Abstract
Both clinical and experimental evidence illustrate that p190 and p210 BCR/ABL oncogenic tyrosine kinases induce resistance to DNA damage and confer an intrinsic genetic instability. Here, we investigated whether BCR/ABL expression could modulate nucleotide excision repair (NER). We found that ectopic expression of p210 BCR/ABL in murine lymphoid BaF3 cell line inhibited NER activity in vitro, promoting hypersensitivity of these cells to ultraviolet (UV) treatment and facilitating a mutator phenotype. However, expression of p210 BCR/ABL in human and murine myeloid cell lines and primary bone marrow cells resulted in the increased NER activity and resistance to UV irradiation. The ABL tyrosine kinase inhibitor STI571 reversed these effects, showing that p210 BCR/ABL tyrosine kinase activity is responsible for deregulation of NER. Hypoactivity of NER in p210 BCR/ABL-positive lymphoid cells was accompanied by the decreased interaction between proliferating cell nuclear antigen (PCNA) and xeroderma pigmentosum group B (XPB); conversely, this interaction was enhanced in p210 BCR/ABL-positive myeloid cells. p190 BCR/ABL did not affect NER in lymphoid and myeloid cells. In summary, our study suggests that p210 BCR/ABL reduced NER activity in lymphoid cells, leading to hypersensitivity to UV and mutagenesis. In contrast, p210 BCR/ABL expression in myeloid cells facilitated NER and induced resistance to UV. (Blood. 2003;102:2632-2637)
Introduction
BCR/ABL oncogenic tyrosine kinase is derived from translocation of the c-abl gene on chromosome 9 to the bcr locus on chromosome 22 (t(9;22); Philadelphia chromosome [Ph]1) and is present in essentially all cases of chronic myelogenous leukemia (CML) and a cohort of patients with acute lymphocytic leukemia (ALL).1,2 The translocation may result in p190, p210, and p230 BCR/ABL kinases, depending on the position of the bcr breakpoint.3
BCR/ABL oncogenic tyrosine kinase exhibits 2 complementary roles in cancer. The first and best known is stimulation of signaling pathways that render cells independent of their environment. BCR/ABL allows cells to proliferate in the absence of growth factors, protects them from apoptosis in the absence of external survival factors, and promotes invasion and metastasis.4 The second role of BCR/ABL in hematologic malignancies, which is only just beginning to be fully recognized, is that it can render cells resistant to genotoxic therapies.5-12 Treatment of CML with chemotherapy leads to a transient reduction of the number of leukemic cells.13 However, the leukemic cells usually recover more quickly than normal counterparts. Therefore, these therapies do not mediate durable remissions in patients with CML. In addition, high-dose chemotherapy treatment combined with stem cell transplantation does not eradicate the disease in most patients as indicated by the poor efficacy of syngeneic twin transplantation in CML.14
Although inhibition of caspase 3-dependent apoptotic pathways and prolongation of G2/M cell cycle phase seem to play important roles in the survival of BCR/ABL leukemia cells after DNA damage,5,8,11 our recent findings indicate that repair of drug-induced DNA lesions may also be essential for drug resistance12,15 and can collaborate with antiapoptotic pathways and G2/M checkpoint to exert drug resistance.16 BCR/ABL-positive hematopoietic cells are resistant to apoptosis induced by many chemotherapeutic agents and γ-radiation,8,11,17-21 which induce a variety of DNA lesions requiring different mechanisms of DNA repair.22,23 Double-strand breaks (DSBs) represent the most serious lesions, which if unrepaired can be lethal,24 but BCR/ABL-transformed cells displayed the enhanced ability to repair DSBs by homologous recombination repair (HRR).12
Nucleotide excision repair (NER) plays a prominent role in the repair of a broad range of other types of DNA lesions such as bulky or helix-altering lesions or base damage.25,26 An interaction between BCR or p210 BCR/ABL proteins (but not p190 BCR/ABL protein) and xeroderma pigmentosum group B (XPB), one of the components of multiprotein complex involved in NER, has been described and proposed to alter NER activity.27,28 Moreover, the functions of core transcription factor IIH (TFIIH), a basal transcription factor complex involved in DNA transactions such as DNA repair and transcription, have been altered in p210 BCR/ABL-expressing cells.10 However, the influence of p190 and p210 BCR/ABL on NER activity has not been extensively investigated. In this study, we demonstrated that p210 BCR/ABL, but not p190 BCR/ABL, regulates NER activity and survival after ultraviolet C (UVC) irradiation in leukemias of myeloid and lymphoid origin.
Materials and methods
Reagents
STI571, generously provided by Novartis (Basel, Switzerland), was prepared as 10-mM stock solution in water and was diluted in culture medium before use. Cells were treated with STI571 for 24 hours in the presence of appropriate growth factors.
Cell lines
The murine interleukin 3 (IL-3)-dependent myeloid and lymphoid cell lines 32Dcl3 and BaF3, respectively, and their p190 BCR/ABL- and p210 BCR/ABL-expressing counterpart clones were maintained in Iscoves modified Dulbecco medium (IMDM) supplemented with 10% fetal calf serum (FCS) and 15% WEHI-conditioned medium as a source of IL-3.16,29 Bone marrow mononuclear cells from mice pretreated with 5-fluorouracil were infected with Mig1 p210BCR/ABL-IRES-GFP (internal ribosome entry site-green fluorescent protein) or Mig1 IRES-GFP retroviral construct as described.29 The mixtures of BCR/ABL-positive and -negative GFP+ cells obtained after sorting were expanded for 2 to 3 weeks in the presence of 15% WEHI- and Chinese hamster ovary cell strain KL (CHO-KL)-conditioned media as the sources of IL-3 and stem cell factor, respectively. Human megakaryocytic leukemia cell line UT7 and p210 BCR/ABL-expressing clone UT-7/9 were cultured in RPMI containing 10% FCS, supplemented with 5 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor.30 Human CML cell line K562 and the acute myelocytic leukemia cell line HL60 were obtained from ATCC (Manassas, VA) and were cultured in RPMI supplemented with 10% FCS.
Growth assay
Cells (105/mL) were treated with UVC light at 254 nm in phosphate-buffered saline (PBS) and resuspended in respective medium. Cell viability was measured by trypan blue exclusion after 48-hour culture in the presence of IL-3. Cell proliferation was assessed by clonogenic assay when 103 cells were plated in methylcellulose in the presence of IL-3, and colonies were counted after 7 days.12
Whole cell extract preparation
Cell extract preparation was performed according to the previously described protocol.31 Briefly, the control and BCR/ABL-transfected cells were washed with ice-cold PBS and harvested by centrifugation, and the cell pellet was suspended in hypotonic buffer (10 mM Tris (tris(hydroxymethyl)aminomethane)-HCl, pH 7.5, 10 mM KCl, 10 mM MgCl2, 1 mM DTT [dithiothreitol]) containing protease inhibitors. The cells were disrupted with a Dounce homogenizer, on ice. Nuclei were harvested by centrifugation, and the nuclear proteins were extracted in hypotonic buffer containing 350 mM NaCl. Cytosolic and nuclear extract proteins were precipitated by addition of ammonium sulfate. The precipitates were resuspended in dialysis buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA (ethylenediaminetetraacetic acid), 100 mM mono-K glutamic acid, and 10% glycerol) and dialyzed for 2 hours at 4°C. The extracts were frozen in liquid nitrogen and stored at -70°C.
Preparation of plasmids and treatment with damaging agents
The 2959-bp pBS (pBluescript KS+; Stratagene, La Jolla, CA) and the related 3738-bp pHM14 plasmid (pHM) were prepared by alkaline lysis method from Escherichia coli JM109. Both plasmids were carefully purified by 1 cesium chloride and 2 neutral sucrose gradient centrifugations, as previously described.32 pBluescript KS+ plasmid was treated with UV light (300 J/m2) at 254 nm. This dose produces about 10 UV lesions per plasmid (pBS-UV).
In vitro repair synthesis assay
The repair synthesis assay was performed as previously described.33 The reaction mixture (50 μL), containing 300 ng damaged pBluescript KS+ and untreated pHM14 closed circular plasmids, 0.074 MBq (2 μCi) α[32P]-deoxycytidine triphosphate (dCTP) (2.96 × 1013 Bq [800 Ci]/mmol), 200 μg cell extract protein, and 70 mM potassium glutamate, was added to the reaction buffer (45 mM HEPES (N-2-hydroxyethylpiperazine-N ′-2-ethanesulfonic acid)-KOH [pH 7.8]; 7.4 mM MgCl2; 0.9 mM DTT; 0.4 mM EDTA; 2 mM adenosine 5′-triphosphate [ATP]; 20 μM each deoxyguanosine triphosphate [dGTP], deoxyadenosine triphosphate [dATP], and deoxythymidine triphosphate [dTTP]; 4 μM dCTP; 40 mM phosphocreatine; 2.5 μg phosphocreatine kinase, 3.4% glycerol; and 18 μg bovine serum albumin) for 3 hours at 30°C, as previously described.34 Before electrophoresis on a 1% agarose gel containing 0.5 μg/mL ethidium bromide, plasmid DNA was purified from reaction mixtures and linearized with EcoRV. Gels were dried and processed on PhosphorImager, and the radioactivity was quantified. Photographs of the ethidium bromide-stained gels were scanned. Specific incorporation of [α32P]-dCMP (deoxycytidine monophosphate) was expressed as radiolabeled incorporation in the damaged versus undamaged plasmid. The value of [α32P]-dCMP incorporation was normalized to the same amount of DNA as determined from the fluorograph.
Immunofluorescence
Nuclear localization of proliferating cell nuclear antigen (PCNA) and XPB proteins was detected by immunofluorescence as described,12 using mouse monoclonal anti-PCNA (PC10; Upstate Biotechnology, Lake Placid, NY) and rabbit polyclonal anti-p89 XPB (S-19; Santa Cruz Biotechnology, Santa Cruz, CA). Rhodamine Red-X goat antimouse immunoglobulin G (IgG) and Alexa Fluor 488 goat antirabbit IgG secondary antibodies (Molecular Probes, Eugene, OR) were applied. Negative controls were performed without primary antibodies. DNA was counterstained with the DNA fluorochrome 4′,6′diamedino-2-phenylindole (DAPI). Specific staining was visualized with an inverted Olympus IX70 fluorescence microscope equipped with a Cook Sensicom ER camera (Olympus America, Melville, NY). A series of 3-dimensional images of each individual picture (cell) were stored in the SlideBook 3.0.1 (Intelligent Imaging Innovations, Denver, CO). Deconvolution was applied to increase resolution and contrast of the images. A collection of 3-dimensional images describing individual cells was converted to a one 2-dimensional picture. At least 20 individual cells were analyzed per experimental group. Pictures were prepared with Adobe Photoshop (San Jose, CA).
Hprt gene mutation assay
Determination of UVC-induced mutants on hprt gene was essentially performed as previously described.35 Immediately after irradiation, an aliquot of cells was removed and seeded on 96-well plates to determine the surviving fraction. Macroscopic colonies (> 50 cells) were scored after 7 days, and relative surviving fraction was calculated. The rest of the cells were grown for a week to allow the expression of hprt mutant phenotype. The cells were then seeded (5000 cells/well) in the presence of 30 μM 6-thioguanine (6-TG) in 96-well plates. After 1 additional week, macroscopic colonies were scored, and mutation frequencies were calculated after correction for plating efficiency (1 cell/well) and UV survival.35
Western analysis
Cell lysates were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described before.29 Western analysis was performed using the primary antibodies recognizing PCNA (PC10; Upstate Biotechnology), p89 XPB (S-19; Santa Cruz), phosphotyrosine (PY20; Oncogene Research Products, Cambridge, MA; and P-Tyr-100; Cell Signaling Technology, Ozyme, France), actin (C11; Santa Cruz), and c-Abl (Ab-3; Oncogene Research).
Results
BCR/ABL regulates NER activity
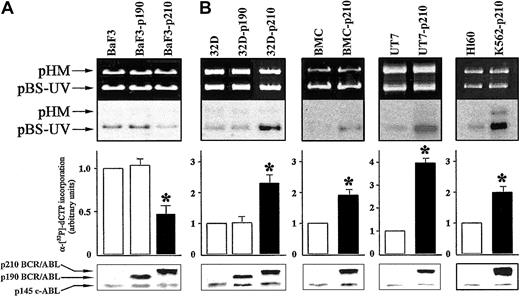
To evaluate DNA repair activity, we used an in vitro DNA repair assay performed with whole cell extracts from the different cell lines expressing p190 or p210 BCR/ABL, compared with the respective control cells. Cell extracts from p210 BCR/ABL-transfected BaF3 lymphoid cells (BaF3-p210) displayed about 2-fold reduction in DNA repair activity compared with the parental BaF3 cells (Figure 1A). This result was observed on 3 independent clones (data not shown). In contrast, p210 BCR/ABL-expressing 32Dcl3 myeloid cells (32D-p210) showed more than 2-fold increased DNA repair activity of UV damage (Figure 1B). In view of these discordant results, we examined DNA repair activity in other myeloid cells: human UT7 cell line and cells transfected with BCR/ABL, murine bone marrow cells (BMCs), and cells infected with BCR/ABL retrovirus, as well as human Ph1-positive K562 (p210 BCR/ABL) and Ph1-negative HL60 cell lines. The in vitro NER activity was increased by 4-fold in BCR/ABL-expressing UT7 and 2-fold in BMCs and K562 cell lines compared with their respective control cells (Figure 1B). Expression of p190 BCR/ABL did not affect NER activity in BaF3 (Figure 1A) and 32Dcl3 (Figure 1B) cells.
p210 BCR/ABL exerts different effects on NER depending on cell origin. DNA repair synthesis activity on UVC-damaged plasmid was measured with the extracts from (A) BaF3, BaF3-p190, and BaF3-p210 lymphoid cells and (B) 32D, 32D-p190, and 32D-p210, BMC and BMC-p210, UT7 and UT7-p210, and HL60 and K562-p210 myeloid cells. Cell extracts were incubated with the damaged (pBS-UV) and undamaged (pHM) plasmids in a repair synthesis reaction. Ethidium bromide-stained gel and autoradiography of the dried gel (upper panels), quantification of the DNA repair activity expressed as the ratio of radiolabel incorporation in the damaged versus undamaged plasmid (middle panels, DNA repair activity in the untreated parental cell extracts was arbitrary set at 1), and the ABL proteins expression (lower panels) are shown. Results represent mean ± SD of 3 separate experiments performed with 3 independent preparations of cell extracts. The * indicates difference between values for control and BCR/ABL-expressing cells at P < .01, as determined by Student t test.
p210 BCR/ABL exerts different effects on NER depending on cell origin. DNA repair synthesis activity on UVC-damaged plasmid was measured with the extracts from (A) BaF3, BaF3-p190, and BaF3-p210 lymphoid cells and (B) 32D, 32D-p190, and 32D-p210, BMC and BMC-p210, UT7 and UT7-p210, and HL60 and K562-p210 myeloid cells. Cell extracts were incubated with the damaged (pBS-UV) and undamaged (pHM) plasmids in a repair synthesis reaction. Ethidium bromide-stained gel and autoradiography of the dried gel (upper panels), quantification of the DNA repair activity expressed as the ratio of radiolabel incorporation in the damaged versus undamaged plasmid (middle panels, DNA repair activity in the untreated parental cell extracts was arbitrary set at 1), and the ABL proteins expression (lower panels) are shown. Results represent mean ± SD of 3 separate experiments performed with 3 independent preparations of cell extracts. The * indicates difference between values for control and BCR/ABL-expressing cells at P < .01, as determined by Student t test.
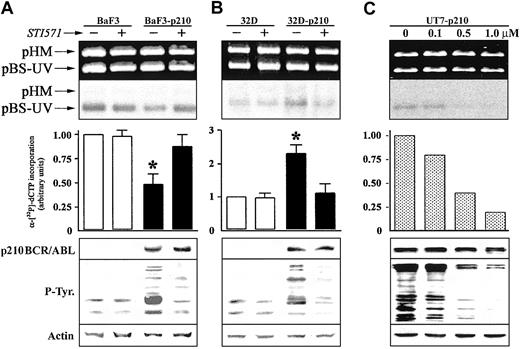
We next investigated whether p210 BCR/ABL tyrosine kinase activity was required for the regulation of NER activity. Pretreatment with 1 μM STI571, a selective inhibitor of the ABL tyrosine kinase activity,36 had no significant effect on NER activity in BaF3 and 32Dcl3 parental cell lines (Figure 2). STI571 totally restored NER function in Ba/F3-p210 cells (Figure 2A); conversely, NER activity was inhibited in 32Dcl3-p210 cells (Figure 2B). These results were similar in the different clones tested. NER activity correlated with p210 BCR/ABL kinase activity as determined by the use of various concentrations of STI571 to achieve different levels of inhibition of the ABL kinase (Figure 2C).
The effect of ABL kinase inhibitor STI571 on DNA repair activity in p210 BCR/ABL-positive cells. DNA repair synthesis activity on UVC-damaged plasmid was examined with the extracts from (A) BaF3 and BaF3-p210 lymphoid cells and (B) 32D and 32D-p210 myeloid cells pretreated (+) or not (-) with 1 μM STI571, and (C) UT7-p210 pretreated for 24 hours with the indicated concentrations of STI571 (upper and middle panels). For more details, see Figure 1 legend. p210 BCR/ABL, phosphotyrosine proteins, and actin (loading control) expression were determined by Western analysis (lower panels). The * indicates difference between values for untreated and STI571-pretreated cells at P < .01, as determined by Student t test.
The effect of ABL kinase inhibitor STI571 on DNA repair activity in p210 BCR/ABL-positive cells. DNA repair synthesis activity on UVC-damaged plasmid was examined with the extracts from (A) BaF3 and BaF3-p210 lymphoid cells and (B) 32D and 32D-p210 myeloid cells pretreated (+) or not (-) with 1 μM STI571, and (C) UT7-p210 pretreated for 24 hours with the indicated concentrations of STI571 (upper and middle panels). For more details, see Figure 1 legend. p210 BCR/ABL, phosphotyrosine proteins, and actin (loading control) expression were determined by Western analysis (lower panels). The * indicates difference between values for untreated and STI571-pretreated cells at P < .01, as determined by Student t test.
p210 BCR/ABL regulates the cellular sensitivity to UVC
In Xeroderma pigmentosum syndrome (XP), an archetypical NER deficiency-associated pathology, altered NER capacity has been correlated with UV hypersensitivity.37 Therefore, we examined UVC-mediated cytotoxicity in p210 BCR/ABL-expressing cells and control cells. Trypan blue exclusion test showed that expression of p210 BCR/ABL in BaF3 cells resulted in significant sensitization to UVC (Figure 3A, left panel). In contrast, p210 BCR/ABL-transfected 32Dcl3 cells were resistant to UVC irradiation compared with parental cells (Figure 3B, left panel). Therefore, UVC sensitivity paralleled in vitro NER activity. Results from clonogenic assay (Figure 3, right panels) were in accordance with those from trypan blue exclusion test (Figure 3, left panels). Preincubation with STI571 resulted in abrogation of hypersensitivity to UVC in BaF3-p210, whereas it reduced the resistance in 32Dcl3-p210 cells. The sensitivity of parental BaF3 and 32Dcl3 cells to UVC was not affected by STI571 treatment.
p210 BCR/ABL kinase affects the sensitivity to UVC cytotoxicity. (A) BaF3 and BaF3-p210 lymphoid cells and (B) 32D and 32D-p210 myeloid cells were pretreated or not with 1 μM STI571 for 24 hours and irradiated with the indicated dose of UVC. Cell viability was assessed 48 hours later using trypan blue exclusion test (left panels). Cell growth was tested in clonogenic assay (right panels). Each experiment was performed in duplicate, and the results are represented as a mean ± SD of 3 separate experiments. The * and ** indicate difference between values for p210 BCR/ABL-positive and -negative cells at P < .05 and P < .01, respectively, as determined by Student t test.
p210 BCR/ABL kinase affects the sensitivity to UVC cytotoxicity. (A) BaF3 and BaF3-p210 lymphoid cells and (B) 32D and 32D-p210 myeloid cells were pretreated or not with 1 μM STI571 for 24 hours and irradiated with the indicated dose of UVC. Cell viability was assessed 48 hours later using trypan blue exclusion test (left panels). Cell growth was tested in clonogenic assay (right panels). Each experiment was performed in duplicate, and the results are represented as a mean ± SD of 3 separate experiments. The * and ** indicate difference between values for p210 BCR/ABL-positive and -negative cells at P < .05 and P < .01, respectively, as determined by Student t test.
p210 BCR/ABL affects the colocalization of XPB and PCNA in response to UVC
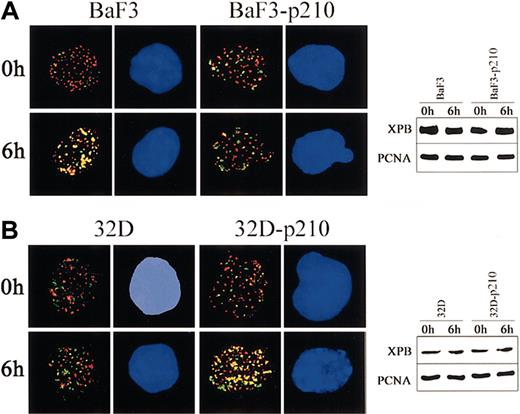
To obtain more insight about the contrasting role of p210 BCR/ABL in regulation of NER and UVC cytotoxicity in lymphoid and myeloid cells, 2-color immunofluorescence was applied to examine the kinetics of assembly of XPB and PCNA proteins after UVC-dependent DNA damage. p210 BCR/ABL abrogated the colocalization of XPB and PCNA in lymphoid BaF3 cells exposed to UVC irradiation (Figure 4A, left panel), whereas it stimulated this reaction in myeloid 32Dcl3 cells (Figure 4B, left panel). These results were not dependent on the differences in protein expression because Western analysis demonstrated similar levels of XPB and PCNA in parental cells and p210 BCR/ABL-transformed counterparts before and after UVC treatment (Figure 4A-B; right panels).
p210 BCR/ABL regulates the nuclear assembly of XPB and PCNA in response to UVC. (A) BaF3 and BaF3-p210 lymphoid cells and (B) 32D and 32D-p210 myeloid cells were irradiated or not with 10 J/m2 UVC. Nuclear localization of PCNA (red) and XPB (green) was detected on the cytospins prepared at time 0 and 6 hours after irradiation. DNA was counterstained by DAPI. Yellow color indicates the possible colocalization sites on PCNA and XPB. Results represent 2 independent experiments. Original magnification, × 1000. Expression of PCNA and XPB in total cell lysates before and after UVC was detected by Western analysis.
p210 BCR/ABL regulates the nuclear assembly of XPB and PCNA in response to UVC. (A) BaF3 and BaF3-p210 lymphoid cells and (B) 32D and 32D-p210 myeloid cells were irradiated or not with 10 J/m2 UVC. Nuclear localization of PCNA (red) and XPB (green) was detected on the cytospins prepared at time 0 and 6 hours after irradiation. DNA was counterstained by DAPI. Yellow color indicates the possible colocalization sites on PCNA and XPB. Results represent 2 independent experiments. Original magnification, × 1000. Expression of PCNA and XPB in total cell lysates before and after UVC was detected by Western analysis.
p210 BCR/ABL facilitates mutagenesis in UVC-treated BaF3 cells
Mutagenic action of UV irradiation is increased in NER-deficient cells.25 Therefore, we determined the frequencies of UVC-induced mutations in Ba/F3-p210 cell lines by examining a conventional target, the hprt gene. As expected from our previous work, nonirradiated Ba/F3-p210 cells exhibited slightly increased mutation rates, as compared with control BaF3 cells (Table 1). However, UVC irradiation dramatically increased mutation rates in a dose-dependent manner in p210 BCR/ABL-expressing cells (65-fold), whereas it has a negligible effect on mutation rates in control cells. At the highest dose, nearly 10% of surviving BaF3-p210 cells carried mutation in hprt gene compared with only 0.06% of control cells.
Discussion
The Philadelphia chromosome, Ph1, results in the bcr/abl translocation that appears to constitutively activate the ABL kinase.38 Leukemia cells containing the BCR/ABL tyrosine kinase have been shown to be resistant to a number of therapeutic drugs as well as γ-irradiation.8,11,17-21 The mechanism(s) of BCR/ABL-mediated drug resistance has not been well characterized. Although the inhibition of caspase-3 activation and induction of a G2/M cell cycle arrest can play a significant role,38 other cellular functions may be equally responsible for therapeutic drug resistance. Our recent studies revealed a novel mechanism of genotoxic resistance in BCR/ABL-positive cells: stimulation of DSB repair by homologous recombination (HR).12 That phenomenon is dependent on BCR/ABL-induced elevation of the expression of RAD51, the mammalian homologue of E coli RecA protein, which plays an essential role in homologous recombination.39 These studies for the first time demonstrated that stimulation of DNA repair mechanisms could be involved in drug resistance of Ph1 leukemias. In addition, BCR/ABL was reported to inhibit the DNA-PK catalytic subunit (DNA-PKcs),40 an important element in repair of DSBs by nonhomologous end-joining (NHEJ),24 but NHEJ repair activity seemed to be elevated in CML cells.41 Thus, the 2 major mechanisms of DSB repair, HR and NHEJ, can be modified by BCR/ABL.
Although DSBs pose a major threat to genomic integrity and cell survival, other types of DNA lesions such as base modifications, adducts, and mismatches may also lead to cell death or mutator phenotype, if not repaired or repaired unfaithfully.42 NER represents a major pathway involved in reparation of these lesions.25,26 The process involves recognition of a lesion and excision of approximately 25 to 30 nucleotides, including the damaged ones, followed by repair by DNA synthesis (repair synthesis) using the opposite, normal DNA strand as a template. Recognition and excision require a multiprotein complex including TFIIH subcomplex, XPA, XPG proteins and replication protein A (RPA), whereas repair synthesis involves another multiprotein complex including PCNA, replication factor C (RFC), RPA, DNA polymerase δ/ϵ, and DNA ligase I.43 The most relevant DNA lesions subject to NER are cyclobutane pyrimidine dimers (CPDs) and [6-4] photoproducts (6-4PPs), 2 major kinds of injury produced by the short-wave UV.26
We found that cells transformed by p210 BCR/ABL tyrosine kinase displayed a remarkably different ability to repair the UVC-mediated DNA damage, depending on cell origin.Although the repair activity was reduced in p210 BCR/ABL-positive lymphoid cells, it was significantly enhanced in p210 BCR/ABL-positive myeloid cells compared with healthy counterparts. The use of several lymphoid cell clones and myeloid cell models (murine and human p210 BCR/ABL cell lines and primary cells, and Ph1-positive leukemia cells, along with the appropriate control cells) excluded the possibility of clonal variations. In addition, inhibition of p210 BCR/ABL tyrosine kinase activity by STI571 reversed these effects, indicating that inhibition or acceleration of NER is a dynamic process dependent on the constitutive kinase activity of p210 BCR/ABL. The ability to repair UVC-dependent DNA lesions directly corresponded to cell survival after UVC irradiation. Neither the repair ability nor survival of UVC-treated parental cells was affected by STI571.
The regulation of NER is specific for p210 BCR/ABL kinase; p190 BCR/ABL and TEL/ABL kinases do not affect NER (Figure 1 and data not shown, respectively). In contrast to p210 BCR/ABL, the latter kinases do not contain the CDC24 motif implicated in the interaction with XPB at least in yeast 2-hybrid screen,27 and do not coimmunoprecipitate with XPB (data not shown). Therefore, p210 BCR/ABL-XPB complex may be important for triggering of the initial steps of NER.
p210 BCR/ABL-mediated inhibition of NER activity in lymphoid cells is supported by previous observations that in vitro TFIIH assembly and activity in repair assay were moderately inhibited by p210 BCR/ABL expression in the UV-sensitive 27-1 cells (CHO 9 derivative). This effect corresponded to the decreased survival of p210 BCR/ABL-positive cells after UV irradiation.10,27 We observed here that p210 BCR/ABL inhibited formation of XPB nuclear foci and abrogated their colocalization with PCNA foci in BaF3 lymphoid cells after UVC irradiation. This finding further supports the negative influence of p210 BCR/ABL on NER in lymphoid cells. The effect is not due to the general disability of p210 BCR/ABL-positive lymphoid cells to repair DNA damage because HR was actually enhanced in these cells, leading to improved survival after treatment with cisplatin and mitomycin C.16 Unrepaired UVC-mediated DNA lesions may provide a strong signal to apoptosis, which cannot be overcome even by p210 BCR/ABL-dependent antiapoptotic mechanisms.9 In addition, the inability to repair UVC-mediated base damage may result in an enhanced mutational burden in cells.37 In accordance, BaF3-p210 cells surviving UVC treatment displayed a mutator phenotype as shown by our research here, as measured by mutation-dependent inactivation of the hprt gene.44 It is tempting to speculate that these mutations arose as a result of efficient replicative bypass of the UVC lesions unrepaired by NER. This hypothesis is supported by our previous observation that error-prone DNA polymerase β, which is overexpressed in BCR/ABL cells,45 can facilitate translesion synthesis of bulky adducts such as cisplatin and UV, thereby enhancing mutagenesis.46,47
In myeloid cells p210 BCR/ABL exerted an opposite effect on NER compared with lymphoid cells. It is not clear at this time why the lineage origin is so crucial for p210 BCR/ABL-mediated regulation of NER activity, but substantial differences in molecular pathogenesis of p210 BCR/ABL-mediated myeloid and lymphoid malignancies have been reported.48,49 One hypothesis is that cell lineage determines the BCR/ABL-mediated assembly of multiprotein complexes involved in NER.25,26 In support of this hypothesis we observed that formation of nuclear complexes involving XPB and their colocalization with PCNA was inhibited in lymphoid cells, but enhanced in myeloid counterparts, in response to UVC damage. However, XPB phosphorylation, its interaction with p210 BCR/ABL and with the members of TFIIH complex (p62, p44, and cyclin H10 ) is not affected by the myeloid or lymphoid origin (data not shown), suggesting that other components of NER machinery and/or those affecting NER indirectly25 may be regulated by p210 BCR/ABL in a lineage-specific manner.
In conclusion, this work suggests that genotoxic agents causing DNA lesions repaired by NER could be more effective against leukemias expressing p190 than the p210 form of BCR/ABL kinase, and against BCR/ABL-positive lymphoid than myeloid leukemias. In addition, therapeutic strategies involving STI571 (imatinib mesylate) and genotoxic agents50-54 should be planned with caution. Imatinib mesylate treatment combined with cytostatic drugs inducing DNA lesions, which require NER for repair, may be beneficiary for most patients with CML, but not for patients with p210 BCR/ABL-positive ALL and CML in lymphoid blast crisis.
Prepublished online as Blood First Edition Paper, June 26, 2003; DOI 10.1182/blood-2002-10-3207.
Supported by grants from la Ligue contre le Cancer 32 (grant 2001) (D.L.), Fondation de France (Comité Leucémie) (Y.C.), and the National Cancer Institute (Public Health Service grant CA89052) (T.S.). T.S. is a Scholar of the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr A. Turhan for providing the BCR/ABL-transfected BaF3 and UT-7 cells.