Abstract
Antigens implicated in the graft-versus-leukemia (GVL) effect in chronic myeloid leukemia (CML) include WT1, PR1, and BCR-ABL. To detect very low frequencies of these antigen-specific CD8+ T cells, we used quantitative polymerase chain reaction (qPCR) to measure interferon-γ (IFN-γ) mRNA production by peptide-pulsed CD8+ T cells from HLA-A*0201+ healthy volunteers and from patients with CML before and after allogeneic stem cell transplantation (SCT). Parallel assays using cytomegalovirus (CMV) pp65 tetramers demonstrated the IFN-γ copy number to be linearly related to the frequency of tetramer-binding T cells, sensitive to frequencies of 1 responding CD8+ T cell/100 000 CD8+ T cells. Responses to WT1 and PR1 but not BCR-ABL were detected in 10 of 18 healthy donors. Responses to WT1, PR1, or BCR-ABL were observed in 9 of 14 patients with CML before SCT and 5 of 6 after SCT, often to multiple epitopes. Responses were higher in patients with CML compared with healthy donors and highest after SCT. These antigen-specific CD8+ T cells comprised central memory (CD45RO+CD27+CD57–) and effector memory (CD45RO–CD27–CD57+) T cells. In conclusion, leukemia-reactive CD8+ T cells derive from memory T cells and occur at low frequencies in healthy individuals and at higher frequencies in patients with CML. The increased response in patients after SCT suggests a quantitative explanation for the greater effect of allogeneic SCT.
Introduction
Recent studies have identified a variety of antigens that elicit CD8+ T-cell responses against myeloid leukemias. They include minor histocompatibility antigens such as HA-1 and HA-2,1 overexpressed self proteins such as proteinase-3,2,3 and Wilms tumor (WT1),4 as well as neoantigens created by chromosomal translocations such as BCR-ABL.5-8 All of these antigens have been implicated in a curative graft-versus-leukemia (GVL) effect of allogeneic stem cell transplantation (SCT) for chronic myeloid leukemia (CML).4,9-11 This evidence is based on the occurrence of leukemia antigen-specific cytotoxic T lymphocyte (CTL) expansions exceeding 10% of circulating T cells and coinciding with the onset of durable molecular remissions of CML.10 CD8+ T cells recognizing PR1, WT1, and BCR-ABL have also been found in patients with myeloid leukemias8,12 and can be elicited from healthy individuals by multiple stimulations with peptide.1,2,4,8
The observation that T cells specific for nonpolymorphic leukemia antigens such as PR1, WT1, and BCR-ABL are involved in the GVL response, but are not capable of eradicating CML in the autologous setting, appears at first sight contradictory. The presumed greater efficiency of alloreactive T cells to these antigens is poorly understood; it could relate to either a qualitative or a quantitative property of the cells themselves, or be due to a synergistic function with the allogeneic stimuli. In addition, the finding of autoreactive T cells in patients with CML raises questions about the origin of autoreactivity to PR1, WT1, and BCR-ABL in healthy individuals. Of the 3, BCR-ABL is a neoantigen, arising only after the formation of a Philadelphia chromosome–positive clone. However, the observation that very low frequencies of BCR-ABL positivity occur in healthy individuals raises the possibility that healthy individuals may already have an immune response to BCR-ABL.13,14 To explore further the nature of T cells specific for leukemia antigens, we studied peripheral blood from a series of healthy individuals and patients with CML before and after SCT. The investigation of very low frequencies of circulating T cells required sensitive techniques to permit the direct examination of circulating lymphocytes before their activation status is modified by in vitro expansion. For this purpose we developed a sensitive quantitative polymerase chain reaction (qPCR) technique capable of detecting antigen-specific T cells at frequencies in the order of 1/100 000. Here, we report the widespread occurrence of low frequencies of WT1- and PR1-specific T cells in both patients with CML and in healthy individuals. These CD8+ T cells were interferon-γ (IFN-γ)–producing, antigen-experienced, central memory, and effector memory T cells. The major difference in antigen-specific T cells between healthy donors and patients was in their frequency, which was lowest in healthy donors, higher in patients, and highest after SCT.
Patients, materials, and methods
Patients and healthy controls
All donors and patients were treated at the National Institutes of Health on protocols approved by the NIH Institutional Review Board. After informed consent, cells from patients with CML as well as their HLA-identical healthy donors were obtained from leukapheresis products (LPs) before stem cell transplantation. Peripheral blood mononuclear cells (PBMCs) were obtained from the apheresis products of other unrelated HLAA*0201+, CMV-seropositive and -seronegative healthy volunteers. The cells were separated using Ficoll-Hypaque density gradient centrifugation (Organon Teknika, Durham, NC) and subsequently frozen in RPMI 1640 complete medium (CM), (Life Technologies, Gaithersburg, MD) supplemented with 20% heat-inactivated fetal calf serum (FCS) and 10% dimethyl sulfoxide (DMSO) according to standard protocols. Before use, frozen cells were thawed, washed, and suspended in RPMI-CM + 10% pooled AB serum (Sigma Chemical, St Louis, MO). High-resolution HLA class I genotyping was performed by sequence-specific PCR using genomic DNA (HLA Laboratory, Department of Transfusion Medicine, Warren G. Magnusson Center, NIH, Bethesda, MD). The presence of immunoglobulin G (IgG) and IgM cytomegalovirus (CMV) antibodies in the donors was analyzed by passive latex agglutination (CMVSCAN kit; Becton Dickinson Microbiology System, Cockeysville, MD).
Cell lines
T2 cells (American Type Culture Collection, Manassas, VA) are HLAA*0201+ but express very low levels of surface HLA-A2.1 unless peptide-pulsed and are unable to present endogenous antigens. These cells were maintained in RPMI-CM-10% FCS.
Peptide synthesis
Peptides used in this study were prepared by Bisosynthesis (Lewisville, TX) to a minimum purity of 95%. The identity of each of the peptides was confirmed by mass spectral analysis. The following peptides were tested: PR1 169-177 (VLQELNVTV), derived from the azurophilic granule protein proteinase 3,2 BCR-ABL 922-930 (GFKQSSKAL) from the b3a2 junctional region,8 WT1 126-134 (RMFPNAPYL),4 CMV peptide 495-503 (NLVPMVATV) derived from the immunodominant pp65 protein,15 HIV-1 p17 Gag 77-85 (SLYNTVATL),16 and the synthetically modified (to enhance HLA-A2 binding) gp100 peptide (209-2M) melanoma self-Ag 209-217 (IMDQVPFSV).17
CD8+ T-cell selection
CD8+ T cells were purified from PBMCs of healthy donors and patients using a CD8+ isolation kit (Dynal, Oslo, Norway). Immunomagnetic beads were detached from isolated cells by using DetachaBead (Dynal) with high purity (> 98%) and viability (> 99%). The purity of positively and negatively selected cells was checked by flow cytometry.
RNA extraction and cDNA synthesis
RNA isolation on test samples was performed using RNeasy mini kits (Qiagen, Valencia, CA). Total RNA was eluted with water and stored at –80°C. For reverse transcription of mRNA and cDNA synthesis, 1 μg total RNA was reverse transcribed and stored at –20°C until qPCR was performed.
Quantitative PCR
Gene expression was measured using an ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA) as described previously.18,19 The feasibility of this approach for the analysis of antigen-specific T-cell responses both in peripheral blood lymphocytes and in tumor tissues has been previously validated.20 Primers for CD8 and IFN-γ and TaqMan probes (Custom Oligonucleotide Factory, Foster City, CA) were designed to span exon–exon junctions to prevent amplification of genomic DNA. To create a standard curve, the cDNA was generated by reverse transcription with the same technique used for the preparation of test cDNA. IFN-γ and CD8 cDNA was amplified by PCR using the same primers designed for the reverse transcriptase (RT)–PCR, purified, and quantified by UV spectrophotometry. The number of cDNA copies was calculated by using the molecular weight of each gene amplicon. Serial dilutions of the amplified genes at known concentrations were tested by RT-PCR. Quantitative RT-PCR reactions of cDNA specimens, cDNA standards, and water as negative control (NTC) were conducted in a total volume of 25 μL with TaqMan Master Mix (Applied Biosystems), 400 nM primers, and 200 nM probe. Primer sequences were as follows: IFN-γ (forward), 5′-AGCTCTGCATCGTTTTGGGTT; IFN-γ (reverse), 5′-GTTCCATTATCCGCTACATCTGAA; IFN-γ (probe), FAM-TCTTGGCTGTTACTGCCAGGACCCA-TAMRA; CD8 (forward), 5′-CCCTGAGCAACTCCATCATGT; CD8 (reverse), 5′-GTGGGCTTCGCTGGCA; and CD8 (probe), FAM-CAGCCACTTCGTGCCGGTCTTC-TAMRA. Thermal cycler parameters included 10 minutes at 95°C and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Standard curve extrapolation of copy number was performed for both IFN-γ and CD8. Sample data were normalized by dividing the number of copies of IFN-γ transcripts by the number of copies of CD8 transcripts. All PCR assays were performed in duplicates and reported as the mean. A 2-fold difference in gene expression was found to be within the discrimination ability of the assay.
Direct PCR assay of peptide-specific CD8+ T-cell reactivity
To screen for peptide-specific CD8+ T cells, we measured IFN-γ mRNA production by CD8+ T cells stimulated with candidate peptides. Preliminary experiments to optimize assay conditions were performed with CD8+ T cells obtained from HLA-A*0201+, CMV-seropositive donors, stimulated with HLA-A*0201–associated CMV peptide pp65 (495-503) pulsed T2 cells. After isolation, 1 × 106 CD8+ T cells were plated per well in a 96-well flat-bottom plate with 200 μL RPMI-CM supplemented with 10% human AB serum and incubated overnight at 37°C (humidity, 90%; CO2, 5%) to minimize background expression of IFN-γ mRNA because of lymphocyte manipulation. CD8+ T cells were then stimulated in vitro with test peptides using a protocol adapted from previous studies.2 In brief, T2 cells were washed in serum-free medium and incubated with individual peptides at concentrations of 0.1, 1, and 10 μM at 37°C in 5% CO2 for 2 hours. The peptide-pulsed T2 cells were then irradiated with 7500 cGy, washed once, and added to CD8+ T cells. Control wells contained CD8+ T cells in the presence of unloaded T2 cells. After 3 hours of coculture at 37°C in 5% CO2, cells were harvested for RNA isolation and cDNA transcription. qPCR was performed for IFN-γ mRNA expression and normalized to copies of CD8 mRNA from the same sample. Additional negative controls included HLAA*0201-negative individuals, CMV-negative HLA-A*0201+ individuals, and T2 cells pulsed with gp100 (209-2M) peptide. Replicate tests on aliquots of a single-cryopreserved apheresis sample from 3 different donors showed a coefficient of variation of 6.9% to 20.8%.
Production of peptide–MHC class I tetrameric complexes
Biotin-tagged HLA-A*0201 heavy chains and β2-microglobulin (β2m) were expressed as insoluble inclusion bodies in Escherichia coli strain BL21(DE3)pLysS as described previously.21 Inclusion bodies were released by repeated freeze/thaw cycles and purified by washing with 0.5% Triton X-100 buffer (Sigma). Soluble biotinylated peptide–major histocompatibility complex class I (pMHCI) monomers were produced as described previously with minor modifications.21,22 Briefly, HLA-A*0201 heavy chain and β2m inclusion body preparations were denatured separately in 8 M urea buffer and refolded at a 1:1 molar ratio in 2-mercaptoethylamine/cystamine redox buffer (Sigma) with added synthetic peptide (BioSynthesis). Individual complexes were produced with the following peptides: (1) NLVPMVATV (CMV), (2) VLQELNVTV (PR1), (3) RMFPNAPYL (WT1), and (4) SLYNTVATL (HIV-1 Gag). After buffer exchange into 10 mM Tris (tris(hydroxymethyl)aminomethane), pH 8.1, refolded monomers were purified by anion exchange and then biotinylated as described previously using d-biotin and BirA enzyme.23 Excess biotin was removed by gel filtration. Biotinylated monomers were conjugated by addition of fluorochrome-labeled streptavidin (ProZyme, San Leandro, CA) at a pMHC-to-streptavidin molar ratio of 4:1 to produce tetrameric complexes. Once prepared, tetramers were stored in the dark at 4°C.
Flow cytometry
CMV tetramer-phycoerythrin (PE) and HIV1 Gag tetramer-PE were used as positive and negative controls, respectively. Sample staining was performed using 3 × 106 PBMCs in 50 μL 1% FCS/PBS (phosphate-buffered saline). Tetramers (1-2 μg per test with respect to the peptide–MHC class I component) were added for 20 to 30 minutes at 37°C. Cells were washed once in PBS and 1% FCS and then stained with a titrated panel of directly conjugated antibodies to CD4, CD19, CD14, CD16, CD57, CD8, CD27 (all PharMingen, San Jose, CA), CD45RO (Dako, Carpinteria, CA), and CD3 (Coulter, Miami, FL). Alexa 430, fluorescein isothiocyanate (FITC), Texas-Red PE, PE, Cy5PE, Cy7PE, and allophycocyanin (APC) were used as fluorophores. The lymphocytes were then washed in PBS, 0.5 mM EDTA (ethylenediaminetetraacetic acid), 1% BSA, and resuspended in 1% paraformaldehyde in PBS. Data acquisition was performed with FACSDiva Calibur (BD/Pharmingen, San Jose, CA). A minimum of 1.5 × 106 gated cells were acquired. Data were analyzed using FlowJo software (TreeStar, San Carlos, CA). Intracellular cytokine detection was performed as described previously.24 In brief, positively selected CD8+ T cells (1 × 106) were incubated with T2 cells loaded without or with peptide at varying doses. After 2 hours, 10 μg Brefeldin A (Sigma) was added, and after 16 additional hours, CD8+ T cells were stained using CD8 peridinin chlorophyll protein (PerCP), and fix/permeablized followed by staining with IFN-γ FITC (all BD/Pharmingen).
Statistical analysis
Evidence of a specific response to stimulation, as determined by qPCR studies, consisted of detection of mRNA for IFN-γ in CD8+ T cells stimulated with relevant peptide versus unloaded antigen-presenting cells (background). A cutoff value of 2.0 for the ratio of IFN-γ mRNA (corrected for CD8 mRNA) obtained from CD8+ T cells stimulated with relevant epitope to that obtained from CD8+ T cells stimulated with unpulsed APC was considered to be evidence of epitope specificity. The cutoff value was derived by analyzing IFN-γ mRNA transcripts detectable in CD8+ T cells from both healthy donors and patients with CML stimulated with gp100 (209-2M) (irrelevant peptide) compared with background. Analysis of these CD8+ T cells identified a mean ratio of 0.96 (range, 0.8-1.7) with 95% and 99% confidence intervals of 0.96 ± 0.76 and 0.96 ± 1.16, respectively, a standard error of 0.09, and a standard deviation of 0.28. The cutoff ratio (stimulation index) was estimated by adding the mean to 3 standard deviations, which equaled 1.8. To minimize the possibility of falsely considering CD8+ T cells immunoreactive, we accepted a 2-fold increase in stimulated-unstimulated IFN-γ transcript ratio as evidence of epitopespecific reactivity.
Wilcoxon's rank sum test was calculated to determine whether there was a statistically significant difference in IFN-γ production in response to test peptides between patients with CML and healthy controls. Statistical significance was achieved when P values were less than .05.
Results
Identification of leukemia peptide-specific CD8+ T cells in healthy individuals and patients with CML
To determine whether PR1, WT1, and BCR-ABL peptide-specific CD8+ T cells exist in healthy donors and patients with CML, we looked for IFN-γ mRNA production by qRT-PCR in antigen-stimulated CD8+ T cells from 18 HLA-A*0201+ healthy donors and 14 patients with CML. As controls, CD8+ T cells were also stimulated with CMV pp65 (positive control) and gp100 (209-2M) (negative control) peptides. A positive response required a threshold of 100 or more IFN-γ mRNA copies/104 CD8 copies and a stimulation index (SI) of 2 or more, where SI = IFN-γ mRNA copies/104 CD8 copies in peptide-pulsed T2 cell cultures/unpulsed cultures.
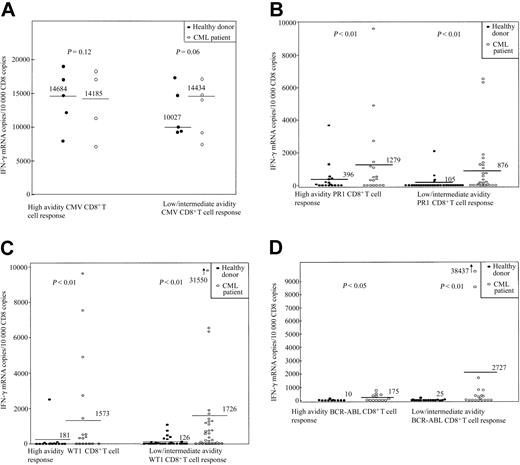
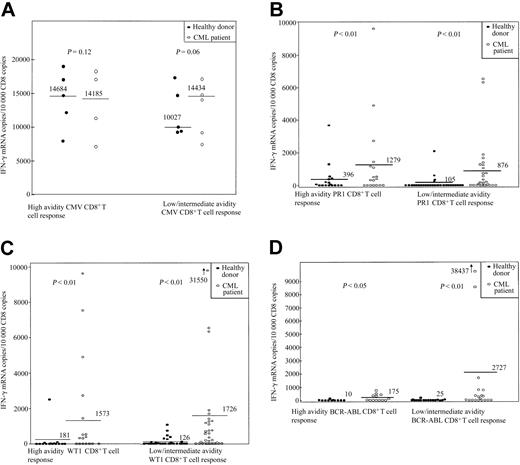
Responses to leukemia antigens were detected in 10 of 18 healthy individuals; the range of IFN-γ mRNAcopies/104 CD8 copies was 145 to 3688 (SI, 2-1298). Five donors responded to stimulation with PR1 (donors 1, 4, 5, 17, and 18), 3 to WT1 (donors 7, 8, and 9), and 2 to both WT1 and PR1 (donors 2 and 3). None responded to BCR-ABL or to gp100 (209-2M) peptide (Table 1; Figure 1).
CD8+ T-cell response to stimulation with the HLAA*0201–restricted peptides CMV pp65, PR1, WT1, and BCR-ABL in 18 healthy donors and 14 patients with CML. (A) CMV pp65. (B) PR1. (C) WT1. (D) BCR-ABL. CD8+-selected T cells were incubated for 3 hours either with unpulsed APC or with APC pulsed with 3 doses of peptide (0.1, 1, and 10 μM). Values represent copies of IFN-γ mRNA per 104 copies of CD8 mRNA. Because of limitation in the amount of PBMCs available, the intermediate dose testing was omitted in certain cases, and the data on intermediate and low avidity are presented together. The CD8+ T-cell response to stimulation with each particular peptide was calculated by subtraction of IFN-γ mRNA copies/104 CD8 copies induced by unpulsed APC (background) from that induced by peptide-pulsed APC. Values more than 100 IFN-γ mRNA copies per 104 copies of CD8 and at least 2 times that of background were defined as positive responses. Bars represent the median number of IFN-γ mRNA copies/104 CD8 copies for each condition. Responses to stimulation with PR1 and WT1 were signifi-cantly higher in the CML group. Responses to stimulation with BCR-ABL were detectable only in the CML group.
CD8+ T-cell response to stimulation with the HLAA*0201–restricted peptides CMV pp65, PR1, WT1, and BCR-ABL in 18 healthy donors and 14 patients with CML. (A) CMV pp65. (B) PR1. (C) WT1. (D) BCR-ABL. CD8+-selected T cells were incubated for 3 hours either with unpulsed APC or with APC pulsed with 3 doses of peptide (0.1, 1, and 10 μM). Values represent copies of IFN-γ mRNA per 104 copies of CD8 mRNA. Because of limitation in the amount of PBMCs available, the intermediate dose testing was omitted in certain cases, and the data on intermediate and low avidity are presented together. The CD8+ T-cell response to stimulation with each particular peptide was calculated by subtraction of IFN-γ mRNA copies/104 CD8 copies induced by unpulsed APC (background) from that induced by peptide-pulsed APC. Values more than 100 IFN-γ mRNA copies per 104 copies of CD8 and at least 2 times that of background were defined as positive responses. Bars represent the median number of IFN-γ mRNA copies/104 CD8 copies for each condition. Responses to stimulation with PR1 and WT1 were signifi-cantly higher in the CML group. Responses to stimulation with BCR-ABL were detectable only in the CML group.
Clinical data for the patients with CML are shown in Tables 2 and 3. In 9 of 14 patients before SCT, a CD8+ T-cell response to WT1, PR1, or BCR-ABL was observed, with a range of IFN-γ mRNA copies/104 CD8 copies of 171 to 6337 (SI, 2-615) (Table 1; Figure 1). Four patients had a positive response to stimulation with both WT1 and PR1 peptides (UPNs 17, 38, 41, 199). Two responded to stimulation with all 3 peptides (UPNs 210 and 254). A CMV-seronegative patient had a positive response to stimulation with WT1, PR1, and BCR-ABL but not to CMV (UPN 210). One patient had a response to PR1 and BCR-ABL (UPN 283), and 2 patients responded to PR1 or WT1 (UPNs 25 and 181). Six patients were tested 3 to 36 months after SCT, all of whom had achieved 100% donor T-cell chimerism at analysis. Five patients had a response to stimulation with one or more peptides. One patient responded to both PR1 and WT1 (UPN 210); 1 to WT1 and BCR-ABL (UPN 327 at day 100); 2 to BCR-ABL alone (UPN 223 and UPN 327 when retested at day 360); and 2 to PR1, WT1, and BCR-ABL (UPNs 181 and 319). Unfortunately, not enough material was available to test all 3 peptide doses in the patient who did not mount a response (UPN 283). In the case of patients UPN 181, 283, 319, and 327, samples before and after SCT and from their respective sibling donors (healthy donors 6, 16, 2, and 18) were available for analysis. For patient 223 material was available after SCT and from his donor (donor 17). Patient 319 had no response to stimulation with PR1, WT1, and BCR-ABL prior to his allogeneic SCT. His sibling donor showed substantial CD8+ T-cell reactivity to stimulation with PR1 and to a lesser extent with WT1 but not BCR-ABL (Table 1). After SCT, the patient had significant responses to stimulation with PR1, WT1, and BCR-ABL, suggesting the transfer of PR1- and WT1-specific CD8+ T cells from donor to recipient. Patient 283 was CMV seropositive before SCT with a CMV-seronegative donor. After SCT he lost the ability to mount a CD8+ T-cell response to stimulation with the CMV peptide. Donors 17 and 18 had activity to stimulation with PR1, although after SCT no PR1 activity could be detected in their respective recipients. Patient 327 was tested at 2 different time points: 100 and 360 days after transplantation. At day 100 he had a significant CD8+ response to stimulation with a high dose of WT1 and smaller BCR-ABL responses. By day 360 the WT1 response had disappeared, whereas the BCR-ABL response had expanded. These data suggest that pre-existing leukemia-specific donor CD8+ T cells can expand in the recipient after SCT, although formal clonotype analysis would be required for conclusive demonstration of this phenomenon; however, leukemia-specific CD8+ T-cell responses that were not detectable in the donor can also develop de novo in the recipient.
In individuals with leukemia the response to stimulation with PR1 and WT1 peptides was increased compared with healthy donors (P < .01). The highest amplitude responses occurred after SCT (390-38 437 IFN-γ mRNA copies/104 CD8 copies; SI, 2.0-9645). Control HLA-A*0201– CMV-seropositive and HLAA*0201+ CMV-seronegative samples stimulated with pp65 (495-503)– and gp100 (209-2M)–pulsed T2 cells were used in each assay run. These controls were consistently negative for IFN-γ production (data not shown).
To further ascertain that the leukemia antigen response was indeed specific to the disease, we studied 5 patients with solid tumors and 6 patients with other hematopoietic tumors (Table 4). None of the 5 patients with solid tumors mounted a CD8+ T-cell response to PR1, WT1, or BCR-ABL. One patient with secondary acute myeloblastic leukemia (AML) mounted a response to all doses of PR1 and WT1 peptides tested, but not to BCR-ABL (Table 4). Therefore, we were able to ascertain that the antigen response is indeed appropriate for the presence of the tumor antigen. Given that more than 50% of the healthy donors have responses to PR1 and/or WT1, we would expect a similar proportion of individuals with illnesses unrelated to CML (or AML), such as those with solid tumors, to respond in a similar way, but in our series this was not observed and may be explained by suboptimal sample size.
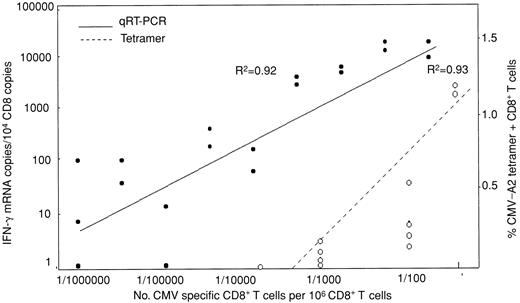
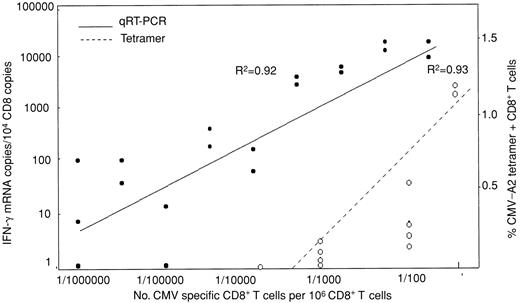
To validate the assumption that IFN-γ mRNA transcription reflects the frequency of peptide-specific CD8+ T cells and IFN-γ protein secretion, samples assayed for CMV pp65 (495-503) response by qRT-PCR were compared with flow cytometric quantification of CD8+ T cells specific for the peptide by tetramer or intracellular IFN-γ (IC-IFN-γ) production. In 6 subjects assayed for IC-IFN-γ and 7 for CMV tetramer, there was a strong correlation of IFN-γ mRNA copies with IC-IFN-γ and tetramer-positive T cells (R2 = 0.92 and 0.78, respectively) (data not shown). To define linearity and limit of detection of the qRT-PCR assay, CD8+ T cells from CMV-seropositive individuals were stimulated with peptide-loaded T2 cells and diluted into autologous unstimulated CD8+ cells. The tetramer technique was used to calibrate the number of CMV-specific T cells in the dilution. In this way the lower limit of detection by qRT-PCR was found to be 1 CMV-specific CD8+ T cell/100 000 CD8+ T cells, equivalent to approximately 100 IFN-γ mRNA copies. The same dilutions assayed by tetramer showed a lower limit of detection of 1/10 000 (Figure 2).
CD8+ T cells (106) were stimulated with pp65 CMV peptide-pulsed T2 cells for 3 hours and then diluted logwise into unstimulated, autologous CD8+ T cells. The number of CMV-specific CD8+ T cells in the starting material determined by tetramer assay was used to calibrate CMV-specific CD8+ T cells in each dilution and correlate this with the number of IFN-γ mRNA copies such that the lowest concentration contained one CMV-positive CD8+ T cell/106 nonstimulated CD8+ T cells. RNA was then extracted for qRT-PCR. The lower limit of detection by the tetramer and PCR assay was 1/10 000 and 1/100 000 CMV-specific CD8+ T cells, respectively.
CD8+ T cells (106) were stimulated with pp65 CMV peptide-pulsed T2 cells for 3 hours and then diluted logwise into unstimulated, autologous CD8+ T cells. The number of CMV-specific CD8+ T cells in the starting material determined by tetramer assay was used to calibrate CMV-specific CD8+ T cells in each dilution and correlate this with the number of IFN-γ mRNA copies such that the lowest concentration contained one CMV-positive CD8+ T cell/106 nonstimulated CD8+ T cells. RNA was then extracted for qRT-PCR. The lower limit of detection by the tetramer and PCR assay was 1/10 000 and 1/100 000 CMV-specific CD8+ T cells, respectively.
CD8+ T-cell response to different peptide concentrations as a measure of functional avidity
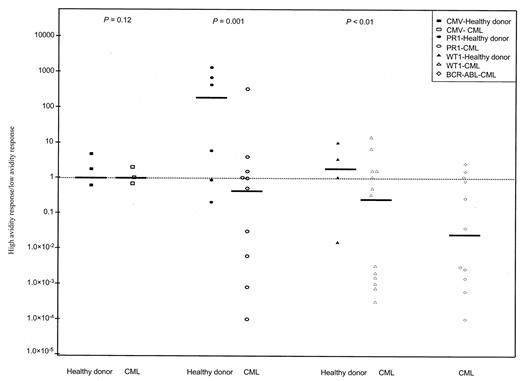
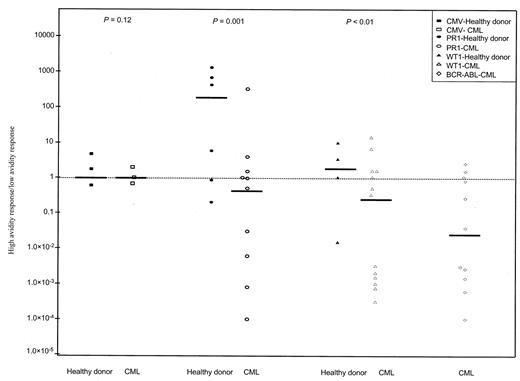
To determine functional avidity, the response of CD8+ T cells to stimulation with 3 concentrations of peptide (0.1, 1, and 10 μM) was measured by qRT-PCR. High- and low-avidity T cells have been previously shown to possess different requirements for both peptide/MHC density and CD8 interaction.25,26 In our experiments, high-avidity CD8+ T cells were defined as those capable of producing IFN-γ in response to a lower concentration of peptide (0.1 μM), whereas intermediate- and low-avidity CD8+ T cells were those that produced IFN-γ in response to a higher concentration of peptide (1 and 10 μM, respectively). To examine the issue of functional avidity, we determined the ratio of high- to lowavidity CD8+ T-cell responses in healthy donors and patients with CML by measuring the number of IFN-γ mRNA copies/104 CD8 copies produced by cells stimulated with low- or higher-peptide concentrations, respectively (Figure 3). We observed that in patients with CML, low-avidity CD8+ T-cell responses to PR1, WT1, and BCR-ABL were more abundant than high-avidity responses (median high-low avidity ratios of 0.73, 0.32, and 0.04, respectively). In contrast, in healthy donors, CD8+ T-cell responses to PR1 and WT1 were mostly high avidity (median ratios, 214 and 2.2, respectively). The difference in response in the 2 groups was statistically significant (P = .01 and P < .05, respectively). In contrast, high- and low-avidity CMV-specific CD8+ T-cell responses were equally represented and were not statistically signifi-cantly different in the 2 groups (P = .12).
High- and low-avidity CD8+ T-cell responses determined by sensitivity to peptide concentration in healthy donors and patients with CML. Stimulating CD8+ T cells with 0.1 μM and 10 μM CMV pp65 (□), PR1 (○), WT1 (▵), and BCR-ABL (⋄) determined high- and low-avidity responses, respectively. Results shown are the ratios of high- to low-avidity CD8+ T-cell responses, calculated for individual healthy donors (filled symbols) and patients with CML (open symbols). Ratios were obtained by the following calculation: (number of IFN-γ mRNA copies/104 CD8 copies with 0.1 μM peptide)/(number of IFN-γ mRNA copies/104 CD8 copies with 10 μM peptide). Bars represent the median high- and low-avidity ratio for each condition. CD8+ T-cell responses to PR1, WT1, and BCR-ABL in patients with CML were mostly low avidity, whereas CD8+ T-cell responses in healthy donors were skewed toward high-avidity responses (P = .01 and P < .05, respectively). CMV responses were not statistically different in the 2 groups (P = .12). Dashed line represents a median ratio of 1.
High- and low-avidity CD8+ T-cell responses determined by sensitivity to peptide concentration in healthy donors and patients with CML. Stimulating CD8+ T cells with 0.1 μM and 10 μM CMV pp65 (□), PR1 (○), WT1 (▵), and BCR-ABL (⋄) determined high- and low-avidity responses, respectively. Results shown are the ratios of high- to low-avidity CD8+ T-cell responses, calculated for individual healthy donors (filled symbols) and patients with CML (open symbols). Ratios were obtained by the following calculation: (number of IFN-γ mRNA copies/104 CD8 copies with 0.1 μM peptide)/(number of IFN-γ mRNA copies/104 CD8 copies with 10 μM peptide). Bars represent the median high- and low-avidity ratio for each condition. CD8+ T-cell responses to PR1, WT1, and BCR-ABL in patients with CML were mostly low avidity, whereas CD8+ T-cell responses in healthy donors were skewed toward high-avidity responses (P = .01 and P < .05, respectively). CMV responses were not statistically different in the 2 groups (P = .12). Dashed line represents a median ratio of 1.
Phenotypic analysis of CMV-, PR1-, and WT1-specific CD8+ T cells
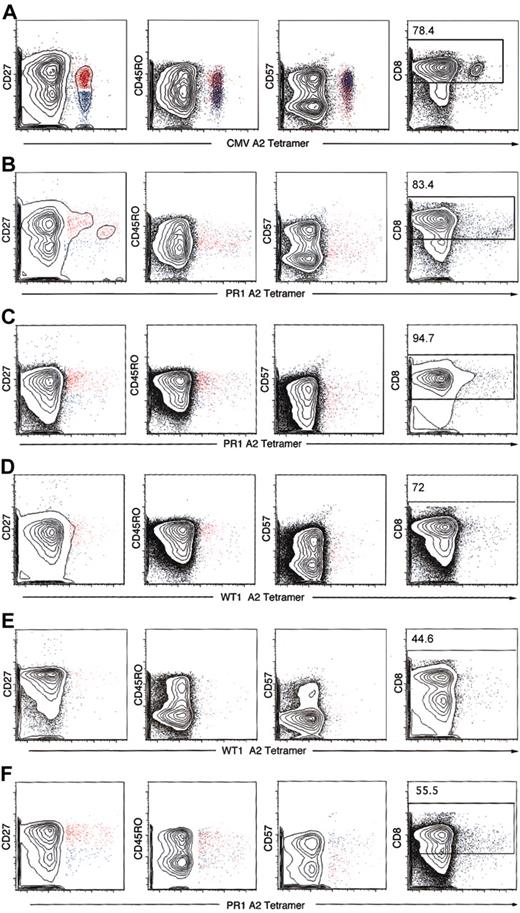
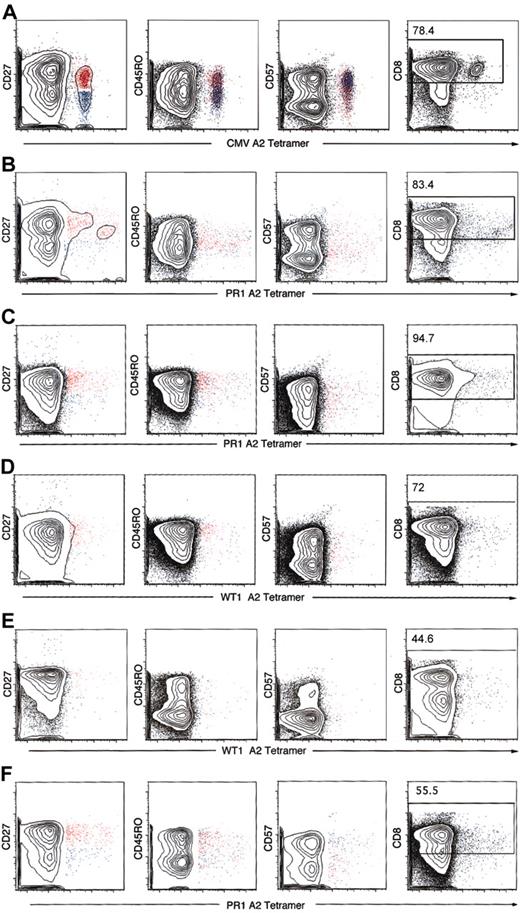
Antigen-specific CD8+ T cells selected by HLA-A*0201 tetramers were analyzed for expression of CD45RO, CD27, and CD57 for characterization of naive, memory, and effector phenotype. In 2 healthy donors, the frequencies of PR1 and WT1 tetramer-positive cells were high enough to permit phenotypic characterization (Figure 4). Because of technical difficulties with the synthesis and quality of the HLA-A*0201 BCR-ABL tetramer, we were unable to study the phenotype of the BCR-ABL–specific CD8+ T cells. CMV tetramer-specific CD8+ T cells had a predominant phenotype of effector memory cells and a minor population of central memory T cells (Figure 4A). In contrast, in healthy donors and patients with CML, PR1, and WT1 tetramer-specific CD8+ T cells displayed 2 phenotypes: central memory (CD45RO+CD27+CD57–) and terminally differentiated effector memory (CD45RO–CD27–CD57+) (Figure 4 and Table 5) with a predominant phenotype of central memory. After stem cell transplantation, a shift toward more effector memory phenotype was noted, probably implying an ongoing GVL effect.
Phenotypic characterization of tetramer-positive CD3+CD8+ T cells. Analysis of PBMCs was performed by 6-color flow cytometry in patients with CML patients before and after SCT and 2 healthy donors whose CD8+ T-cell frequencies to PR1 and WT1 were high enough to be visualized by tetramer staining. CD45RO, CD27, and CD57 phenotype of CD3/CD8-gated tetramer-positive lymphocytes was analyzed. Cells were gated on CD3+ lymphocytes, and the number in the top subpanel represents the percentage of CD3+ T cells that express CD8. Most of the CMV tetramer CD8+ T cells were CD45RO–CD27–CD57+ (blue), consistent with an effector memory phenotype (A). PR1 and WT1 tetramer-positive CD3+CD8+ T cells in 2 patients with CML before SCT and 2 patients after allogeneic SCT showed a mixture of CD45RO+CD27+CD57– (red) and CD45RO–CD27–CD57+ (blue), with most of the tetramer-gated cells showing the former phenotype (B-C). The same held true when PR1 and WT1 tetramer-positive T cells in 2 healthy donors were studied. Representative data are presented here. (A) CMV-positive control (UPN 283). (B) Patient with CML before SCT PR1 (UPN 210). (C) Patient with CML after SCT PR1 (UPN 319). (D) Patient with CML after SCT WT1 (UPN 319). (E) Healthy donor (UPN 12) WT1. (F) Healthy donor (UPN 2) PR1.
Phenotypic characterization of tetramer-positive CD3+CD8+ T cells. Analysis of PBMCs was performed by 6-color flow cytometry in patients with CML patients before and after SCT and 2 healthy donors whose CD8+ T-cell frequencies to PR1 and WT1 were high enough to be visualized by tetramer staining. CD45RO, CD27, and CD57 phenotype of CD3/CD8-gated tetramer-positive lymphocytes was analyzed. Cells were gated on CD3+ lymphocytes, and the number in the top subpanel represents the percentage of CD3+ T cells that express CD8. Most of the CMV tetramer CD8+ T cells were CD45RO–CD27–CD57+ (blue), consistent with an effector memory phenotype (A). PR1 and WT1 tetramer-positive CD3+CD8+ T cells in 2 patients with CML before SCT and 2 patients after allogeneic SCT showed a mixture of CD45RO+CD27+CD57– (red) and CD45RO–CD27–CD57+ (blue), with most of the tetramer-gated cells showing the former phenotype (B-C). The same held true when PR1 and WT1 tetramer-positive T cells in 2 healthy donors were studied. Representative data are presented here. (A) CMV-positive control (UPN 283). (B) Patient with CML before SCT PR1 (UPN 210). (C) Patient with CML after SCT PR1 (UPN 319). (D) Patient with CML after SCT WT1 (UPN 319). (E) Healthy donor (UPN 12) WT1. (F) Healthy donor (UPN 2) PR1.
Discussion
This study provides direct evidence that memory CD8+ T-cell responses recognizing several leukemia-associated self-antigens exist at low frequencies in healthy individuals and to a greater extent in patients with CML. Using qRT-PCR, we found that CD8+ T cells recognizing leukemia-associated antigens WT1 and PR1 are present in low frequencies in more than 50% of healthy individuals. In contrast, none had a response to the BCR-ABL junctional peptide. The presence of such CD8+ T cells in healthy subjects confirms WT1 and PR1 to be self-antigens and concords with the ability to elicit PR1- and WT1-specific CTLs from healthy individuals by repeated antigen stimulation,3,4 the finding of antibodies to WT1 in up to 16% of healthy subjects,27,28 and the occurrence of proteinase-3–specific antibody and T-cell responses in most healthy individuals and in patients with Wegener granulomatosis.29-31 Indeed, natural antibodies reacting with a variety of self-antigens have been detected in serum of healthy individuals.32 Nevertheless, the frequency of WT1- and PR1-specific CD8+ T cells in our study was extremely low and (with 2 exceptions) below the limit of detection using tetramers. Thus, WT1 and PR1 appear to behave as tissue- but not leukemia-specific antigens in the same category as antigens identified in solid tumors.33 The absence of a response to BCR-ABL suggested that at least in HLA-A*0201–positive individuals this particular BCR-ABL sequence was not antigenic. Although this finding could indicate that BCR-ABL is not antigenic because it is a true neoantigen absent from the normal antigenic environment, we cannot exclude the possibility that antigenic BCR-ABL sequences exist in the peripheral T-cell repertoire in individuals of other HLA types.
Although the frequencies of responses to CMV antigen were comparable in healthy subjects and in patients, we found that CD8+ T cells to WT1, PR1, and BCR-ABL circulate in patients with CML at significantly higher frequencies. Even higher frequencies to some of these antigens (> 0.5%) were found after allogeneic SCT, suggesting that these leukemia antigen-associated CD8+ T cells expand in the recipient after transplantation and contribute to remission. Responses to BCR-ABL occurred in 3 of 9 patients before and 5 of 6 patients after SCT. In all, 7 of 14 patients before SCT and 4 of 6 after SCT had responses to multiple tumor antigens. In contrast with previous reports10 PR1-specific CD8+ T cells were detectable even in patients not treated with interferon-α. This finding may be explained by the higher sensitivity of the qRT-PCR assay compared with tetramer staining.
The qRT-PCR IFN-γ mRNA assay allowed for the identification of very low frequencies of circulating antigen-specific CD8+ T cells following brief in vitro exposure to candidate peptides. This assay showed a linear relationship between IFN-γ mRNA copy number and peptide-specific CD8+ T-cell frequencies. With the use of a CMV pp65 peptide, the IFN-γ copy number detected by qRT-PCR correlated well with IFN-γ protein production by IC staining (R2 = 0.98) and with antigen-specific CD8+ T cells measured by tetramer staining (R2 = 0.78). Furthermore, we demonstrated that qRT-PCR was at least 10 times more sensitive in the detection of CMV-specific T cells than tetramer staining. qRT-PCR has the advantage over other techniques of being faster, more sensitive, and requiring fewer cells. Moreover, because cell expansion is not required to detect T-cell reactivity, the method allows the detection of functional antigen-specific T cells unmodified in frequency or functional state by in vitro expansion.
In addition to quantitative differences in antigen-specific T cells between healthy subjects and patients with CML, we sought qualitative differences in T-cell responses between patients and healthy donors. Using 3 peptide concentrations, we characterized functional avidity by measuring the ability of CD8+ T-cell populations to respond to the stimulation provided by 3 logarithmically different peptide concentrations.34 We detected both high- and low-avidity antigen-specific CD8+ T cells in healthy donors and patients with CML irrespective of their transplantation status. Patients showed an increase in low-avidity CD8+ T-cell responses in both autologous and allogeneic settings compared with healthy donors, and the difference was statistically significant. These results suggest that the repertoire of leukemia antigen-specific CD8+ T cells is diverse both in terms of clonal composition and efficiency of peptide recognition. This observation is further supported by a recent paper by Molldrem et al35 who showed that distinct high- or low-avidity PR1 CTLs can be expanded from the peripheral circulation of healthy individuals. However, only lowavidity CTLs were detected in the peripheral blood of patients with CML, and it was not possible to elicit high-avidity PR1 CTLs from untreated patients with CML. The tendency for a preponderance of low-avidity CD8+ T cells in patients with CML may be explained by the loss of high-avidity CD8+ T cells by apoptosis following exposure to supra-optimal antigen density on leukemic antigen-presenting cells. When a malignant cell overexpressing tumorassociated self-antigens expands, high-avidity T cells with specificity for those tumor antigens might be selectively eliminated over time through clonal deletion. This is similar to the process of clonal exhaustion of high-avidity CD8+ T cells that occurs during viral infection,36 also shown for CD4+ cells.37
The finding of low frequencies of IFN-γ–producing WT1- and PR1-specific CD8+ T cells in healthy individuals suggested that they belonged to an antigen-experienced memory cell population, because naive T cells characteristically produce cytokines other than IFN-γ.38 To better characterize the functional status of these cells in patients and healthy individuals, we used multiparametric flow cytometry to study the surface phenotype of PR1 and WT1 tetramer-specific CD8+ T cells. Two healthy donors had suffi-ciently high frequencies of PR1- and WT1-specific CD8+ T cells to allow the study of their functional phenotype. Overlapping markers such as CD45RA, CD45RO, CD28, CD27, CD57, and CCR7 have been used to identify the differentiation state of antigen-specific CD8+ T cells.38-41 On the basis of CD45RO, CD27, and CD28 expression and analysis of the replicative history and clonality of the T-cell populations,42 phenotypically distinct and sequential stages of CD8+ T-cell differentiation have been proposed.43 We found that, in both patients and healthy subjects, leukemia-reactive CD8+ T cells displayed a similar mixed CD45ROdim/CD27–/CD57+ and CD45RObright/CD27+/CD57– phenotype, corresponding to terminally differentiated effector memory and central memory phenotypes, respectively. Following allogeneic transplantation, there was a shift toward more effector phenotype, implying an ongoing graft-versus-leukemia effect.
Taken together, our data confirm the presence of circulating CD8+ T cells recognizing several leukemia-associated antigens both in CML patients before and after transplantation and in healthy subjects. The finding of CD8+ T cells specific for up to 3 antigens in patients with CML suggests that the leukemia presents a number of epitopes that could render it susceptible to T-cell attack, for example in GVL responses. IFN-γ production and surface phenotype confirmed leukemia antigen-reactive CD8+ T cells to be memory cells distributed between the central memory and effector memory pools. The presence of terminally differentiated effectors suggests a persisting antigenic stimulus, even in healthy individuals, as also occurs in chronic infections with CMV, Epstein-Barr virus (EBV), and HIV.44 Although there were significant differences in the functional avidity of CD8+ T-cell responses between patients and healthy donors, and a tendency toward more effector CD8+ T cells following allogeneic transplantation, the major difference was the much higher frequency of leukemia antigen-specific CD8+ T cells in patients with CML and the occurrence of BCR-ABL–specific CD8+ T cells only in these individuals. It is thus possible that BCR-ABL represents a true leukemia neoantigen absent from the antigenic environment of healthy individuals. In this study differences in antigen-specific CD8+ T-cell frequencies were the only explanation reconciling the apparent failure of an antileukemic effect in patients with CML not receiving transplants, with the presumed success of functionally similar but higherfrequency T cells exerting a GVL effect after transplantation. The implication of this finding is that increasing the T-cell frequency, for example, by peptide vaccination, may be an important goal when using immunotherapy as treatment for CML. Furthermore, it should be feasible to boost existing memory T-cell responses to leukemia antigens both in patients and in healthy stem cell donors.
Prepublished online as Blood First Edition Paper, June 26, 2003; DOI 10.1182/blood-2003-01-0150.
Supported by grants from the Leukaemia Research Fund, United Kingdom, and the Medical Research Council, United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Simone Mocellin for technical advice, Mrs Leslie Wehrlen for providing patient samples and data, and Professor John Goldman for his support and encouragement.