Abstract
The mechanisms involved in anti-inflammatory action of transforming growth factor β (TGFβ) have been examined by evaluating its effect on chemokine gene expression in mouse macrophages. Lipopolysaccharide (LPS)–stimulated expression of the CXC chemokines KC and MIP-2 was selectively reduced by TGFβ in a time- and protein synthesis–dependent process. While TGFβ had a modest effect on transcription of the KC and MIP-2 mRNAs as measured by nuclear run-on, it had no effect on LPS-stimulated luciferase expression driven by the KC promoter nor on the activation of nuclear factor κB (NFκB) DNA-binding activity and transactivation function. Interestingly, KC mRNA levels were markedly reduced by TGFβ treatment in cells transfected with KC genomic or cDNA constructs driven from either the KC or cytomegalovirus (CMV) promoters, demonstrating the importance of sequences within the mature mRNA and suggesting that suppression may involve a posttranscriptional mechanism. In support of this possibility, LPS stimulation prolonged the half-life of KC mRNA and this stabilization response was blocked in cells treated with TGFβ. Examination of KC mRNA expressed under control of a tetracycline-responsive promoter demonstrated that TGFβ prevented stabilization of KC mRNA, in response to LPS but did not alter KC mRNA half-life directly. KC mRNA stabilization by LPS was dependent on activation of p38 mitogen-activated protein kinase (MAPK) activity, and TGFβ treatment inhibited p38 MAPK activation. These findings support the hypothesis that TGFβ-mediated suppression of chemokine gene expression involves antagonism of LPS-stimulated KC mRNA stabilization via inhibition of p38 MAPK.
Introduction
Macrophages play a key role in detection and elimination of pathogens mediated through recognition of and response to microbial products such as lipopolysaccharide (LPS), peptidoglycan, and lipoteichoic acid.1-3 This response is mediated in part through the modulation of cytokine and chemokine gene expression necessary for communication between various cell types found at sites of inflammation and critical for the successful orchestration and resolution of the reaction.4,5 Because inflammatory responses exhibit significant potential for tissue damage, it is necessary to have effective anti-inflammatory control, and this is often mediated by a set of inhibitory cytokines that include transforming growth factor β (TGFβ).5,6 Indeed, multiple roles for TGFβ in regulating inflammatory and adaptive immune responses have been documented, which on balance, suggest that this cytokine can provide both positive and negative control of inflammation.7,8 For example, while loss-of-function studies of TGFβ family ligands in mice have revealed their critical roles in embryonic development and tissue homeostasis during adult life,7-9 these mice also develop clinical symptoms of an autoimmune syndrome and exhibit deregulated macrophage function.8,10 It has been reported recently that the anti-inflammatory potential of apoptotic cell debris is mediated through the production of TGFβ and the subsequent inhibitory actions of this cytokine.11,12 Although TGFβ has been reported to modulate expression of genes at both transcriptional and posttranscriptional levels, the mechanistic basis for these inhibitory effects of TGFβ is incompletely understood.11,13-18
Expression of chemokine genes at sites of injury and inflammation represents a major source of chemoattractant activity critical for regulation of the trafficking of inflammatory leukocytes into such tissues. TGFβ has been previously shown to suppress the expression of a select subset of inducible chemokines.11 While LPS-induced expression of KC and MIP-2 was suppressed by TGFβ, MCP-1 expression was unaltered. Furthermore, some changes in KC and MIP-2 production involved alterations in mRNA levels, while inhibition of TNFα production appeared to involve regulation of mRNA translation.11 In the present study we undertook analysis of both endogenous and transfected KC genes in order to identify the mechanisms through which TGFβ promotes such selective anti-inflammatory function. The results demonstrate that TGFβ-mediated inhibition is achieved at least in part through alteration of the rate of KC mRNA decay. In this regard, several CXC chemokine mRNAs including KC are known to be regulated by modulation of mRNA stability,19,20 and our findings support the hypothesis that TGFβ antagonizes LPS-mediated stabilization of KC mRNA.
Materials and methods
Reagents
Dulbecco modified Eagle medium (DMEM), RPMI-1640, and Dulbecco phosphate-buffered saline (PBS) were obtained from the Central Cell Service Media Lab of the Lerner Research Institute at the Cleveland Clinic Foundation. Antibiotics, agarose, Tris (tris(hydroxymethyl)aminomethane), and RNAse inhibitor were purchased from Invitrogen (Carlsbad, CA). Formamide, dextran sulfate, salmon sperm DNA, actinomycin D (ActD), cycloheximide (CHX), MOPS (3-[N-Morpholino]propanesulphonic acid), LPS (prepared from the Escherichia coli serotype 0111:B4), leupeptin, aprotinin, phenylmethylsulfonylfluoride (PMSF), and diethylpyrocarbonate were from Sigma-Aldrich (St Louis, MO). Highly purified lipoprotein-free LPS was provided by Dr Stephanie Vogel (University of Maryland Medical School). Fetal bovine serum (FBS) was obtained from Biosource International (Rockville, MD) and was heat-inactivated before use. All cell culture reagents were specified to be endotoxin-free. Brewer thioglycollate broth (TG) was obtained from Difco Laboratories (Detroit, MI). SuperFect transfection reagent and DNA plasmid preparation kits were obtained from Qiagen (Valencia, CA). TRI-reagent was purchased from Molecular Research Center (Cincinnati, OH). Guanidine thiocyanate and cesium chloride were purchased from Fisher Biotech (Fair Lawn, NJ). Doxycyclin (Dox) was obtained from Clontech (Palo Alto, CA). Random priming kits were purchased from Stratagene (Cedar Creek, TX). RNAse-free DNAse was obtained from Promega (Madison, WI). Nylon transfer membrane was purchased from Micron Separation (Westboro, MA). Dupont NEN Research Products (Boston, MA) is the source of α-[32P] deoxycytosine triphosphosphate. Poly (dI-dC) and deoxynucleoside triphosphates were purchased from Pharmacia (Uppsala, Sweden). Restriction enzymes and bovine serum albumin were purchased from Boehringer Mannheim (Indianapolis, IN). All chemokine enzyme-linked immunosorbent assay (ELISA) kits, antibodies against interleukin-10 (IL-10), and TGFβ1 were obtained from R&D Systems (Minneapolis, MN). Chloramphenicol acetyl transferase (CAT) ELISA kits were obtained from Roche (Indianapolis, IN). Antibodies against p38 mitogen-activated protein kinase (MAPK) and phosphorylated p38 MAPK were obtained from New England Biolabs (Beverly, MA). Protogel, sequagel (acrylamide, N, N-methylene bisacrylamide, urea), and related buffers were obtained from National Diagnostics (Atlanta, GA). SB203580 and PD980589 were purchased from Calbiochem (San Diego, CA). Protein assay reagents were purchased from Bio-Rad Laboratories (Richmond, CA).
Preparation of peritoneal exudate macrophages
Specific pathogen-free, inbred C57BL6 female mice, 6 to 8 weeks of age, were purchased from Charles River Laboratories (Wilmington, MA) and housed in microisolator cages with autoclaved food and bedding to minimize exposure to viral and microbial pathogens. TG-elicited macrophages were obtained as reported previously.21 Peritoneal exudate cells (5 × 106) were plated in 100-mm Petri dishes, incubated for 2 hours at 37°C in an atmosphere of 5% CO2, and then washed 3 times with Hanks balanced salt solution to remove nonadherent cells. The macrophages were cultured overnight in RPMI-1640 at 37°C in an atmosphere of 5% CO2 and then cultured in the presence or absence of stimuli for the indicated times.
Cell culture and transient or stable transfection
The RAW264.7 mouse macrophage-like cell line was maintained in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 1% penicillin/streptomycin, and subcultured twice weekly, as described previously.22 RAW264.7 cells were transfected using SuperFect transfection reagent. Briefly, the cells were seeded at a density of 1 × 106 cells per 100-mm Petri dish 24 hours before transfection. Specific plasmid DNA (10 μg) was mixed with 20 μL SuperFect and incubated for 20 minutes at room temperature. The cells were washed with PBS once and then incubated in complete medium containing SuperFect with 10 μg plasmid DNA for 3 hours. In transient transfections, the cells were then washed with PBS, pooled, and seeded to dishes at 1 × 106 cells/100-mm Petri dish. The cells were cultured in complete medium for 24 hours before treatment. CAT levels were measured by ELISA according to the manufacturer's instructions. For stable transfection, the cells were washed with PBS once after 3 hours with plasmid DNA and subcultured in complete medium for 48 hours before adding selection medium with G418 antibiotic (500 μg/mL). A number of stable cell lines were selected for characterization of chemokine mRNA expression and protein production.
Preparation of plasmid
The plasmids encoding KC, MIP-2, MCP-1, IP-10, GAPDH (glyceraldehyde-3′-phosphate dehydrogenase), and α-tubulin cDNA fragments were as described previously.23-26 Plasmids containing fragments of the KC promoter linked to the CAT reporter gene were described previously.27 An expression construct encoding the genomic form of the KC gene (pgKC-CMV) was prepared in pcDNA3 (Promega) by insertion of a 2200 nucleotide sequence encoding the full KC gene at the EcoRV site. This clone contained the full transcription unit of KC beginning at nucleotide position 1 and continuing 326 nucleotides beyond the 3′ terminus of the mRNA.27 A second genomic KC expression construct (pgKC-104) was prepared in which a fragment containing 104 nucleotides of the KC gene promoter27 was inserted in place of the cytomegalovirus (CMV) promoter in pgKC-CMV. pTRE2 and pTRE2-luciferase vectors were purchased from Clontech Laboratories. Full-length KC cDNA (952 nucleotide)20 was subcloned into pTRE2 at EcoR1 sites downstream of the tet-responsive element (TRE) to create pTRE-KCcDNA. Plasmids were prepared using Qiagen kits according to the manufacturer's instructions.
Preparation of RNA and Northern hybridization analysis
Total cellular RNA was extracted from primary peritoneal exudate macrophages by the guanidine thiocyanate-cesium chloride method28 and from RAW264.7 cells using the TRI-reagent according to manufacturer's instructions. Equal amounts of RNA (20 μg) were analyzed by Northern hybridization as described previously.26,29 Blots were quantified by phosphorimage analysis using an instrument from Molecular Dynamics (Sunnyvale, CA). Specific chemokine mRNA levels were normalized to levels of GAPDH mRNA measured in the same RNA sample.
Western blot analysis
RAW264.7 cells were lysed in 50 mM TrisHCl, 150 mM NaCl, 10 μM NaF, 5 μM Na Pyrophosphate, 10 μM Na Vanadate, 1 μM PMSF, 10% glycerol, and 1% Triton X-100. Extract protein (20 μg) was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (10% acrylamide) and transfered to nylon membrane using a semidry transfer cell (Bio-Rad) for one hour at 10 V constant voltage in transfer buffer (48 mM Tris, 39 mM glycine, and 20% methanol, pH 9.2). Blots were blocked with 5% nonfat milk in TBS-T (0.15 M NaCl, 0.5% Tween 20, 50 mM Tris, pH 7.4) for 1 hour at 4°C, then incubated for an additional 18 hours at 4°C with rabbit polyclonal antibodies (1 μg/mL) against phosphorylated p38 MAPK or p38 MAPK in 5% nonfat milk TBS-T solution. After washing 3 times in TBS-T, filters were incubated at room temperature for one hour with goat anti–rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase and then washed again as above. Antibody binding was detected using the enhanced chemiluminescence kit from Amersham (Arlington Heights, IL).
Nuclear run-on assay
TG-elicited macrophages (5 × 107) were treated as indicated in the text and nuclei isolated as described previously.29,30 Transcription initiated in intact cells was allowed to complete in the presence of α-[32P] uridine triphosphate (UTP), and the RNA was isolated and hybridized to slot-blotted plasmids containing specific cDNA (7 μg DNA/slot). The α-tubulin gene was used as an internal standard. The expression of specific transcripts was quantified by phosphorimage analysis. Numeric values for specific transcripts were normalized to α-tubulin transcript level in the same sample. This ratio in untreated samples was arbitrarily set to unity. Experimental values are presented as fold induction relative to untreated samples.
Preparation of cell extracts and electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from treated cell cultures as described previously.27,31 The following complementary oligonucleotides were used as probes for nuclear factor κB (NFκB) binding activity: 5′-GGT TGC AGG GAA ACA CCC TGT ACT CCG GGA ATT TCC CTG GCC-3′; 5′-GGC CAG GGAAAT TCC CGG AGT ACA GGG TGT TTC CCT GCA ACC-3′. For binding reactions, nuclear extracts (10 μg protein) were incubated in 25 μL total reaction volume containing 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.9), 80 mM NaCl, 0.1 mM EDTA (ethylenediaminetetraacetic acid), 1 mM dithiothreitol (DTT), 8% glycerol, and 80 ng/mL poly (dI-dC) for 10 minutes at 4°C. The 32P-labeled oligonucleotide (0.5 ng, 5 × 105 cpm) was then added to the reaction mixture and incubated for 20 minutes at room temperature. The reaction products were analyzed by electrophoresis in a 6% nondenaturing polyacrylamide gel with 0.25 × TBE buffer (22.3 mM Tris, 22.2 mM borate, and 0.5 mM EDTA). The gel was dried and analyzed by autoradiography.
Results
TGFβ selectively affects LPS-induced chemokine gene expression in mouse macrophages
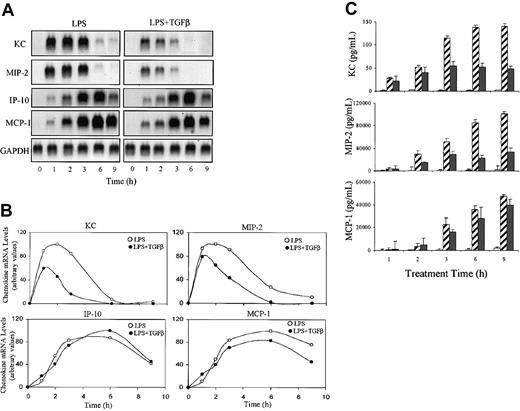
We first examined whether exogenous TGFβ affected LPS-induced expression of a set of chemokine mRNAs (KC, MIP-2, IP-10, and MCP-1) in TG-elicited primary mouse macrophages. LPS induced strong expression of all 4 chemokines and the inclusion of TGFβ in the treatment protocol resulted in selective inhibition of KC and MIP-2 without altering expression of IP-10 and MCP-1 (Figure 1A). Although LPS stimulated optimal expression of both chemokine mRNAs within one hour, TGFβ-mediated suppression of KC or MIP-2 mRNA levels was not maximal until 2 to 4 hours after initiation of stimulation. Similar inhibitory effects were observed on production of KC and MIP-2 secreted protein levels (Figure 1C). The decreased levels of KC and MIP-2 proteins paralleled the alteration in mRNA, indicating that TGFβ did not effect mRNA translation. Similar results were obtained using RAW264.7 cells stimulated with either commercial LPS or highly purified lipoprotein-free LPS (not shown).
The effect of TGFβ on LPS-induced chemokine mRNA and protein levels. Adherent TG-elicited macrophages (1 × 107 cells/100-mm Petri dish) were stimulated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for the indicated times. (A) Total RNA was prepared and levels of KC, MIP-2, IP-10, and MCP-1 mRNA were analyzed by Northern hybridization using 20 μg total RNA in each lane. Blots were hybridized with the indicated radiolabeled cDNA probes. Similar results were obtained in 3 separate experiments. (B) The blot in panel A was quantified by phosphorimage analysis. Values presented are in arbitrary units where each chemokine level is normalized to the level of GAPDH mRNA in the same sample. (C) The levels of chemokine protein in the culture medium were analyzed by ELISA. Values are means ± SEM of 3 separate experiments. □ indicates no treatment (UT); ▨, LPS only; and ▪, TGF + LPS.
The effect of TGFβ on LPS-induced chemokine mRNA and protein levels. Adherent TG-elicited macrophages (1 × 107 cells/100-mm Petri dish) were stimulated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for the indicated times. (A) Total RNA was prepared and levels of KC, MIP-2, IP-10, and MCP-1 mRNA were analyzed by Northern hybridization using 20 μg total RNA in each lane. Blots were hybridized with the indicated radiolabeled cDNA probes. Similar results were obtained in 3 separate experiments. (B) The blot in panel A was quantified by phosphorimage analysis. Values presented are in arbitrary units where each chemokine level is normalized to the level of GAPDH mRNA in the same sample. (C) The levels of chemokine protein in the culture medium were analyzed by ELISA. Values are means ± SEM of 3 separate experiments. □ indicates no treatment (UT); ▨, LPS only; and ▪, TGF + LPS.
The effect of TGFβ on LPS-induced KC mRNA expression is time- and protein synthesis–dependent
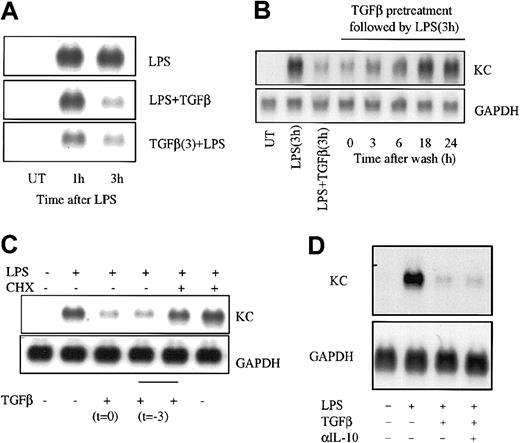
Because TGFβ addition at the time of LPS treatment did not achieve maximal suppression of KC and MIP-2 mRNA levels until 2 hours after stimulation (Figure 1), we reasoned that the effects of TGFβ may be time-dependent. To test this possibility, RAW264.7 macrophages were treated with TGFβ either 3 hours prior to or simultaneously with stimulation by LPS and the levels of KC mRNA were determined 1 and 3 hours later (Figure 2A). As expected, addition of LPS and TGFβ together had little effect at 1 hour but produced suppression at 3 hours. When cells were pretreated with TGFβ for 3 hours, the inhibitory effect was evident even at 1 hour. In addition, the suppression of KC mRNA expression by TGFβ was relatively long lived; when cells were washed free of TGFβ after a 3-hour treatment period and stimulated with LPS following further incubation intervals, KC expression remained suppressed for at least 6 hours but returned to normal by 18 hours (Figure 2B).
The effect of TGFβ on LPS-induced KC mRNA expression is time- and protein synthesis–dependent. (A) RAW264.7 cells were treated or not with TGFβ (20 ng/mL) for 3 hours before adding LPS (10 ng/mL) and/or TGFβ for a further 1 or 3 hours of incubation. Total RNA was prepared and levels of chemokine mRNA were determined as described in the legend to Figure 1. (B) RAW264.7 cells were cultured in the presence of TGFβ (20 ng/mL) for 3 hours and then washed and placed in culture medium without TGFβ for the indicated times prior to the addition of LPS for 3 hours. KC mRNA levels were determined as above. Controls included no treatment (UT), LPS alone for 3 hours, or LPS and TGFβ at the same time for 3 hours. (C) RAW264.7 cells were treated or not with TGFβ (20 ng/mL) in the presence or absence of CHX (10 μg/mL) for 3 hours. Cultures were washed and fresh medium without TGFβ or CHX was added, and the cells were stimulated with LPS for an additional 3 hours prior to analysis of KC mRNA levels. (D) RAW264.7 cells were treated with LPS (10 ng/mL) alone or with TGFβ (20 ng/mL) for 3 hours in the presence of neutralizing antibody against IL-10 as indicated. Total RNA was prepared and analyzed for KC and GAPDH mRNA levels by Northern hybridization. Similar results were obtained in 3 separate experiments.
The effect of TGFβ on LPS-induced KC mRNA expression is time- and protein synthesis–dependent. (A) RAW264.7 cells were treated or not with TGFβ (20 ng/mL) for 3 hours before adding LPS (10 ng/mL) and/or TGFβ for a further 1 or 3 hours of incubation. Total RNA was prepared and levels of chemokine mRNA were determined as described in the legend to Figure 1. (B) RAW264.7 cells were cultured in the presence of TGFβ (20 ng/mL) for 3 hours and then washed and placed in culture medium without TGFβ for the indicated times prior to the addition of LPS for 3 hours. KC mRNA levels were determined as above. Controls included no treatment (UT), LPS alone for 3 hours, or LPS and TGFβ at the same time for 3 hours. (C) RAW264.7 cells were treated or not with TGFβ (20 ng/mL) in the presence or absence of CHX (10 μg/mL) for 3 hours. Cultures were washed and fresh medium without TGFβ or CHX was added, and the cells were stimulated with LPS for an additional 3 hours prior to analysis of KC mRNA levels. (D) RAW264.7 cells were treated with LPS (10 ng/mL) alone or with TGFβ (20 ng/mL) for 3 hours in the presence of neutralizing antibody against IL-10 as indicated. Total RNA was prepared and analyzed for KC and GAPDH mRNA levels by Northern hybridization. Similar results were obtained in 3 separate experiments.
These findings further suggested that TGFβ might itself induce expression of a new gene product as a part of the inhibitory mechanism. To test this latter idea, macrophages were treated with the reversible protein synthesis inhibitor CHX during the TGFβ pretreatment period, washed free of inhibitor and TGFβ, and subsequently stimulated with LPS for 3 hours (Figure 2C). Pretreatment with CHX alone did not alter the response to LPS, while pretreatment with TGFβ led to suppression comparable with that seen in Figure 2A-B. When protein synthesis was blocked with CHX during the TGFβ pretreatment period, however, the inhibitory effects of TGFβ were abrogated. This indicates that the inhibitory action of TGFβ on LPS-induced KC mRNA expression depends upon protein synthesis that may reflect the participation of a TGFβ-induced gene product. To determine if the effects of TGFβ might involve enhanced production of IL-10, a known anti-inflammatory cytokine that can also diminish KC and MIP-2 gene expression,32 RAW264.7 cells were treated with LPS and/or TGFβ in the presence of neutralizing antibody against IL-10 and KC mRNAlevels were assessed (Figure 2D). While TGFβ treatment produced marked suppression of LPS-stimulated KC mRNA levels, this was not blocked by inclusion of the anti–IL-10 antibody.
Effects of TGFβ on chemokine gene transcription
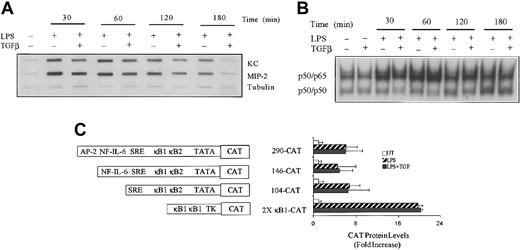
To identify whether the rates of LPS-induced KC and MIP-2 gene transcription were affected by TGFβ, nuclear run-on experiments were performed. Cultures of peritoneal macrophages were treated with LPS for various times in the absence or presence of TGFβ, the nuclei were harvested, and the RNA transcripts initiated in vivo were allowed to elongate in vitro in the presence of 32P-UTP. The radiolabeled RNA products were hybridized to slot-blotted cDNAs encoding KC, MIP-2, and α-tubulin (Figure 3A). Transcription of the KC and MIP-2 mRNAs was elevated within 30 minutes, appeared to be optimal after stimulation with LPS for 60 minutes, and declined significantly between 2 and 3 hours after stimulation. Although cotreatment with TGFβ produced modest reduction in LPS-stimulated transcription of both KC and MIP-2 genes, this was not evident until 2 hours after LPS addition, consistent with the delayed effect seen on mRNA levels (Figure 1). The control of KC transcription is largely dependent upon 2 juxtaposed NFκB sites located between 70 and 90 nucleotides from the start site.27 However, the activation of nuclear NFκB DNA-binding activity in response to LPS stimulation was not altered by TGFβ (Figure 3B). To further explore the impact of TGFβ treatment on LPS-induced chemokine gene transcription, a series of KC gene promoter fragments linked with the CAT reporter cDNA were studied in transiently transfected RAW264.7 macrophages. Fragments containing 290, 146, and 104 nucleotides immediately upstream of the KC transcription start site were coupled with the CAT reporter cDNA in the pCATbasic plasmid as described previously27 and used to transfect RAW264.7 cells. At 24 hours after transfection the cells were stimulated or not with LPS for an additional 18 hours and cell extracts prepared to measure levels of CAT activity. As previously demonstrated,27 all 3 fragments were capable of driving enhanced reporter gene expression in response to stimulation with LPS (Figure 3C). In the presence of TGFβ, however, there was no reduction in response. When 2 copies of the most active site (κB1) were placed in a heterologous promoter context (pTK), a similar strong response to LPS stimulation was obtained that was, like the KC promoter fragment, insensitive to suppression by cotreatment with TGFβ. Hence, the inhibitory effects of TGFβ on LPS-induced KC mRNA levels are not mediated by inhibition of the activity of the KC promoter or the activation and transactivation function of NFκB.
The effect of TGFβ on LPS-induced KC and MIP-2 gene transcription. (A) TG-elicited macrophages were cultured in medium alone or with LPS (10 ng/mL) in the absence or presence of TGFβ (20 ng/mL) for the indicated times. Nuclei were isolated and used for determination of transcription for KC, MIP-2, and tubulin by nuclear run-on analysis as described in “Materials and methods.” (B) RAW264.7 cells were either untreated or treated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for the times indicated before the preparation of nuclear extracts. Of each nuclear extract, 5 μg was analyzed for NFκB DNA-binding activity by EMSA using radiolabeled oligonucleotides containing the κ B1 sequence from the KC promoter. (C) RAW264.7 cells were transiently transfected with the indicated constructs, and after 24 hours of rest, the cells were either untreated or stimulated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for 18 hours before the analysis of CAT protein production by ELISA. Different KC promoter fragments linked to CAT are illustrated schematically. Values presented are the means ± SEM of 3 separate experiments. Similar results were obtained in 2 separate experiments.
The effect of TGFβ on LPS-induced KC and MIP-2 gene transcription. (A) TG-elicited macrophages were cultured in medium alone or with LPS (10 ng/mL) in the absence or presence of TGFβ (20 ng/mL) for the indicated times. Nuclei were isolated and used for determination of transcription for KC, MIP-2, and tubulin by nuclear run-on analysis as described in “Materials and methods.” (B) RAW264.7 cells were either untreated or treated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for the times indicated before the preparation of nuclear extracts. Of each nuclear extract, 5 μg was analyzed for NFκB DNA-binding activity by EMSA using radiolabeled oligonucleotides containing the κ B1 sequence from the KC promoter. (C) RAW264.7 cells were transiently transfected with the indicated constructs, and after 24 hours of rest, the cells were either untreated or stimulated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for 18 hours before the analysis of CAT protein production by ELISA. Different KC promoter fragments linked to CAT are illustrated schematically. Values presented are the means ± SEM of 3 separate experiments. Similar results were obtained in 2 separate experiments.
TGFβ-mediated suppression of KC expression depends upon its mRNA sequence
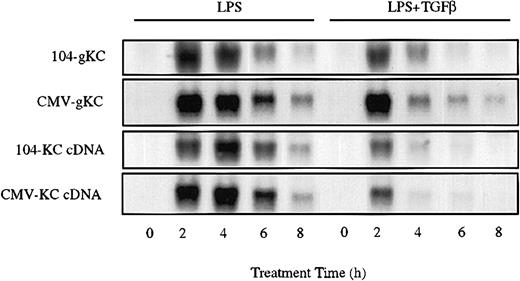
While KC promoter constructs transfected in RAW264.7 cells were insensitive to the inhibitory effects of TGFβ, LPS-stimulated expression of KC mRNA derived from stably transfected KC transgenes exhibited marked sensitivity (Figure 4). A variant of RAW264.7 cells deficient in expression of the endogenous KC gene was stably transfected with either a genomic clone of KC or the full-length KC cDNA (either under control of the 104 nucleotide KC promoter or the CMV promoter in the pcDNA3 vector). In the absence of LPS stimulation, expression of KC mRNA in these cells was modest (CMV promoter) or not detectable (KC promoter). In response to LPS, however, expression was strongly elevated within 2 hours. Cotreatment with LPS and TGFβ resulted in marked inhibition of the accumulation of mRNAs derived from all 4 constructs. As the response of the isolated KC promoter to LPS was unaffected by TGFβ, these findings suggest that sequences within the mRNA can confer sensitivity to the suppressive action of TGFβ. Furthermore, since the promoters alone do not show sensitivity to TGFβ, these results suggest that suppressive function may operate, at least in part, through posttranscriptional mechanisms.
KC mRNA sequence confers sensitivity to TGFβ. RAW264.7 cell lines stably transfected with the indicated genomic or cDNA clones of KC were stimulated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for the indicated times. Total RNA was prepared and used to determine the levels of transgenic KC mRNA by Northern hybridization. Similar results were obtained in 3 separate experiments.
KC mRNA sequence confers sensitivity to TGFβ. RAW264.7 cell lines stably transfected with the indicated genomic or cDNA clones of KC were stimulated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for the indicated times. Total RNA was prepared and used to determine the levels of transgenic KC mRNA by Northern hybridization. Similar results were obtained in 3 separate experiments.
TGFβ antagonizes LPS-mediated stabilization of KC mRNA
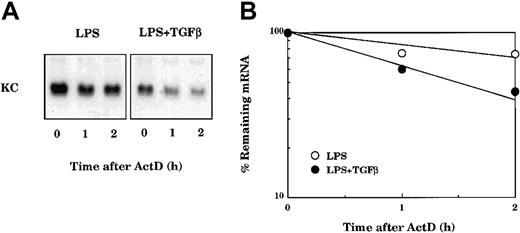
The effects of LPS and TGFβ treatment on KC mRNA stability were evaluated in primary macrophages. Cultures were stimulated with LPS for 3 hours either in the presence or absence of TGFβ. ActD was added to prevent further transcription, and the decay of KC mRNA was followed by Northern hybridization analysis (Figure 5). KC mRNA in LPS-stimulated cells remained stable over 2 hours after treatment with ActD. In contrast, the stability of KC mRNA in cells cotreated with LPS and TGFβ was markedly reduced. Hence the action of TGFβ appears to reduce levels of KC mRNA in part by increasing the rate of mRNA decay.
TGFβ blocks LPS-mediated stabilization of KC mRNA. (A) TG-elicited macrophages were stimulated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for 3 hours. Each culture was subsequently treated with ActD (5 μg/mL), and at the indicated times cultures were used to prepare total RNA for analysis of KC mRNA by Northern hybridization (B). The autoradiographs were quantified using the IMAGE software from the National Institutes of Health (NIH, Rockville, MD). Similar results were obtained in 2 separate experiments.
TGFβ blocks LPS-mediated stabilization of KC mRNA. (A) TG-elicited macrophages were stimulated with LPS (10 ng/mL) in the presence or absence of TGFβ (20 ng/mL) for 3 hours. Each culture was subsequently treated with ActD (5 μg/mL), and at the indicated times cultures were used to prepare total RNA for analysis of KC mRNA by Northern hybridization (B). The autoradiographs were quantified using the IMAGE software from the National Institutes of Health (NIH, Rockville, MD). Similar results were obtained in 2 separate experiments.
Previous work has demonstrated that KC mRNA is short lived by virtue of the presence of adenosine-uridine (AU)–rich sequences found in the 3′ untranslated region (UTR) of the molecule.20,33,34 Moreover, multiple unstable cytokine and chemokine mRNAs are known to be stabilized in response to stimulation by extracellular agents including LPS.19,35-37 Therefore, we reasoned that the effect of TGFβ could be either to directly modulate the decay of KC mRNA or to interfere with stabilization induced in response to LPS. It is difficult to measure the decay rate of KC mRNA in the absence of LPS (because LPS is required for its transcription), and thus we chose to examine the effects of both LPS and TGFβ on decay of KC mRNA driven from a tetracycline-regulated promoter as recently described.38 Initially we prepared a line of RAW264.7 cells stably transfected to express the tet repressor protein tetR-VP16 in which transcription from a TRE can be negatively controlled by the addition of Dox. In this setting the decay of specific mRNAs under TRE control can then be followed without the use of transcriptional poison such as ActD. To create the stable tetR-VP-16–transfected macrophages, we used as the parental strain the derivative of RAW264.7 cells referred to above that does not express the endogenous KC gene. This allowed detection of transfected full-length KC mRNA in the absence of endogenous KC mRNA background.
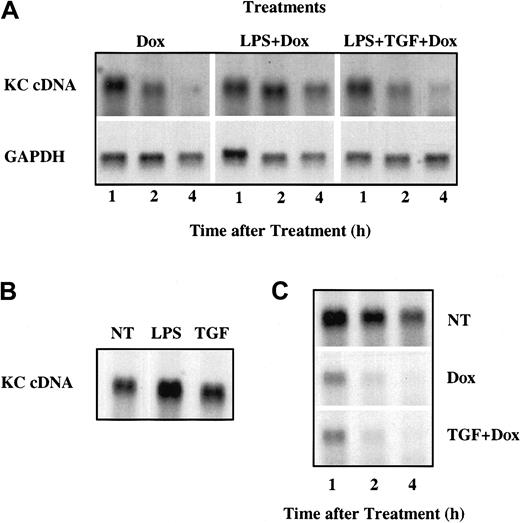
A pool of tet-off RAW264.7 cells was transfected with a plasmid encoding the full-length KC mRNA under control of a TRE promoter (pTRE-KCcDNA). At 3 hours after transfection, the cells were divided into separate culture dishes for different treatment conditions and rested for 24 hours. Individual culture dishes were treated with Dox alone or with LPS in the presence or absence of TGFβ. KC mRNA was highly unstable following addition of Dox and decayed to background levels within 4 hours (Figure 6A). LPS treatment reduced this decay rate substantially and this effect of LPS was blocked in the presence of TGFβ (Figure 6A). The reduction of KC mRNA stability seen in cells cotreated with LPS and TGFβ could be a direct effect of TGFβ on the rate of KC mRNA decay or could result from inhibition of the stabilization response to LPS. To discriminate between these possibilities, the effects of TGFβ alone on KC mRNA levels were determined both in the presence and absence of Dox. LPS treatment of tet-off RAW264.7 cells transfected with pTRE-KCcDNA in the absence of Dox resulted in elevated levels of KC mRNA, while treatment with TGFβ alone had no effect (Figure 6B). Furthermore, KC mRNA decayed rapidly but with comparable kinetics in cells treated either with Dox alone or Dox in combination with TGFβ (Figure 6C). These findings collectively demonstrate that TGFβ impacts on the stability of KC mRNA only indirectly, by interfering with the stabilization response to LPS.
TGFβ inhibits LPS-mediated KC mRNA stabilization in tet-off RAW264.7 cells. (A) tet-off RAW264.7 cells were transiently transfected with pTRE2/KCcDNA and after 3 hours were subcultured into separate dishes for individual treatments. After an additional 18 hours of rest, separate cultures were treated with Dox (100 ng/mL), LPS (10 ng/mL), or TGFβ (20 ng/mL) alone or in combination as indicated for 1, 2, or 4 hours. Total RNA was prepared and KC or GAPDH mRNA levels were determined by northern hybridization. (B) tet-off RAW264.7 cells transfected as above with pTRE2/KCcDNA were untreated (NT) or treated with LPS (10 ng/mL) or TGFβ (20 ng/mL) for 4 hours prior to analysis of KC mRNA levels. (C) tet-off RAW264.7 cells transfected with pTRE2/KCcDNA were untreated or treated with Dox or Dox + TGFβ for 1, 2, or 4 hours prior to analysis of KC mRNA levels by northern hybridization. Similar results were obtained in 2 separate experiments.
TGFβ inhibits LPS-mediated KC mRNA stabilization in tet-off RAW264.7 cells. (A) tet-off RAW264.7 cells were transiently transfected with pTRE2/KCcDNA and after 3 hours were subcultured into separate dishes for individual treatments. After an additional 18 hours of rest, separate cultures were treated with Dox (100 ng/mL), LPS (10 ng/mL), or TGFβ (20 ng/mL) alone or in combination as indicated for 1, 2, or 4 hours. Total RNA was prepared and KC or GAPDH mRNA levels were determined by northern hybridization. (B) tet-off RAW264.7 cells transfected as above with pTRE2/KCcDNA were untreated (NT) or treated with LPS (10 ng/mL) or TGFβ (20 ng/mL) for 4 hours prior to analysis of KC mRNA levels. (C) tet-off RAW264.7 cells transfected with pTRE2/KCcDNA were untreated or treated with Dox or Dox + TGFβ for 1, 2, or 4 hours prior to analysis of KC mRNA levels by northern hybridization. Similar results were obtained in 2 separate experiments.
Role of p38 MAPK in KC mRNA stability
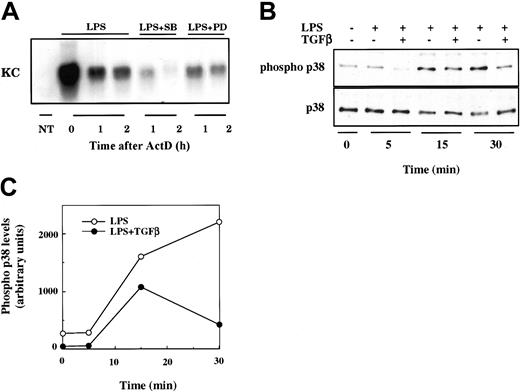
The stabilization of some AU-rich mRNAs by extracellular stimulus has been shown to depend upon the activation of p38 MAPK.35,37 Furthermore, TGFβ has been reported to interfere with the transcription of LPS-induced inflammatory genes in macrophages by indirect inhibition of p38 activity.39 Hence we wished to determine if p38 MAPK activation is necessary for stabilization of KC mRNA. The decay of LPS-induced KC mRNA in RAW264.7 cells was markedly enhanced in the presence of the specific p38 MAPK inhibitor SB203580 (Figure 7A). This effect is relatively specific for p38, as an inhibitor of the extracellular signal-related kinase 1/2 (ERK1/2) MAPKs (PD980589) had only a modest effect on the stability of LPS-induced KC mRNA. We next determined if TGFβ treatment could antagonize LPS-stimulated activation of p38 MAPK. RAW264.7 cells were pretreated or not with TGFβ for 3 hours prior to stimulation with LPS for various times. Total cell lysates were prepared and analyzed for total p38 protein and phosphorylated p38 (Figure 7B). LPS stimulated the activation of p38 as evidenced by the increase in levels of the phosphorylated form present at 15 and 30 minutes. In TGFβ-pretreated cells, however, the amount of phosphorylated p38 was markedly reduced (especially at 30 minutes). TGFβ did not alter the levels of total p38. In combination, these findings suggest that TGFβ can block LPS-mediated stabilization of KC mRNA via interference with the activation of p38 MAPK.
Role of p38 MAPK activity in control of KC mRNA stability. (A) RAW264.7 cells were stimulated with nothing (NT) or LPS (10 ng/mL) for 2 hours prior to the addition of ActD in the presence or absence of SB203580 (2 μM) or PD980589 (50 μM) as indicated. Total RNA was prepared at 0, 1, and 2 hours after ActD treatment and analyzed by Northern hybridization for KC mRNA levels. Similar results were obtained in 2 separate experiments. (B). RAW264.7 cells were treated or not with TGFβ for 3 hours and subsequently stimulated with LPS (10 ng/mL) for the indicated times. Total cell lysates were prepared and analyzed for both phosphorylated p38 and total p38 levels by Western blot. The film was quantified using the NIH IMAGE software package. Similar results were obtained in separate experiments.
Role of p38 MAPK activity in control of KC mRNA stability. (A) RAW264.7 cells were stimulated with nothing (NT) or LPS (10 ng/mL) for 2 hours prior to the addition of ActD in the presence or absence of SB203580 (2 μM) or PD980589 (50 μM) as indicated. Total RNA was prepared at 0, 1, and 2 hours after ActD treatment and analyzed by Northern hybridization for KC mRNA levels. Similar results were obtained in 2 separate experiments. (B). RAW264.7 cells were treated or not with TGFβ for 3 hours and subsequently stimulated with LPS (10 ng/mL) for the indicated times. Total cell lysates were prepared and analyzed for both phosphorylated p38 and total p38 levels by Western blot. The film was quantified using the NIH IMAGE software package. Similar results were obtained in separate experiments.
Discussion
The anti-inflammatory activity of TGFβ1 is well recognized and is derived, in part, from its ability to suppress the production of proinflammatory products in mononuclear phagocytes.6,7,11 The intracellular molecular mechanisms through which this negative function is accomplished are, however, only partially understood. The present study explored the steps in LPS-stimulated chemokine gene expression that may be sensitive to the action of TGFβ. This work has demonstrated that the suppressive function of TGFβ is selective for a subset of LPS-responsive chemokine genes. Furthermore, the inhibitory effects of TGFβ on chemokine expression require protein synthesis and operate, at least in part, by antagonisim of LPS-mediated mRNA stabilization. Finally, the stabilization by LPS requires the activation of p38 MAPK and this is blocked by TGFβ. These conclusions are based upon the following experimental findings. (1) While TGFβ inhibits the induction of KC and MIP-2 mRNAs, it has no effect on the expression of either IP-10 or MCP-1. (2) The reduction in expression of both KC and MIP-2 is time-dependent and the protein synthesis inhibitor CHX blocks the development of suppression. (3) Although TGFβ modestly inhibits LPS-induced KC and MIP-2 transcription, it has no effect on NFκB activation nor NFκB-dependent transcription. (4) Sensitivity to TGFβ-mediated suppression is conferred by sequences within the mature mRNA. (5) LPS treatment stabilizes KC mRNA and TGFβ antagonizes this response. (6) Inhibition of p38 MAPK activation in LPS-stimulated macrophages enhances KC mRNA decay, and TGFβ dampens LPS-mediated activation of p38.
The present results suggest that TGFβ-mediated inhibition of KC mRNA expression depends upon TGFβ-induced expression of a new gene product or products that subsequently execute the suppressive mechanism. In support of this hypothesis, TGFβ-mediated suppression is not maximal for at least 2 hours when TGFβ and LPS are added at the same time. Suppression is not delayed, however, if cells are treated with TGFβ for 3 hours before exposure to LPS. Furthermore, the pretreatment efficacy is lost when protein synthesis is inhibited with CHX during the pretreatment period. This could reflect a requirement for continuous synthesis of a short-lived protein or for the induced expression of a new gene in response to TGFβ. This is not a result, however, of TGFβ-mediated enhancement of IL-10 production since neutralizing antibody to IL-10 had no effect on the suppressive function.
KC and MIP-2 gene transcription was modestly reduced in the presence of TGFβ, but there was no apparent impact on the activation or function of NFκB, the primary transcription factor involved in LPS-regulated expression of these genes.27,40 This finding is consistent with the selectivity of TGFβ-mediated suppression; IP-10 and MCP-1 mRNA expression are unaffected by TGFβ, though transcription of both depends upon activation of NFκB.22,41 Indeed, the kinetic patterns of expression of KC and MIP-2 mRNAs versus IP-10 and MCP-1 mRNAs are quite distinct, suggesting the mechanistic difference (Figure 1). Prior studies have shown that TGFβ-mediated suppression of interferon γ–, IL-1α–, or TNFα-stimulated transcription does not involve interference with signaling responses or transcription factor activation to any of these stimulatory agents.16,42,43 In addition, transcription from a previously defined LPS-responsive KC promoter fragment was not suppressed by TGFβ. This result suggests that the effects of TGFβ on KC transcription might involve a site or sites located outside of the immediate promoter region.
Although the KC promoter did not show any sensitivity to TGFβ, constructs containing the full KC gene in the context of either the homologous KC promoter or a heterologous promoter (CMV) showed sensitivity nearly comparable with the endogenous gene. This could indicate that sequences within the transcription unit itself are necessary for transcriptional suppression and this possibility has not been ruled out. Alternatively, this behavior could reflect regulation at a posttranscriptional level, and the finding that TGFβ decreased the stability of LPS-induced KC mRNA supports this possibility. While many studies have reported that TGFβ suppresses inflammatory gene transcription, alterations in the stability of the target mRNA have also been demonstrated.17,18 Furthermore, a number of CXC chemokine mRNAs are known to exhibit altered mRNA decay rates in response to extracellular stimuli.19,20,32,34,36 For example, IL-8, Gro, and KC mRNAs are unstable because of the presence of multiple AU-rich sequences within their 3′UTRs but can be stabilized in response to IL-1α. Finally, IL-10, another anti-inflammatory cytokine, achieves suppression of cytokine and chemokine expression at least in part by enhancing specific mRNA decay.32,33,44 Because LPS has also been reported to stabilize inflammation-related mRNAs,35,37 the effect of TGFβ could result either from a direct effect on the stability of KC mRNA or from the ability of TGFβ to interfere with stabilization in response to LPS. Distinguishing between these alternative hypotheses required examination of KC mRNA decay in the absence of LPS, which is necessary for its transcription. This was accomplished using the “tet-off” system to control the transcription of a KC transgene. LPS prolonged the half-life of KC mRNA transcribed from the tet-responsive promoter, while TGFβ had no effect when used alone. Inclusion of TGFβ in the incubation along with LPS prevented the ability of LPS to stabilize KC mRNA, demonstrating clearly that TGFβ achieves the posttranscriptional effect indirectly by interfering with the response to LPS.
A recent report has suggested that the inhibitory effects of TGFβ on chemokine gene expression in mouse macrophages may involve the activation of MPK-1 protein phosphatase.39 This mechanism proposes that TGFβ inhibits LPS-activated p38 MAPK through the activation of ERK and MPK-1 and ultimately results in reduced activation of NFκB. Because LPS stimulation of mRNA stability has been reported to depend upon p38 protein kinase activity,35,37 this mechanism could account for the ability of TGFβ to antagonize KC mRNA stabilization. Indeed, LPS-mediated stimulation of KC mRNA stability can be blocked more effectively by treatment with the p38 MAPK–specific inhibitor SB203580 than by the ERK1/2 inhibitor PD980589. Furthermore, pretreatment of cells with TGFβ inhibited LPS-stimulated p38 MAPK activity measured as phosphorylated p38 protein. These results are consistent with the possibility that the ability of TGFβ to prevent stabilization of chemokine mRNA is a result of the inhibition of p38 activity.
The present study documents the physiologic complexity involved in the action of TGFβ as an endogenous anti-inflammatory regulator through suppression of proinflammatory gene expression. The inhibition of CXC chemokine production is likely to be an important contributor to this function by reducing the recruitment of inflammatory neutrophils to sites of tissue injury. The selective effects of TGFβ on KC and MIP-2 compared with IP-10 and MCP-1 do not necessarily reflect selective antagonism of neutrophil recruitment in vivo. Furthermore, TGFβ is one of several endogenous anti-inflammatory cytokines that also include IL-4/IL-13, IL-10, and IL-9.5,14,45-47 Multiple studies have illustrated the spectrum of inflammation-associated genes that can be inhibited by these individual agents and demonstrated the mechanistic diversity involved. These include variable dependence on intermediate protein synthesis and their differential targeting of transcriptional or posttranscriptional activities.17,48,49 In addition, the cell type, the nature of the stimulus inducing the primary inflammatory response, and the specific gene under study are all important parameters influencing the final outcome. While this complexity confounds our understanding of TGFβ function, it suggests that detailed knowledge may ultimately provide the opportunity to manipulate the inflammatory process in a very selective fashion.
Prepublished online as Blood First Edition Paper, April 24, 2003; DOI 10.1182/blood-2002-12-3771.
Supported by United States Public Health Service grants CA62220 and CA39621.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.