs) were prepared by annealing complementary single-stranded ODNs containing 4 phosphorothioate bonds at each end (Integrated DNA Technologies, Coralville, IA). Single-stranded STAT1 ODNs were tagged with the fluorescent marker 6-fam ((di-O-pivaloyl-fluorescein)-3-aminopropyl) and mixed with unlabeled single-stranded STAT1 ODNs at a ratio of 1:10, respectively, prior to annealing to the complementary ODNs. The sequences of the ODNs were as follows (phosphorothioate-bonded nucleotides are in bold): STAT1 decoy, 5′-CATGTTATGCATATTCCGGGAAGTG-3′; mismatch control STAT1 ODNs (S1m), 5′-CATGTTATGCAGACCGTGGTTAGTG-3′. CLL (8 × 106) cells were resuspended in 300 μL RPM1-1640 containing 10% FBS and incubated with 10 μM STAT1 decoy ODNs or 10 μM S1m ODNs on ice for 10 minutes. Electroporation was performed in an ECM 830 BTX Electro Square Porator (BTX Corporation, San Diego, CA) using 4-mm gap cuvettes, 440 V, and a pulse length of 10 milliseconds. Cells were vortexed, incubated on ice for 15 minutes, washed with PBS/1% BSA, and then resuspended in RPMI-1640 containing 10% FBS, 10 mM HEPES, 50 IU/mL penicillin, and 50 μg/mL streptomycin. Transduction efficiency was monitored by flow cytometric analysis of 6-fam fluorescence and was routinely 75% to 100%. Viability was monitored by trypan blue exclusion and was routinely 60% to 80% after 24 hours of cell culture.
RNA isolation and microarray analysis
CLL cells were left untreated or were treated with 10 nM Bryostatin 1 or 10 U/mL IFNγ (R&D Systems) for 3 hours. Total RNA was isolated using Trizol (Invitrogen Life Technologies, Carlsbad, CA) and further purified using RNEasy spin columns (Qiagen). Preparation of cRNA, oligonucleotide array hybridization to HG-U133A GeneChip arrays (Affymetrix, Santa Clara, CA), and scanning of the arrays was performed by the Dana-Farber Microarray Core Facility. Data analysis was performed using the DNA-Chip Analyzer (dChip) software.18 Gene array data were normalized, and perfect-match only model-based expression intensities were obtained in dChip. To exclude those genes that lacked sufficient variability across groups, gene filtering was also performed in dChip. The metabolic quiescence of CLL cells resulted in relatively low magnitude changes in the most genes. Therefore, to identify those genes whose expression patterns best distinguished 2 groups, we required the fold change between the group means to exceed a threshold of 1.5.
Results
Bryostatin 1 induces activation of STAT1
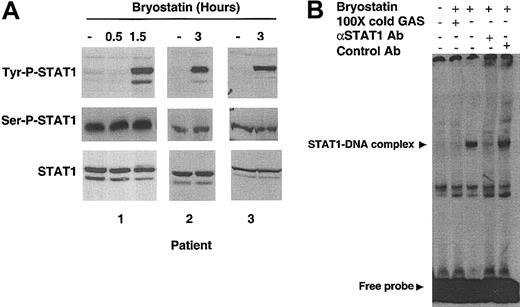
To determine whether Bryostatin 1 can modulate STAT signaling, primary CLL cells were incubated with Bryostatin 1 and then analyzed for changes in STAT1 tyrosine or serine phosphorylation. With the use of an antibody specific for the phosphorylation of tyrosine 701, the amino acid residue necessary for activation of STAT1, Western blot analysis revealed that STAT1 was tyrosine phosphorylated within 1.5 hours after addition of Bryostatin 1 (Figure 1A). Bryostatin 1 had no effect on the constitutive serine phosphorylation of STAT1 in CLL cells. To confirm the ability of tyrosine-phosphorylated STAT1 to bind to its cognate DNA sequence, electrophoretic mobility shift assay (EMSA) was performed using a radiolabeled interferon gamma activation (GAS) probe, containing the STAT-binding motif of the IRF-1 gene. An inducible protein-DNA complex was detectable in nuclear extracts from CLL cells stimulated with Bryostatin 1 for 1.5 hours (Figure 1B). The specificity of this complex was confirmed by competition of the binding activity with unlabeled GAS. Addition of an antibody against STAT1, but not an irrelevant control antibody, abolished the complex, verifying that the complex contains STAT1. Supershift analysis was performed using antibodies against all other STAT proteins, and no STAT transcription factor other than STAT1 was found to be activated by Bryostatin 1 (data not shown).
Bryostatin 1 induces tyrosine phosphorylation and DNA binding activity of STAT1. (A) Whole cell lysates from CLL cells treated with 10 nM Bryostatin 1 for 0.5 to 3 hours were analyzed by SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) followed by Western blotting using antibodies specific for the tyrosine phosphorylated form of STAT1 (Tyr-P-STAT1), serinephosphorylated form of STAT1 (Ser-P-STAT1), or total STAT1 (bottom panel). Data from the 3 patients' cells shown are representative of results from 21 patients' cells. (B) DNA binding activity of nuclear extracts from CLL cells was examined by EMSA using a radiolabeled probe containing a GAS sequence. Cells were treated with 10 nM Bryostatin 1 as indicated. One hundred molar excess of unlabeled probe (100 × cold GAS) was used for competition experiments. Anti-STAT1 antibody was added to the binding reaction mix to verify that the protein-DNA complex contained STAT1. A control antibody (anti-STAT6) was used to verify specificity.
Bryostatin 1 induces tyrosine phosphorylation and DNA binding activity of STAT1. (A) Whole cell lysates from CLL cells treated with 10 nM Bryostatin 1 for 0.5 to 3 hours were analyzed by SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) followed by Western blotting using antibodies specific for the tyrosine phosphorylated form of STAT1 (Tyr-P-STAT1), serinephosphorylated form of STAT1 (Ser-P-STAT1), or total STAT1 (bottom panel). Data from the 3 patients' cells shown are representative of results from 21 patients' cells. (B) DNA binding activity of nuclear extracts from CLL cells was examined by EMSA using a radiolabeled probe containing a GAS sequence. Cells were treated with 10 nM Bryostatin 1 as indicated. One hundred molar excess of unlabeled probe (100 × cold GAS) was used for competition experiments. Anti-STAT1 antibody was added to the binding reaction mix to verify that the protein-DNA complex contained STAT1. A control antibody (anti-STAT6) was used to verify specificity.
Activation of STAT1 is dependent on PKC activation
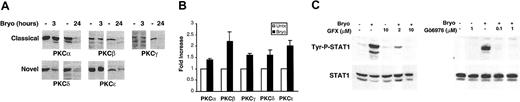
Bryostatin 1 is known to be a potent agonist/antagonist of the family of phospholipid-dependent serine/threonine protein kinases known as protein kinase C (PKC).19,20 This family of proteins is divided into 3 major subgroups, classical (α, β, and γ), novel (ϵ, δ, η, θ, and μ), and atypical (ζ and λ/ι), based on their activation mechanisms, cellular function, and expression patterns.21 To begin to define the role of PKC in Bryostatin 1–induced STAT signaling, the PKC isoforms activated by Bryostatin 1 in CLL cells were defined. Prolonged activation of PKC results in proteasomal degradation and loss of expression of activated PKC isoforms.22 Western blot analysis revealed that both short-term (3 hours) and long-term (24 hours) treatment of CLL cells with Bryostatin 1 resulted in the loss of expression of PKCα, PKCβ, PKCγ, PKCδ, and PKCϵ (Figure 2A). Therefore, both classical and novel forms of PKC were affected by Bryostatin 1 treatment in CLL cells. CLL cells were also analyzed for Bryostatin 1–induced effects on PKC η, θ, and λ/ι but were found to express little or none of these PKC isoforms (data not shown). To confirm that Bryostatin 1 caused a concomitant increase in enzymatic activity of each of these PKC isoforms, PKC kinase activity was measured by immunoprecipitating specific PKC isoforms and performing immune complex kinase assays on each of the immunoprecipitates. The kinase activity of PKCα, PKCβ, PKCγ, PKCδ, and PKCϵ each increased by 40% to 100% following Bryostatin 1 treatment, indicating that the change in expression of these isoforms reflected enhanced activity (Figure 2B).
Bryostatin 1–induced STAT1 activation is dependent on PKC activation in CLL cells. (A) CLL cells were left untreated or treated with 10 nM Bryostatin 1 for 3 or 24 hours. Whole cell lysates were analyzed by Western blotting for the presence of different PKC isoforms. Data are representative of 3 experiments using 3 different patients' cells. (B) CLL cells were treated with 10 nM Bryostatin 1 for 30 minutes. Cells were lysed, and PKC kinase activity was measured after immunoprecipitation in an in vitro kinase assay. Values represent the means ± SEMs of assays performed on CLL cells from 3 patients. (C) CLL cells were pretreated with the indicated concentrations of GF109203X (GFX) or Gö6976 for 1 hour prior to addition of 10 nM Bryostatin 1 for 3 hours. Whole-cell lysates were analyzed by Western blotting for tyrosine phosphorylation of STAT1 (Tyr-P-STAT1). Blots were stripped and reprobed for total STAT1. Data are representative of 4 experiments using 3 different patients' cells.
Bryostatin 1–induced STAT1 activation is dependent on PKC activation in CLL cells. (A) CLL cells were left untreated or treated with 10 nM Bryostatin 1 for 3 or 24 hours. Whole cell lysates were analyzed by Western blotting for the presence of different PKC isoforms. Data are representative of 3 experiments using 3 different patients' cells. (B) CLL cells were treated with 10 nM Bryostatin 1 for 30 minutes. Cells were lysed, and PKC kinase activity was measured after immunoprecipitation in an in vitro kinase assay. Values represent the means ± SEMs of assays performed on CLL cells from 3 patients. (C) CLL cells were pretreated with the indicated concentrations of GF109203X (GFX) or Gö6976 for 1 hour prior to addition of 10 nM Bryostatin 1 for 3 hours. Whole-cell lysates were analyzed by Western blotting for tyrosine phosphorylation of STAT1 (Tyr-P-STAT1). Blots were stripped and reprobed for total STAT1. Data are representative of 4 experiments using 3 different patients' cells.
Although PKC-independent activation of STATs has been reported,23,24 other studies have demonstrated PKC-dependent activation of STAT proteins,25,26 indicating that STAT phosphorylation may be either upstream or downstream of PKC activation. To determine whether Bryostatin 1–induced STAT1 activation is dependent on PKC signaling, CLL cells were analyzed for Bryostatin 1–induced STAT1 activation following treatment with selective pharmacologic inhibitors of PKC. One-hour pretreatment of CLL cells with GF109203X, which selectively inhibits both novel and classical forms of PKC, or Gö6976, which selectively inhibits classical forms of PKC, followed by a 3-hour treatment with Bryostatin 1 revealed that inhibition of PKC activation prevents STAT1 tyrosine phosphorylation (Figure 2C). Therefore, STAT1 activation is dependent on PKC activation.
Activation of STAT1 is dependent on MAPK activation and new protein synthesis
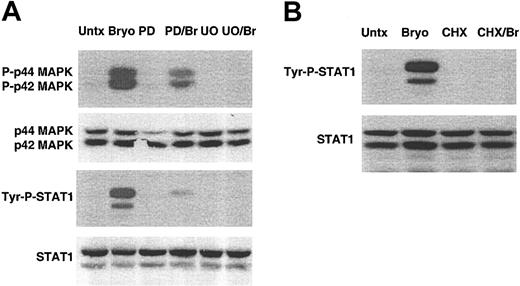
Both Bryostatin 1 and PKC activation are known to stimulate mitogen-activated protein kinase (MAPK) activity.27-30 MAPK is known to be required for Bryostatin 1–induced differentiation of acute lymphoblastic leukemia cells.28 We reasoned that MAPK and its associated downstream signaling events could be important in Bryostatin 1–induced differentiation of CLL cells. We examined whether Bryostatin 1 stimulated MAPK in CLL cells and whether MAPK activation was required for STAT1 activation. Following treatment of CLL cells with Bryostatin 1, phosphorylation of MAPK was induced within 10 minutes (Figure 3A). To determine whether phosphorylation of MAPK was required for STAT1 activation, CLL cells were pretreated with inhibitors of MEK1/2 (mitogen-activated protein [MAP]/extracellular signal-regulated kinase [ERK] 1 and 2), which is the kinase upstream of MAPK. Cells were incubated with either PD98059 or U0126 for 1 hour and then treated with Bryostatin 1 for 3 hours. Western blot analysis confirmed that PD980509 (partially) and U0126 (completely) blocked phosphorylation of MAPK. Both MAPK inhibitors blocked Bryostatin 1–induced STAT1 activation, demonstrating that STAT1 activation is dependent on the activation of MAPK.
Bryostatin 1–induced STAT1 activation requires MAPK activation and new protein synthesis. (A) CLL cells were pretreated with pharmacologic inhibitors of MEK1/2, PD98059 (PD) or U0126 (U0), for 1 hour and then treated with 10 nM Bryostatin 1 (Br) for 3 hours. Whole cell lysates were prepared and analyzed by Western blotting using antibodies specific for the tyrosine phosphorylated form of STAT1 (Tyr-P-STAT1) or the phosphorylated forms of the MAP kinases, P-p44 and P-p42 MAPK. Blots were stripped and reprobed for total STAT1 and total p44 and p42 MAPK. The results are representative of 5 patients' cells analyzed. Untx indicates untreated. (B) CLL cells were pretreated with 10 μM cycloheximide for 1 hour and then treated with 10 nM Bryostatin 1 for 3 hours. Whole cell lysates were prepared and analyzed by Western blotting using antibodies specific for Tyr-P-STAT1, and blots were stripped and reprobed for total STAT1. Similar results were found in experiments using 4 additional patients' cells.
Bryostatin 1–induced STAT1 activation requires MAPK activation and new protein synthesis. (A) CLL cells were pretreated with pharmacologic inhibitors of MEK1/2, PD98059 (PD) or U0126 (U0), for 1 hour and then treated with 10 nM Bryostatin 1 (Br) for 3 hours. Whole cell lysates were prepared and analyzed by Western blotting using antibodies specific for the tyrosine phosphorylated form of STAT1 (Tyr-P-STAT1) or the phosphorylated forms of the MAP kinases, P-p44 and P-p42 MAPK. Blots were stripped and reprobed for total STAT1 and total p44 and p42 MAPK. The results are representative of 5 patients' cells analyzed. Untx indicates untreated. (B) CLL cells were pretreated with 10 μM cycloheximide for 1 hour and then treated with 10 nM Bryostatin 1 for 3 hours. Whole cell lysates were prepared and analyzed by Western blotting using antibodies specific for Tyr-P-STAT1, and blots were stripped and reprobed for total STAT1. Similar results were found in experiments using 4 additional patients' cells.
Cytokine-induced STAT activation in lymphocytes does not require new protein synthesis.31 However, the delay in STAT1 phosphorylation following Bryostatin 1 treatment raised the possibility that protein synthesis is required in this pathway. To test this hypothesis, CLL cells were incubated with the protein synthesis inhibitor cycloheximide. STAT1 activation was abolished when CLL cells were pretreated with 10 μM cycloheximide prior to Bryostatin 1 treatment (Figure 3B). The absence of tyrosine phosphorylation in the presence of both cycloheximide and Bryostatin 1 demonstrates that Bryostatin 1–induced STAT1 activation is sensitive to inhibition of protein synthesis, indicating that intermediate proteins are necessary for activation of STAT1.
Activation of STAT1 is caused by Bryostatin 1–induced IFNγ production by CLL cells
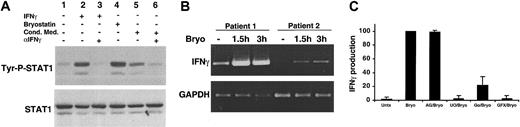
Given that STAT1 activation is dependent on PKC activation, MAPK activation, and requires new protein synthesis, we considered the possibility that Bryostatin 1 could induce a secreted factor that was responsible for the STAT1 activation. Conditioned medium was collected from CLL cells that had been stimulated with Bryostatin 1 for 3 hours, then added to untreated CLL cells, which were analyzed for STAT1 activation occurring within 15 minutes. This time point was picked because it is sufficient to detect cytokine-induced STAT1 activation but it is too early for Bryostatin 1–induced STAT1 phosphorylation. The conditioned medium from Bryostatin 1–treated cells induced immediate STAT1 phosphorylation (Figure 4A, lane 5), indicating that a secreted factor was responsible for the STAT activation. The slightly lower level of STAT1 phosphorylation induced in the cells treated with conditioned medium is attributable to the weaker response to cell stimuli typically observed after CLL cells are placed in culture for several hours (D.A.F. and T.E.B., unpublished observations, June 25, 2001). The only STAT activated in response to Bryostatin 1 is STAT1. Because one of the only cytokines known to exclusively activate STAT1 is IFNγ, we tested whether IFNγ was the factor in the conditioned medium responsible for STAT1 activation. The addition of a neutralizing antibody to IFNγ to Bryostatin 1–conditioned medium inhibited STAT1 activation (Figure 4A, lane 6), indicating that IFNγ was in fact being secreted by Bryostatin 1–treated CLL cells and acting in an autocrine manner to stimulate STAT1 activation.
STAT1 activation by Bryostatin 1 is mediated by IFNγ. (A) CLL cells were untreated (lane 1) or treated with 10 nM Bryostatin 1 (lane 4) for 3 hours. Conditioned medium from CLL cells stimulated with 10 nM Bryostatin 1 for 3 hours was incubated with untreated CLL cells for 15 minutes (lane 5). To test whether IFNγ was responsible for STAT1 activation, 0.5 μg/mL anti-IFNγ neutralizing antibody (αIFNγ) was incubated with conditioned medium for 20 minutes prior to addition to untreated CLL cells (lane 6). CLL cells were stimulated with 500 U/mL IFNγ for 15 minutes as a positive control (lane 2). To verify the neutralizing activity of the anti-IFNγ antibody, 50 U/mL IFNγ were incubated with 0.5 μg/mL anti-IFNγ antibody for 10 minutes prior to incubation with CLL cells for 15 minutes (lane 3). Whole cell lysates were prepared and analyzed for STAT1 tyrosine phosphorylation (Tyr-P-STAT1) by Western blotting. (B) RT-PCR for IFNγ was performed on mRNA from CLL cells left untreated for 1.5 hours or treated with 10 nM Bryostatin 1 for 1.5 to 3 hours. Amplification of GAPDH was performed as a control. (C) IFNγ ELISA was performed on culture supernatants collected from CLL cells untreated or pretreated with 100 μM AG490, 100 μM U0126, 500 μM Gö6976, or 10 μM Gö6850 for 1 hour prior to stimulation with 10 nM Bryostatin 1 for 3 hours. The relative IFNγ production is represented by normalizing the level of IFNγ produced under each condition to that of the Bryostatin 1–treated cells and is expressed as the mean percentage ± SEM from 4 patients.
STAT1 activation by Bryostatin 1 is mediated by IFNγ. (A) CLL cells were untreated (lane 1) or treated with 10 nM Bryostatin 1 (lane 4) for 3 hours. Conditioned medium from CLL cells stimulated with 10 nM Bryostatin 1 for 3 hours was incubated with untreated CLL cells for 15 minutes (lane 5). To test whether IFNγ was responsible for STAT1 activation, 0.5 μg/mL anti-IFNγ neutralizing antibody (αIFNγ) was incubated with conditioned medium for 20 minutes prior to addition to untreated CLL cells (lane 6). CLL cells were stimulated with 500 U/mL IFNγ for 15 minutes as a positive control (lane 2). To verify the neutralizing activity of the anti-IFNγ antibody, 50 U/mL IFNγ were incubated with 0.5 μg/mL anti-IFNγ antibody for 10 minutes prior to incubation with CLL cells for 15 minutes (lane 3). Whole cell lysates were prepared and analyzed for STAT1 tyrosine phosphorylation (Tyr-P-STAT1) by Western blotting. (B) RT-PCR for IFNγ was performed on mRNA from CLL cells left untreated for 1.5 hours or treated with 10 nM Bryostatin 1 for 1.5 to 3 hours. Amplification of GAPDH was performed as a control. (C) IFNγ ELISA was performed on culture supernatants collected from CLL cells untreated or pretreated with 100 μM AG490, 100 μM U0126, 500 μM Gö6976, or 10 μM Gö6850 for 1 hour prior to stimulation with 10 nM Bryostatin 1 for 3 hours. The relative IFNγ production is represented by normalizing the level of IFNγ produced under each condition to that of the Bryostatin 1–treated cells and is expressed as the mean percentage ± SEM from 4 patients.
To verify that IFNγ was being produced by Bryostatin 1–treated CLL cells, semiquantitative RT-PCR was performed on untreated CLL cells and those treated with Bryostatin 1 for 1.5 to 3 hours. Although low levels of IFNγ mRNA were occasionally seen at baseline, Bryostatin 1 treatment led to an induction of IFNγ mRNA within 1.5 hours (Figure 4B). Likewise, IFNγ secretion was induced in CLL cells (range, 4-16 pg/mL) by Bryostatin 1 after 3 hours of exposure (Figure 4C). It is unlikely that contaminating cells, such as peripheral blood T lymphocytes, were the source of IFNγ, considering that the cells used in the assay were greater than or equal to 95% CD5/CD19+. In addition, Curiel et al32 recently demonstrated that Bryostatin 1 alone, in the absence of interleukin 2 (IL-2), does not induce IFNγ in human T lymphocytes.
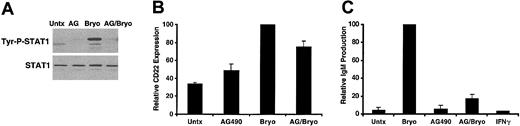
If STAT1 activation is mediated by IFNγ in Bryostatin 1–treated CLL cells, then, like STAT1 activation, IFNγ production should be dependent on PKC activation and MAPK activation. To determine whether PKC activation and MAPK activation were necessary for IFNγ production, CLL cells were pretreated with the MAPK inhibitor U0126 or the PKC inhibitors GFX and Gö6976. Consistent with the hypothesis that IFNγ-activated STAT1 requires MAPK and PKC activation, inhibitors of PKC or MAPK signaling prevented IFNγ production (Figure 4C). This finding indicates that IFNγ, and consequently STAT1, require PKC and MAPK activation. Furthermore, pretreatment of CLL cells with the JAK inhibitor AG490 blocked STAT1 phosphorylation (Figure 5A) but did not affect IFNγ production, indicating that STAT signaling is downstream of IFNγ production in Bryostatin 1–treated CLL cells.
AG490 blocks Bryostatin 1–induced CLL differentiation. (A) CLL cells were untreated or pretreated with 100 μM AG490 for 1 hour and then treated with 10 nM Bryostatin 1 for 3 hours. Whole cell lysates were prepared and analyzed by Western blotting using antibodies specific for the tyrosine phosphorylated form of STAT1 (Tyr-P-STAT1). Blots were stripped and reprobed for total STAT1. Results are representative of experiments performed on 5 patients' cells. (B) AG490 inhibits Bryostatin 1–induced CD22 up-regulation. CLL cells were untreated or pretreated with 100 μM AG490 (AG) and then treated with 10 nM Bryostatin 1 for 24 hours. CD5/CD19+ cells were analyzed for CD22 expression by immunostaining and flow cytometric analysis. Values of CD22 expression were normalized to the level of CD22 expression in the Bryostatin 1–treated cells and are expressed as the mean percentage ± SEM from 3 patients. (C) IgM ELISA was performed on culture supernatants from CLL cells untreated or pretreated with 100 μM AG490 and then treated with 10 nM Bryostatin 1 or 500 U IFNγ for 48 hours. The values of IgM production were normalized to the level of IgM produced by the Bryostatin 1–treated cells and are the mean percentage ± SEM from 3 patients.
AG490 blocks Bryostatin 1–induced CLL differentiation. (A) CLL cells were untreated or pretreated with 100 μM AG490 for 1 hour and then treated with 10 nM Bryostatin 1 for 3 hours. Whole cell lysates were prepared and analyzed by Western blotting using antibodies specific for the tyrosine phosphorylated form of STAT1 (Tyr-P-STAT1). Blots were stripped and reprobed for total STAT1. Results are representative of experiments performed on 5 patients' cells. (B) AG490 inhibits Bryostatin 1–induced CD22 up-regulation. CLL cells were untreated or pretreated with 100 μM AG490 (AG) and then treated with 10 nM Bryostatin 1 for 24 hours. CD5/CD19+ cells were analyzed for CD22 expression by immunostaining and flow cytometric analysis. Values of CD22 expression were normalized to the level of CD22 expression in the Bryostatin 1–treated cells and are expressed as the mean percentage ± SEM from 3 patients. (C) IgM ELISA was performed on culture supernatants from CLL cells untreated or pretreated with 100 μM AG490 and then treated with 10 nM Bryostatin 1 or 500 U IFNγ for 48 hours. The values of IgM production were normalized to the level of IgM produced by the Bryostatin 1–treated cells and are the mean percentage ± SEM from 3 patients.
Inhibition of STAT1 activation blocks CLL differentiation
Bryostatin 1 is known to induce differentiation of CLL cells to antibody-secreting plasmalike cells both in vitro and in vivo on the basis of morphologic changes, expression of CD22 and CD11c cell surface markers, and immunoglobulin production.8-10 The functional significance of Bryostatin 1–induced STAT1 activation was examined by addressing whether STAT1 activation plays a role in Bryostatin 1–induced CLL differentiation. To determine whether STAT signaling was important for CLL differentiation, CLL cells were treated with the JAK inhibitor AG490 and examined for levels of CD22 cell surface expression, a mature B-cell marker, and IgM secretion, a measure of functional B-cell differentiation. As noted, AG490 completely inhibits Bryostatin 1–induced STAT1 activation (Figure 5A). CLL cells exhibit variable levels of CD22 expression. Bryostatin 1 induced further up-regulation of CD22, which was inhibited by pretreatment with AG490 (Figure 5B). Similarly, Bryostatin 1 induced the production of IgM from CLL cells, and this production was completely inhibited by AG490 (Figure 5C). Because AG490 inhibits CD22 up-regulation and IgM production, this finding indicates that STAT1 is required for Bryostatin 1–induced differentiation of CLL cells. To determine whether STAT1 activation was sufficient for CLL differentiation, CLL cells were treated with IFNγ, a well-known, potent stimulator of STAT1 activation. However, IFNγ alone did not induce CD22 (data not shown) or IgM secretion (Figure 5C). Taken together, these results indicate that STAT1 activation is necessary for Bryostatin 1–induced CLL differentiation, but other signaling pathways must contribute to the differentiation-inducing effects of Bryostatin 1 as well.
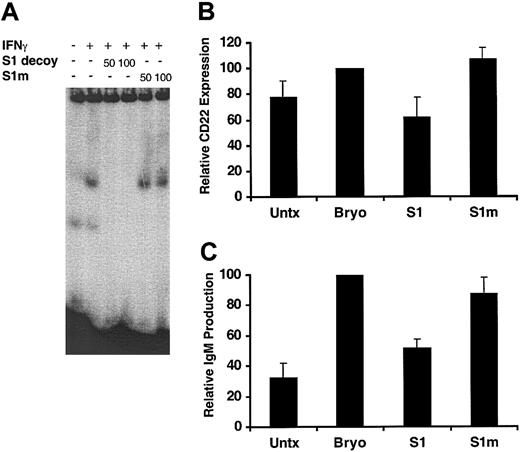
To directly assess the functional role of STAT1 in Bryostatin 1–induced CLL differentiation and to exclude the possibility that a JAK-dependent, STAT-independent mechanism was involved, a double-stranded decoy oligonucleotide designed to mimic a STAT1 target promoter sequence was used to specifically inhibit STAT1-dependent gene induction. The decoy oligonucleotide is designed to disrupt STAT1 transcriptional activity by competing for STAT1 DNA binding. The ability of this STAT1 decoy to block STAT1 DNA binding was demonstrated by using EMSA (Figure 6A). Addition of 50 to 100 molar excess of the STAT1 decoy to nuclear extracts from IFNγ-treated CLL cells completely blocked STAT1 DNA binding, whereas a STAT1 mismatch control oligonucleotide had no effect. Following introduction of the STAT1 decoy or mismatch control oligonucleotide by electroporation, CLL cells were stimulated with Bryostatin 1, and their ability to differentiate was examined by measuring CD22 expression and IgM production. Bryostatin 1 induced further up-regulation of CD22, albeit to a lesser extent than previously observed, likely reflecting interpatient variability (Figure 6B). The STAT1 decoy inhibited Bryostatin 1–induced CD22 expression by 62% compared with electroporated Bryostatin 1–treated cells, whereas the mismatch oligo had no effect. The STAT1 decoy also inhibited IgM production by 54% compared with electroporated Bryostatin 1–treated cells (Figure 6C). The level of IgM was reduced to near background levels in CLL cells transduced with the STAT1 decoy. Inhibition of CD22 expression and IgM production by the STAT1 decoy demonstrates that STAT1 transcriptional activity is necessary for Bryostatin 1–induced CLL differentiation.
Bryostatin 1–induced CLL differentiation requires STAT1 activation. (A) DNA binding activity of nuclear extracts from CLL cells was examined by EMSA using a radiolabeled probe containing a GAS sequence. Cells were untreated or treated for 15 minutes with 500 U/mL IFNγ. For competition experiments, 50 to 100 molar excess of STAT1 decoy ODNs (S1 decoy) or mismatch control STAT1 ODNs (S1m) was incubated with nuclear extracts from IFNγ-treated cells prior to incubation with the radiolabeled probe. (B) CLL cells were transduced with or without 10 μM STAT1 decoy (S1) or 10 μM mismatch control STAT1 ODNs (S1m) by electroporation. All cells were electroporated and then treated with or without (Untx) 10 nM Bryostatin 1 for 24 hours. CD5/CD19+ cells were analyzed for CD22 expression by immunostaining and flow cytometric analysis. Values of CD22 expression were normalized to the level of CD22 expression in the Bryostatin 1–treated cells and are expressed as the mean percentage ± SEM from 3 patients. (C) IgM ELISA was performed on 200 μL culture supernatants from CLL cells treated with or without (Untx) 10 nM Bryostatin 1 for 48 hours following transduction with STAT1 decoy (S1) or mismatch control STAT1 ODNs (S1m) as described in panel B. The values of IgM production were normalized to the level of IgM produced by the Bryostatin 1–treated cells and are the mean percentage ± SEM from 3 patients.
Bryostatin 1–induced CLL differentiation requires STAT1 activation. (A) DNA binding activity of nuclear extracts from CLL cells was examined by EMSA using a radiolabeled probe containing a GAS sequence. Cells were untreated or treated for 15 minutes with 500 U/mL IFNγ. For competition experiments, 50 to 100 molar excess of STAT1 decoy ODNs (S1 decoy) or mismatch control STAT1 ODNs (S1m) was incubated with nuclear extracts from IFNγ-treated cells prior to incubation with the radiolabeled probe. (B) CLL cells were transduced with or without 10 μM STAT1 decoy (S1) or 10 μM mismatch control STAT1 ODNs (S1m) by electroporation. All cells were electroporated and then treated with or without (Untx) 10 nM Bryostatin 1 for 24 hours. CD5/CD19+ cells were analyzed for CD22 expression by immunostaining and flow cytometric analysis. Values of CD22 expression were normalized to the level of CD22 expression in the Bryostatin 1–treated cells and are expressed as the mean percentage ± SEM from 3 patients. (C) IgM ELISA was performed on 200 μL culture supernatants from CLL cells treated with or without (Untx) 10 nM Bryostatin 1 for 48 hours following transduction with STAT1 decoy (S1) or mismatch control STAT1 ODNs (S1m) as described in panel B. The values of IgM production were normalized to the level of IgM produced by the Bryostatin 1–treated cells and are the mean percentage ± SEM from 3 patients.
IFNγ activates a subset of genes induced by Bryostatin 1
Although Bryostatin 1 and IFNγ both activate STAT1 in CLL cells, only Bryostatin 1 can induce differentiation. Thus, it is likely that Bryostatin 1 induces the expression of genes beyond that induced by IFNγ alone. To address this directly, gene expression profiles were compared from CLL cells treated with Bryostatin 1 or IFNγ for 3 hours, a time at which STAT1 activation is known to occur with both treatments. Microarray analysis was performed on cells from 2 patients using oligonucleotide arrays to analyze changes in global patterns of gene expression. Normalized microarray data revealed that a set of genes regulated by IFNγ is also regulated by Bryostatin 1 (Table 1). Among the 1289 genes up-regulated or down-regulated by Bryostatin 1, 78 genes were also regulated by IFNγ. The set of genes included in this overlap group represented nearly one third of all IFNγ-regulated genes and 6.1% of the Bryostatin 1–regulated genes. Overlap between the Bryostatin 1–regulated and IFNγ-regulated genes was confirmed in a second patient's cells in which 34 genes regulated by IFNγ were also regulated among the 995 genes regulated by Bryostatin 1. This finding provides further evidence for an IFNγ autocrine loop that activates STAT1 following exposure to Bryostatin 1. These data indicate that Bryostatin 1 induces differentiation of CLL cells because of its broader effect on gene expression compared with IFNγ.
Discussion
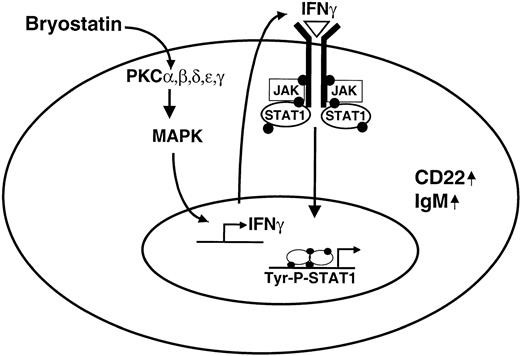
The present study demonstrates that the proximal signaling event in Bryostatin 1–treated CLL cells is the activation of PKC isozymes, which then promote signal transduction events leading to STAT1 activation, which, in turn, is necessary for further differentiation of the CLL cells. The data presented support a model for activation of the STAT signaling pathway in which Bryostatin 1 activates classical and novel PKC isozymes leading to MAPK activation (Figure 7). MAPK activation promotes the transcription and production of IFNγ (also reported by Dumont et al,33 Egerton et al,34 and Mainiero et al35 ), which, in turn, acts in an autocrine manner on the CLL cell leading to activation of the JAK-STAT1 signaling pathway. Tyrosine phosphorylation of STAT1 promotes further B-cell maturation of the CLL cell through a transcription-based mechanism leading to the up-regulation of CD22 expression and IgM production. Further evidence for an operative IFNγ autocrine loop was evident by the notable overlap between Bryostatin 1–regulated genes and IFNγ-regulated genes. Because Bryostatin 1 regulates a larger number of genes compared with IFNγ, and because some of the genes regulated by IFNγ represent a subset of the Bryostatin 1–regulated genes, the difference in phenotypic effects caused by these stimuli is likely explained by the broader effect on gene expression caused by Bryostatin 1.
Model for Bryostatin 1–induced STAT1 activation. Bryostatin 1 activates novel and classical forms of PKC, which leads to MAPK activation. MAPK activation promotes IFNγ transcription and release from the CLL cell. IFNγ acts in an autocrine manner to stimulate STAT1 signaling, which is required for further maturation of the CLL cell through a transcriptional mechanism leading to up-regulation of CD22 expression and IgM production.
Model for Bryostatin 1–induced STAT1 activation. Bryostatin 1 activates novel and classical forms of PKC, which leads to MAPK activation. MAPK activation promotes IFNγ transcription and release from the CLL cell. IFNγ acts in an autocrine manner to stimulate STAT1 signaling, which is required for further maturation of the CLL cell through a transcriptional mechanism leading to up-regulation of CD22 expression and IgM production.
Specific inhibition of STAT1 signaling using a decoy oligonucleotide directly implicates STAT1 in Bryostatin 1–induced maturation of CLL cells. The ability of Bryostatin 1 to induce CD22 up-regulation and IgM production was significantly impaired in CLL cells transduced with the STAT1 decoy oligonucleotide compared with those transduced with a mismatch control oligonucleotide. Because IFNγ alone, which activates STAT1 upon Bryostatin 1 treatment of CLL cells, does not induce CLL differentiation, additional signaling molecules, such as transcription factors or coactivators, must interact with the STAT1 signaling pathway to promote differentiation of CLL cells. The data, therefore, suggest that Bryostatin 1 induces CLL differentiation through STAT1-dependent and STAT1-independent mechanisms.
Possible mechanisms of STAT1-mediated differentiation by Bryostatin 1 include either the direct or indirect transcriptional regulation of CD22 and IgM. The up-regulation of CD22 expression may be a direct transcriptional effect of STAT1 activation because there are a number of putative STAT binding sites located in the CD22 promoter, albeit in the distal promoter region.36 In contrast, increased IgM production is not likely a direct transcriptional effect. IgM production is regulated posttranscriptionally by increased cytoplasmic accumulation and mRNA stability of the IgM heavy chain.37 In the case of Bryostatin 1–induced IgM production, it is more likely that one or more intermediate genes are targeted by STAT1, and that these gene products are part of the repertoire of accessory proteins responsible for posttranscriptional up-regulation of IgM. It is worth noting that, unlike IgM production, which is detectable between 24 and 48 hours following Bryostatin 1 treatment, the kinetics of CD22 up-regulation closely follow the kinetics of STAT1 activation. STAT1 activation occurs within 1.5 hours of Bryostatin 1 treatment and is maintained through 24 hours, the time when CD22 up-regulation is detectable, and returns to basal levels by 48 hours (data not shown).
Although IFNγ is typically produced by T lymphocytes and natural killer (NK) cells, it is known to be produced by malignant B cells38 and has been shown to have either positive or negative physiologic effects on CLL cells, depending on the concentration used. Previous studies have demonstrated that IFNγ can protect CLL cells from apoptosis in vitro and that serum from CLL patients contains increased levels of IFNγ compared with that of healthy donors, suggestive of a role for IFNγ in suppressing apoptosis in vivo as well.38-41 These studies used or detected much higher levels of IFNγ compared with the levels detected in Bryostatin 1–treated CLL cells in the present study. Thus, the level of IFNγ needed to induce STAT1-mediated differentiation may be very low, and there may be a threshold above which IFNγ has antiapoptotic effects. IFNγ may also have inhibitory effects on CLL cells. IFNγ was shown to inhibit the expression of matrix metalloproteinase 9 (MMP-9), a protease responsible for the degradation of the extracellular matrix and possibly associated with tissue infiltration of CLL cells.42 IFNγ inhibited MMP-9 expression even at very low concentrations (1 U/mL). Thus, in addition to activating STAT1, low concentrations of IFNγ may have antiangiogenic effects in CLL similar to that observed in endothelial cells.43,44
Although inappropriate STAT activation is present in a number of acute and chronic leukemias and may contribute to their pathogenesis,45 Bryostatin 1 activation of STAT1 may be an example in which STAT activation is part of a beneficial therapeutic response. STAT1 is constitutively phosphorylated on serine residues in CLL cells in the absence of tyrosine phosphorylation. Although the nature of this constitutive serine phosphorylation is not well understood, serine-phosphorylated STAT1 is localized in the cytoplasm (data not shown) where STAT proteins can associate with other cytoplasmic proteins.46-48 Serine-phosphorylated STAT1 may be disrupting apoptotic pathways by interacting with other cytoplasmic signaling molecules in an abnormal manner, thereby conferring a survival advantage to the CLL cells. By inducing tyrosine phosphorylation of STAT1, Bryostatin 1 may alter aberrant signaling pathways, for example by interfering with the abnormal signaling behavior of serine-phosphorylated STAT1. Translocation of the dually phosphorylated STAT1 transcription factor to the nucleus, demonstrated by the presence of functionally active STAT1 in nuclear lysates from Bryostatin 1–treated cells (Figure 1B), may then regulate the expression of target genes involved in CLL differentiation. Thus, the STAT1 signaling pathway may be an attractive target for therapeutic intervention in CLL. Agents that induce tyrosine phosphorylation of STAT1, such as Bryostatin 1, may elicit a beneficial therapeutic response. Analysis of STAT1 activation could be included as a possible indicator of therapeutic response in future clinical trials of Bryostatin 1. An association between STAT1 activation and therapeutic response was found in patients who were given an immune-based treatment (CD154 gene therapy) for CLL in a phase 1 clinical trial.49 Patients who received immunotherapy had increased STAT1 activation within 24 hours in their CLL cells, which was associated with 2 beneficial therapeutic endpoints, decreased peripheral blood CLL cells and increased TH1 cytokines.
In summary, the present results demonstrate that the physiologic effects of Bryostatin 1 in CLL cells require the activation of the STAT1 signaling pathway. STAT1 activation is dependent on PKC activation, MAPK activation, and release of IFNγ from the CLL cell. Further studies exploring the role of STAT1 in CLL therapeutic regimens in which Bryostatin 1 is combined with other cytotoxic drugs may contribute to the development of this promising agent.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2002-09-2972.
Supported by the National Institutes of Health (grants CA93053, CA79547, CA81534), the Friends of the Dana-Farber Cancer Institute, and the Brent Leahey Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank John Daley and Suzan Lazo for their assistance with FACS analysis, Dr John Gribben for generously providing peripheral blood from patients with CLL, and Bin Zhang and Denise Scholtens for their assistance with the statistical analysis of the microarray data.