Abstract
The Notch family of receptors plays an important role in regulation of cell differentiation via direct contact between hematopoietic progenitor cells (HPCs) and bone marrow stroma (BMS). However the precise contribution of Notch in dendritic cell (DC) differentiation is controversial. In 2 different experimental systems using Notch-1–null embryonic stem cells and Notch-1–deficient HPCs we have found that Notch-1 is necessary for DC differentiation. However, activation of Notch-1 and Notch-2 with cell-bound Notch ligand did not result in differentiation of mature DCs or macrophages. Instead, it caused accumulation of immature myeloid cells. Removal of feeder cells resulted in rapid differentiation of DCs and macrophages. Addition of interleukin 4 (IL-4) into the culture dramatically increased accumulation of functionally potent DCs. Lipopolysaccharide was not able to reproduce this effect. Thus, these data indicate that Notch signaling prevents differentiation of mature myeloid cells. Instead, it results in accumulation of precursors readily able to differentiate into mature DCs once the Notch signal is stopped (eg, after cell emigration from bone marrow) and in the presence of other additional differentiation signals provided by IL-4. Thus, Notch is required but not sufficient for DC differentiation.
Introduction
Dendritic cells (DCs) are professional antigen-presenting cells critically important for induction of immune responses.1-3 Several subsets of these cells including lymphoid, myeloid, and plasmacytoid DCs have been recently identified.4 Differentiation of all DC subtypes is taking place in the bone marrow and it is tightly controlled by a complex network of different soluble growth factors and cytokines and by direct cell-cell contact between hematopoietic progenitor cells (HPCs) and bone marrow stroma (BMS). To date, several growth factors important for DC differentiation have been identified (for a review, see Vuckovic et al5 ). However, the role of direct cell-cell contact in DC differentiation remains unclear.
In recent years Notch signaling emerged as a critical element of the development and maintenance of hematopoiesis. The Notch family is a group of highly conserved molecules that are expressed as receptors on the cell surface and directly regulate gene transcription. There is accumulating evidence that the Notch pathway affects survival, proliferation, and cell fate choices at various stages of hematopoietic cell development, including the decisions of HSCs to self-renew or differentiate and of common lymphoid precursors to undergo T- or B-cell differentiation.6-8 Notch ligands in vertebrates include Jagged-1 and -2, and Delta-1, -2, -3, and -4.8 Jagged-1 and Delta-1 Notch ligands are expressed on BMS cells. Notch-1 is activated following binding of appropriate ligands on adjacent cells to the extracellular domain of Notch-1 on the surface of HPCs. This results in a 2-stage proteolytic cleavage, release, and nuclear translocation of the Notch intracellular domain. This domain interacts with a number of cytoplasmic and nuclear proteins, permitting signal transduction through several pathways that include activation of CBF-1/Rbp-Jk transcription factor and E(spl)/HES genes, which work as negative regulators of lineage-specific gene expression.
An important role of Notch signaling in differentiation of lymphocytes is well established.7-10 The role of Notch signaling in myeloid cell differentiation is much more controversial. In vivo experiments with a conditional knockout of the Notch-1 gene showed no effect on myeloid development.9,11 Differentiation of plasmacytoid DCs was also not affected.12 Immobilized Notch ligand Delta-1 inhibited differentiation of monocytes into mature macrophages with granulocyte-macrophage colony-stimulating factor (GM-CSF). However, it permitted differentiation into mature DCs.13 These data might suggest that Notch is not involved in differentiation of myeloid cells in general and DCs in particular. However, a number of studies suggest possible involvement of Notch in myeloid cell differentiation. Constitutive expression of the activated intracellular domain of mouse Notch-1 in 32D myeloid progenitors inhibits granulocytic differentiation and permits expansion of undifferentiated cells.14 Overexpression of intracellular domain of Notch-1 in these cells inhibited differentiation induced by granulocyte colony-stimulating factor (G-CSF), whereas overexpression of corresponding domain of Notch-2 inhibited differentiation induced by GM-CSF.15 Another group has shown that conditional induction of the constitutively active intracellular domain of murine Notch-1 promoted myeloid differentiation via RBP-J transactivation.16 Recently, Varnum-Finney and colleagues have demonstrated that incubation of murine bone marrow precursors with Notch ligand and growth factors inhibited myeloid differentiation and promoted an increase in the number of precursors capable of short-term lymphoid and myeloid repopulation.17 In a different study a soluble form of Notch ligand Jagged-1 was able to induce maturation of monocyte-derived human DCs.18
Taken together these data depict a rather convoluted picture of the possible involvement of Notch in DC differentiation. In this study we have tried to clarify the potential role of Notch signaling in myeloid cell differentiation in general, and DCs, in particular.
Materials and methods
Mice
Female Swiss mice (6-8 weeks old) were purchased from Charles River Laboratories (Wilmington, MA). Female BALB/c and C57BL/6 mice (6-8 weeks old) were obtained from Harlan (Indianapolis, IN). All mice were housed in specific pathogen-free units of the Division of Comparative Medicine at University of South Florida (Tampa, FL). Notch-1 antisense transgenic (Notch-AS-Tg) mice were described elsewhere.19 Briefly, mice were generated using a Notch-1 antisense construct expressed under the control of the mouse mammary tumor virus long terminal repeat promoter. The genetic background of the founders was C57BL/6:SJL F1, and the mice were then back-crossed for 4 generations with C57BL/6 mice. Hemizygous transgenic mice were then bred to each other, selecting homozygous transgenic and negative mice until 2 syngeneic strains derived from the same original litter were obtained: one homozygous antisense Notch-1 transgenic (Notch-AS-Tg) and one nontransgenic (control). The specificity of Notch-1 down-regulation was confirmed by Western blotting.19
Reagents
The following antibody-producing hybridomas were purchased from the American Type Culture Collection (Manassas, VA) and used as supernatants: anti-CD4 (TIB-207); anti-CD8 (TIB-210); and anti–MHC II (TIB-120). Anti–TER-119, B220, Gr-1 (anti–Ly-6G), IAb, IAq, CD11b, CD86 (B7-2), CD45, CD11c, CD34, Sca-1, c-Kit, and isotype control antibodies were obtained from PharMingen (San Diego, CA). Anti-F4/80 antibody was purchased from Serotec (Raleigh, NC); antiphycoerythrin (anti-PE) or streptavidin microbeads were from Miltenyl Biotec (Auburn, CA). Antibodies against Notch-1 (bTAN20) and Notch-2 (C651.6DbHN) were purchased from Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA); anti–Notch-3 and anti–Notch-4 were from Santa Cruz Biotechnology (Santa Cruz, CA). Low-Tox rabbit complement, and Lympholyte M were purchased from Cedarlane Laboratories (Hornby, ON, Canada). Recombinant human FLT-3 ligand (FL), recombinant murine GM-CSF, G-CSF, macrophage colony-stimulating factor (M-CSF), interleukin 4 (IL-4), IL-3, and tumor necrosis factor α (TNF-α) were obtained from Research Diagnostics (Flanders, NJ), lipopolysaccharide (LPS) from Sigma (St Louis, MO), and poly(dI-dC) from Pharmacia (Piscataway, NJ).
Preparation of enriched HPCs
Bone marrow cells were harvested from the femurs and tibias of mice and enriched for HPCs by depletion of lineage-specific cells as described earlier.19 Briefly, bone marrow cells were incubated with mixture of antibodies (TIB-207, TIB-210, TIB-120, anti–TER-119, anti–Gr-1, anti-B220) for 30 minutes on ice, washed, and treated with complement for 1 hour at 37°C. Dead cells were then removed by centrifugation over a Lympholyte M gradient. The resulting fraction contained less than 20% of lineage-positive cells as detected by flow cytometry.
NIH-3T3-Jagged-1–expressing cell line
NIH-3T3-Jagged-1–expressing cell line (3T3-Jagged) was a gift from Irwin D. Bernstein (Fred Hutchinson Cancer Center, Seattle, WA) and the generation of the cell line was described previously.20 A control fibroblast cell line NIH-3T3-vector (3T3-MSCV) was generated by infecting parental NIH-3T3 cells with empty retroviral vector MSCV, and selected with 1 mg/mL G418.
Generation of DCs from HPCs
DCs were generated from HPCs using FL as described earlier with minor modification.21 Briefly, 5 × 105 HPCs per well of 24-well plates were cultured for 10 days in 2 mL complete medium supplemented with 200 ng/mL FL and 10% of conditioned medium from splenocytes. On day 9, 1 μg/mL LPS was added, and cells were cultured for additional 24 hours, collected, washed, and used for further analysis. For generation of DCs and other myeloid cell on fibroblasts control and Jagged 3T3 fibroblasts were irradiated with 25 Gy and cultured overnight in 24-well plates (1.5 × 105 cells/well). Half million HPCs were placed into each well in 2 mL 10% fetal bovine serum (FBS) RPMI supplemented with 20 ng/mL GM-CSF or M-CSF for 7 or 10 days. In some experiments to generate DCs, IL-4 (10 ng/mL) was also added. Every 3 days, cells were transferred onto new fibroblasts and medium was replaced with growth factors. For DC activation, TNF-α (5 ng/mL) was added 48 hours prior to cell analysis. On day 7 or day 10, all cells were collected and analyzed for DC, macrophage, or granulocyte phenotype.
DC differentiation of Notch-1-/-ES cells
Differentiation of DCs from embryonic stem (ES) cells was performed as described earlier22 with modifications. Notch-1+/+ and Notch-1-/- ES cell lines were kind gifts from Dr Conlon (Case Western Reserve University, Cleveland, OH). They were made in R1-S3 ES cell line and were described in detail elsewhere.23 ES cells were grown on gelatinized plates in complete knockout Dulbecco modified Eagle medium (DMEM) supplemented with 15% ES-qualified FBS (Gibco, Rockville, MD), 0.1 mM nonessential amino acid, 100 μM 2-mecaptoethenol, 2 mM l-glutamine, and leukocyte inhibitory factor (LIF; 1000 U). To induce formation of embryonic bodies hematopoietic cell ES cells were transferred on bacterial-grade Petri dishes with LIF-free knockout DMEM supplemented with 15% ES-qualified FBS and 100 μM 2-mercaptothenol and incubated for 14 days. Medium was changed every other day. On day 14, one embryonic body was transferred to each well of a 24-well plate and incubated in 2 mL/well 15% FBS RPMI medium with 20 ng/mL GM-CSF and 10 ng/mL IL-3 for 30 days. Half of the medium was changed every 3 days. TNF-α (5 ng/mL) was added for the last 5 days before DC analysis.
Isolation of CD45+ and Gr-1+ cells from HPCs cocultured with 3T3 fibroblasts
CD45+ cell or Gr-1+ cell isolation was performed using MiniMACS microbeads according to the manufacturer's protocol (Miltenyi Biotec). After incubation with fibroblasts, cells were collected, washed twice with phosphate-buffered saline (PBS), and incubated with either antimouse CD45 PE-conjugated antibody or antimouse Gr-1 biotinylated antibody on ice for 10 minutes. Then, cells were washed and incubated with either anti-PE microbeads or streptavidin microbeads on ice for 10 minutes. After washing, cells were separated on MACS columns and used in experiments. Purity for CD45+ or Gr-1+ cells was more than 93% in all samples.
Analysis of the phenotype of myeloid cells
The phenotype of DCs and myeloid progenitors was analyzed on FACSCalibur flow cytometer (BD Biosciences, Mountain View, CA) using a CellQuest program (Becton Dickinson, Mountain View, CA). A combination of antibodies conjugated with different fluorochromes was used in multicolor analysis.
Allogeneic MLR
Allogeneic mixed leukocyte reaction (MLR) was used to determine DC function. Briefly, DCs were sorted using FACSVantage SE cell sorter (BD Biosciences) and were cultured for 4 days in triplicate in U-bottom 96-well plates with lymph node cells (105/well) obtained from allogeneic BALB/c mice. Then, 1 μCi (0.037 MBq) thymidine 24 was added to each well 18 hours before cell harvest. T-cell proliferation was measured by [3H]-thymidine incorporation using a liquid scintillation counter (Packard Instrument, Meriden, CT).
EMSA
Electrophoretic mobility shift assay (EMSA) was performed as previously described.24 Briefly, double-stranded oligonucleotides containing the specific binding site for CBF-1 were made by annealing the appropriate single-stranded oligonucleotides at 65°C for 10 minutes. The probes were labeled with α32P deoxyadenosine-5′ triphosphate (dATP) (6000 μCi/mmol [222.0 MBq]; Amersham Life Sciences, Arlington Heights, IL) using Klenow DNA polymerase. The 2 probes used were wild-type 5′-TGGTGTAAACACGCCGTGGGAAAAAATTTA-3′) and mutant 5′-TGGTGTAAACACGCCGTTGGAAAAAATTTA-3′).
HPCs were cultured overnight with 20 ng/mL GM-CSF, washed, and starved for 2 hours in serum-free medium. Two million HPCs per well of 24-well plates were placed on the irradiated 3T3-control or 3T3-Jagged-1 fibroblasts. After coculture CD45+ cells were isolated and EMSA was performed using nuclear extracts.25 Nuclear extract (5 μg) was incubated with labeled probe in binding buffer containing 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 5% glycerol, 0.2 mM EDTA (ethylenediaminetetraacetic acid), 1 mM dithiothreitol (DTT), 5 mM MgCl2, and 4 μg poly(dI-dC). Specific competition assays were performed with a 50-fold excess of unlabeled probes. For CBF-1–binding blockage, 4 μg of monoclonal antibodies (mAbs) against Notch-1 or Notch-2 were incubated with nuclear proteins for 30 minutes on ice prior to addition of α32P-labeled CBF-1 probe. The samples were separated on 4% polyacrylamide gels, and bands were visualized by overnight exposure to x-ray films (Fuji, Stamford, CT) at -70°C.
Western blot assay
Cells were lysed for 30 minutes on ice in lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 1% Triton X-100, 0.5% sodium deoxycholate, 100 mM Na3VO4, 20 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/mL aprotinin, 10 μg/mL leupeptin, 10 μg/mL pepstatin). Samples were subjected to electrophoresis on 10% sodium dodecyl sulfate–polyacrylamide gels followed by transfer to a polyvinylidene difluoride membrane and probing with specific antibody. The bands were visualized by an enhanced chemiluminescence (ECL) detection kit (Amersham Life Sciences).
HPC proliferation on fibroblasts with cytokines
Irradiated fibroblasts (5 × 104/well) were cultured overnight in flat-bottom 96-well plates. HPCs (105/well) were plated on the fibroblasts for 2 or 4 days in triplicates with 20 ng/mL GM-CSF or GM-CSF and 10 ng/mL IL-4. Then 1 μCi (0.037 MBq) [3H]-thymidine, was added to each well 18 hours before cell harvest. HPC proliferation was measured by [3H]-thymidine incorporation using a liquid scintillation counter.
Expression of GM-CSF receptor, IL-4 receptor, and Flt-3–specific mRNA
HPCs were cultured with GM-CSF on 3T3-MSCV or 3T3-Jagged fibroblasts for 48 hours or 72 hours. CD45+ cells were isolated and the total RNA was extracted using the TRIZOL reagent (Life Technologies). Traces of DNA were removed by treatment with DNase I. The cDNA was synthesized from 1 μg total RNA using random hexamers and Superscript II reverse transcriptase (Life Technologies). Samples were subjected to initial denaturation at 94°C for 3 minutes and 28 cycles of polymerase chain reaction (PCR; 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 45 seconds) with final extension for 7 minutes at 72°C. The number of cycles was selected after preliminary experiments to avoid saturation of the PCR products.
The PCR primer pairs used in this study: GM-CSFRα25 : forward, 5′-GCGGGCGACACGAGGATGAAGCAC; reverse, 5′-CTAGGGCTGCAGGAGGTCCTTCCT. IL-4R26 : forward, 5′-GCTAGTTGTCATCCTGCTC; reverse, 5′-GTGATGTGGACTTGGACTC; Flt-3: forward, 5′GGCTCTGTCTCCCCTTCATTG; reverse, 5′-GCCCCCAGCAGATTCACG; hprt: forward, 5′-GATTCAACTTGCGCTCATCTTAGGC; reverse, 5′-GTTGGATACAGGCCAGACTTTGTTG.
The PCR products were visualized on 1% agarose gel. The sizes of PCR products were 263 bp, 352 bp, 393 bp, and 164 bp, respectively. PCR products were transferred in an alkaline transfer buffer (0.4 N NaOH, 1 M NaCl) onto Hybond N+ nylon transfer membranes (Amersham, Highland Park, IL) and probed with 32P-labeled oligonucleotide probes: GM-CSFRα, 5′-TGTCCTCAGCCTCGAGAGGATG-3′; IL-4R, 5′-TGCCAAACGTCCTCACAGC-3′; Flt-3, 5′-CAGGTGGCGGTGAAGATGC-3′; hprt, 5′-GTTGTTGGATATGCCTTGAC-3′.
Statistical analysis
Statistical analysis was performed using parametric methods and JMP statistical software (SAS Institute, Cary, NC).
Results
Notch-1 is necessary for differentiation of DCs
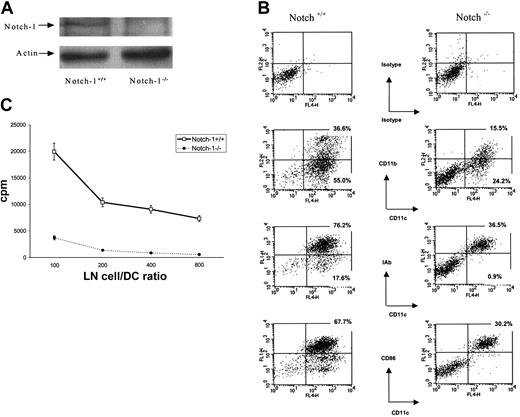
To address the question whether Notch signaling is necessary for DC differentiation we generated DCs from control (Notch-1+/+) and Notch-1-/- ES cells using a recently established experimental system described in “Materials and methods.” Lack of Notch-1 protein in Notch-1-/- cells was confirmed by Western blotting (Figure 1A). This protocol uses GM-CSF and IL-3 and promotes differentiation of immature DCs. To activate DCs, 5 ng/mL TNF-α was added for the last 5 days before the analysis. Around 40% of embryonic bodies from both Notch-1+/+ and Notch-1-/- ES cells were able to produce clusters of loosely attached cells. Analysis of the cell phenotype demonstrated that more than 90% of the cells generated from control ES cells expressed the CD11c marker of DCs. Both myeloid-related (CD11c+CD11b+) and lymphoidrelated (CD11c+CD11b-) DC populations were present (Figure 1B), whereas plasmacytoid DCs (Gr-1+CD11c+B220+) were not detectable (data not shown). More than 75% of these DCs expressed major histocompatibility complex (MHC) class II and almost 70% expressed B7-2 molecules (Figure 1B). These cells were potent stimulators in allogeneic MLRs (Figure 1C). These data were consistent with a previously published observation22 and confirmed the fact that this experimental protocol allows for generation of mature DCs from ES cells. Differentiation of DCs from Notch-1-/- ES cells was significantly impaired. Although the total number of cells generated from individual Notch-1+/+ and Notch-1-/- embryonic bodies was the same, the proportion of both myeloid and lymphoid DCs was reduced more than 2-fold (Figure 1B). The proportion of cells expressing MHC class II and costimulatory B7-2 (CD86) molecules was also more than 2-fold lower than in cells generated from control ES cells (Figure 1B). To test whether these changes in phenotype were associated with functional changes, these cells were used as stimulator in allogeneic MLRs, the function specifically attributed to DCs. The ability of cells generated from Notch-1-/- ES cells to stimulate allogeneic T cells was significantly lower than in their control counterparts, which reflects the decreased presence of functionally competent DCs (Figure 1C). These data demonstrated that Notch-1 is critically important for differentiation of DCs from ES cells.
Notch-1-/- ES cells have reduced capacity to differentiate into DCs. (A) Whole cell lysates were prepared from Notch-1+/+ and Notch-1-/- ES cells, and the presence of Notch-1 protein was determined by Western blotting as described in “Materials and methods.” (B) Embryonic bodies were cultured with GM-CSF and IL-3 for 25 days, followed by a 5-day incubation with TNF-α. Cells were labeled with allophycocyanin (APC)–conjugated anti-CD11c, PE-conjugated anti-CD11b, and fluorescein isothiocyanate (FITC)–conjugated anti-IAb or anti–B7-2 antibodies. Three experiments with the same results were performed. (C) DCs were generated from ES cells as described, irradiated at 150 Gy, and cultured with lymph node (LN) cells from control allogeneic BALB/c mice at different ratios. Cell proliferation was measured in triplicates as described in “Materials and methods.” Values are the average ± SE from 2 experiments.
Notch-1-/- ES cells have reduced capacity to differentiate into DCs. (A) Whole cell lysates were prepared from Notch-1+/+ and Notch-1-/- ES cells, and the presence of Notch-1 protein was determined by Western blotting as described in “Materials and methods.” (B) Embryonic bodies were cultured with GM-CSF and IL-3 for 25 days, followed by a 5-day incubation with TNF-α. Cells were labeled with allophycocyanin (APC)–conjugated anti-CD11c, PE-conjugated anti-CD11b, and fluorescein isothiocyanate (FITC)–conjugated anti-IAb or anti–B7-2 antibodies. Three experiments with the same results were performed. (C) DCs were generated from ES cells as described, irradiated at 150 Gy, and cultured with lymph node (LN) cells from control allogeneic BALB/c mice at different ratios. Cell proliferation was measured in triplicates as described in “Materials and methods.” Values are the average ± SE from 2 experiments.
As shown in Figure 1 a large number of cells generated from Notch-1-/- ES cells lacked expression of DC-specific markers. Using flow cytometry and antibodies specific for different cell lineages we analyzed the population of CD11c- cells. About one third (32.4% ± 4.5%) of CD11c- cells generated from Notch-1+/+ ES cells were CD45+ hematopoietic cells. C-kit+Sca-1+ hematopoi- etic stem cells represented 3.5% ± 0.3% and Gr-1+ CD11b+ immature myeloid cells 3.1% ± 1.1% of CD11c- cells. The rest of the CD45+ cells were Gr-1+CD11b- granulocytes and CD11b+F4/80+ macrophages. In sharp contrast, only 2.0% ± 0.5% of CD11c- cells generated from Notch-1-/- ES cells expressed CD45. About 1% of the cells were c-kit+Sca-1+. Other myeloid cells represented less than 1% of CD11c- cells (data not shown).
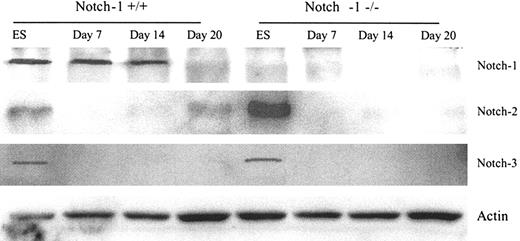
Next, we checked the expression of Notch proteins during DC differentiation from ES cells. Notch-1 was expressed in wild-type ES cells and embryonic bodies and disappeared when cells started differentiation into DCs (Figure 2). Notch-2 expression in Notch- 1-/- ES cells was substantially higher than in Notch-1+/+ ES cells. This may represent a compensatory mechanism, which occurs in Notch-1–deficient cells. Notch-2 expression became undetectable during cell differentiation into embryonic bodies (next 2 weeks) and then slightly increased when cells started differentiation into DCs (day 20; Figure 2). Notch-3 was expressed in both wild-type and Notch-1-/- ES cells, but not at later stages of cell differentiation (Figure 2). Notch-4 was undetectable at all stages of cell differentiation (data not shown).
Notch family members expressed on different stages of ES cell differentiation. Whole cell lysates were prepared from ES cells or cells at different stages of differentiation. Western blot was performed using anti–Notch-1, -2, -3, and β-actin antibodies as described in “Materials and methods.” ES indicates embryonic stem cells cultured with LIF; day 7 and day 14, embryonic body development from ES cells for 14 days; day 20, ES cell DC generation for 6 days with GM-CSF and IL-3 after 14-day embryonic body development.
Notch family members expressed on different stages of ES cell differentiation. Whole cell lysates were prepared from ES cells or cells at different stages of differentiation. Western blot was performed using anti–Notch-1, -2, -3, and β-actin antibodies as described in “Materials and methods.” ES indicates embryonic stem cells cultured with LIF; day 7 and day 14, embryonic body development from ES cells for 14 days; day 20, ES cell DC generation for 6 days with GM-CSF and IL-3 after 14-day embryonic body development.
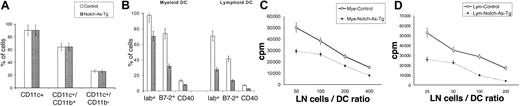
To verify the effect of Notch-1 deficit on DC differentiation we used the other experimental system that used FL and allowed for generation of both myeloid-related and lymphoid-related DCs from HPCs.21 Enriched HPCs were isolated from bone marrow of control or Notch-deficient (Notch-As-Tg) mice. As we reported earlier, the levels of Notch-1 protein in HPCs from Notch-As-Tg mice was 2-fold lower than that in HPCs from control mice.19 Cells were cultured with FL as described in “Materials and methods.” To activate DCs, 1 μg/mL LPS was added to the culture 24 hours prior to analysis. No differences in the total number of cells (data not shown) or in the proportion of populations of CD11c+CD11b+ or CD11c+CD11b- DCs were found between cells isolated from these 2 groups of mice (Figure 3A). These data might indicate that the decrease in Notch-1 protein in HPCs was not sufficient to cause a significant decrease in DC differentiation as was the case in Notch-1–null ES cells. However, when we evaluated the expression of markers attributed to mature DCs, dramatic differences were found within both populations of cells. In DCs generated from HPCs of Notch-As-Tg mice the expression of MHC class II, B7-2, and CD40 molecules was significantly reduced. It manifested in a significant decrease in the proportion of cells positive for MHC class II, B7-2, and CD40 (Figure 3B) and in more than a 2-fold decrease in the intensity of fluorescence (data not shown). The defect in differentiation of both populations of DCs was confirmed in a functional test. DCs generated from HPCs as described were sorted into 2 populations: CD11c+CD11b+ myeloid-related and CD11c+CD11b- lymphoid-related DCs, and used in allogeneic MLRs. Both populations of DCs generated from Notch-1-AS-Tg mice had a significantly lower ability to stimulate allogeneic T-cell proliferation than the cells generated from control mice (Figure 3C-D). Less than 2% of DCs generated with FL expressed markers of plasmacytoid DCs (Gr-1+CD11c+B220+). No differences were observed between control and Notch-As-Tg mice (data not shown).
HPCs from Notch-1–deficient mice had reduced capacity to differentiate into DCs. (A-B) HPCs were isolated from control or Notch-1–deficient Notch-AS-Tg mice and incubated with 200 ng/mL FL and 10% splenocyte-conditioned medium for 10 days. LPS (1 μg/mL) was added 24 hours before cell phenotype analysis. Cells were labeled with APC-conjugated anti-CD11c, PE-conjugated anti-CD11b, and FITC-conjugated anti-IAb, anti–B7-2, or CD40 antibodies. Proportions of IAb+, B7-2+, or CD40+ cells were calculated within the populations of CD11c+CD11b+ myeloid DCs and CD11c+CD11b-lymphoid DCs. Results of 3 performed experiments are shown. Differences between control and Notch-As-Tg mice within populations of lymphoid and myeloid DCs were statistically significant (P < .05). (C-D) DCs were generated from HPCs as described. The same 2 populations of DCs were sorted using FACSVantage SE cell sorter, irradiated at 150 Gy, and cultured with lymph node (LN) cells isolated from control allogeneic BALB/c mice. Cell proliferation was measured in triplicate. Values are the average ± SE from 3 experiments. Differences between values in control and Notch-AS-Tg mice were statistically significant at all LN cell/DC ratios (P < .05).
HPCs from Notch-1–deficient mice had reduced capacity to differentiate into DCs. (A-B) HPCs were isolated from control or Notch-1–deficient Notch-AS-Tg mice and incubated with 200 ng/mL FL and 10% splenocyte-conditioned medium for 10 days. LPS (1 μg/mL) was added 24 hours before cell phenotype analysis. Cells were labeled with APC-conjugated anti-CD11c, PE-conjugated anti-CD11b, and FITC-conjugated anti-IAb, anti–B7-2, or CD40 antibodies. Proportions of IAb+, B7-2+, or CD40+ cells were calculated within the populations of CD11c+CD11b+ myeloid DCs and CD11c+CD11b-lymphoid DCs. Results of 3 performed experiments are shown. Differences between control and Notch-As-Tg mice within populations of lymphoid and myeloid DCs were statistically significant (P < .05). (C-D) DCs were generated from HPCs as described. The same 2 populations of DCs were sorted using FACSVantage SE cell sorter, irradiated at 150 Gy, and cultured with lymph node (LN) cells isolated from control allogeneic BALB/c mice. Cell proliferation was measured in triplicate. Values are the average ± SE from 3 experiments. Differences between values in control and Notch-AS-Tg mice were statistically significant at all LN cell/DC ratios (P < .05).
These data demonstrate that even a partial decrease of Notch-1 protein in HPCs inhibits differentiation of mature, functionally potent DCs. Taken together, these data indicate that Notch-1 is necessary for normal differentiation of both myeloid and lymphoid DCs.
Activation of Notch in HPCs by fibroblasts expressing Jagged-1
These data indicated that deficit of Notch-1 in stem cells or HPCs resulted in impaired differentiation of DCs. Next we asked whether ligand-mediated activation of Notch is sufficient for DC differentiation. HPCs isolated from control mice were cultured on a monolayer of fibroblasts transfected with either MSCV vector (control) or Notch ligand Jagged-1 expression vector (Jagged-1). Different growth factors were used to generate different populations of myeloid cells. HPCs from Swiss mice were used in these experiments to match the haplotype of fibroblasts.
First, we have determined whether these conditions result in Notch activation. Notch-1 and Notch-2 are both expressed on mouse HPCs and can bind to the ligand Jagged-1. After binding, the intercellular domain of Notch is released and translocates to the nuclei where it binds to CBF-1 transcription factor and releases it from the repressor complex with histone deacetylase (HADC) and nuclear corepressor (CoR). CBF-1 in complex with Notch acts as a transcription activator.
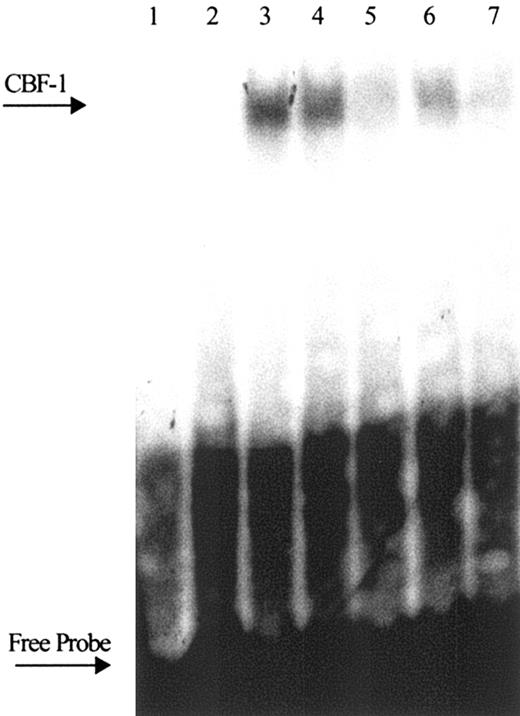
To evaluate Notch activation by Jagged-1 we performed EMSA with CBF-1–specific oligonucleotides. HPCs were cultured for 5 hours on control or Jagged-1 fibroblasts, CD45+ cells were isolated, and nuclear proteins were extracted and used in EMSA. Jagged-1 induced substantial activation of CBF-1, which was manifested in increased specific binding to DNA compared with control (Figure 4, lanes 2-3). To further determine whether Notch-1 and Notch-2 are equally involved in the Jagged-1–mediated activation, we used antibodies against Notch-1 or Notch-2. Nuclear extracts were pretreated with these antibodies and then used in EMSA. Specific binding of this antibody should prevent a Notch/CBF-1 complex from binding to DNA. In our experiments control antibody (TIB-120 against mouse I-Ad,k) did not affect CBF-1 binding (Figure 4, lane 4), whereas anti–Notch-1 antibody inhibited it (Figure 4, lane 5). Similarly, inhibition of CBF-1–binding activity was observed with antibody against Notch-2 (Figure 4, lane 6). However, the effect was slightly lower than that of anti–Notch-1 antibody. Combination of 2 antibodies completely blocked the CBF-1 binding (lane 7). These results demonstrated that in HPC Jagged-1 activated both Notch-1 and Notch-2.
Activation of Notch-1 and Notch-2 by ligation with Jagged-1. HPCs were placed on 3T3-MSCV or 3T3-Jagged-1 fibroblasts for 5 hours; CD45+ cells were isolated and nuclear protein was extracted. CBF-1 binding to DNA was determined by EMSA. For blockade of CBF-1 binding 2 μg/lane nuclear protein was preincubated with specific antibodies against Notch-1 or Notch-1, for 30 minutes on ice prior to incubation with a α-32P probe. Fifty-fold excess of unlabeled “cold” wild-type probe competitor in sample from cells incubated with Jagged-1 fibroblasts (lane 1); samples from cells cocultured with control (lane 2) and Jagged-1 fibroblasts (lane 3); blockade of CBF-1–binding activity with control antibody against mouse I-Ad, k (lane 4), antibody against Notch-1 (lane 5), antibody against Notch-2 (lane 6), and combination of antibodies against Notch-1 and Notch-2 (lane 7). Two experiments with the same results were performed.
Activation of Notch-1 and Notch-2 by ligation with Jagged-1. HPCs were placed on 3T3-MSCV or 3T3-Jagged-1 fibroblasts for 5 hours; CD45+ cells were isolated and nuclear protein was extracted. CBF-1 binding to DNA was determined by EMSA. For blockade of CBF-1 binding 2 μg/lane nuclear protein was preincubated with specific antibodies against Notch-1 or Notch-1, for 30 minutes on ice prior to incubation with a α-32P probe. Fifty-fold excess of unlabeled “cold” wild-type probe competitor in sample from cells incubated with Jagged-1 fibroblasts (lane 1); samples from cells cocultured with control (lane 2) and Jagged-1 fibroblasts (lane 3); blockade of CBF-1–binding activity with control antibody against mouse I-Ad, k (lane 4), antibody against Notch-1 (lane 5), antibody against Notch-2 (lane 6), and combination of antibodies against Notch-1 and Notch-2 (lane 7). Two experiments with the same results were performed.
Effect of Notch activation on DC differentiation from mouse HPCs
Bone marrow cells enriched for HPCs were placed in 24-well plates containing a monolayer of either control or Jagged-1 fibroblasts and cultured with FL for 10 days. CD45+ leukocytes were analyzed using flow cytometry. To our surprise incubation on Jagged-1 fibroblasts did not affect the total number of cells (data not shown) but generated significantly lower proportion of DCs (Figure 5A). Because generation of DCs depends on several different growth factors, we tested the effect of Notch activation in the presence of the other cytokine able to support DC differentiation, specifically GM-CSF. After a 7-day culture on control or Jagged-1 fibroblasts with GM-CSF alone the number of CD45+ cells was equal (data not shown). The cell proliferation assay confirmed that there was no difference in proliferation between these 2 groups (Figure 5B). As in case of FL, HPCs grown on NIH 3T3-Jagged-1 fibroblasts generated fewer DCs than HPCs grown on control fibroblasts. Instead, accumulation of cells with the phenotype of immature myeloid cells (ImCs; Gr-1+CD11b+) was observed (Figure 5C). To test whether this phenomenon is limited to DC differentiation M-CSF was used to differentiate macrophages from HPCs. The total number of CD45+ cells generated in the presence of M-CSF was also not different between the groups (data not shown). However, as in case of GM-CSF a substantially higher proportion of ImCs was generated from HPCs grown on Jagged-1 than on control fibroblasts (Figure 5D). The proportion of mature F4/80+ macrophages was slightly decreased (Figure 5D). It was possible that in an experimental system containing fibroblasts myeloid cell differentiation might be delayed, which would explain the decreased proportion of mature DCs and macrophages. To test this possibility, we extended culture period from 7 days to 10 days. As shown in Figure 5C-D, an extended 10-day culture with GM-CSF or M-CSF did not result in generation of more mature DCs or macrophages and did not decrease the proportion of ImCs. On the contrary, extension of incubation resulted in decreased proportion of DCs (Figure 5C). Thus, our results indicate that Notch activation through cell-associated Jagged-1 ligation promotes accumulation of ImCs rather than differentiation of mature macrophages or DCs.
Accumulation of immature myeloid cells after activation of Notch signaling in HPCs. (A) HPCs were cultured on fibroblasts in 24-well plate with FL and supernatants from control splenocytes for 10 days as described in “Materials and methods.” Cells were then labeled with APC-conjugated anti-CD11c or anti–Gr-1 antibodies, PE-conjugated anti-CD45 antibody, peridinin chlorophyll protein (PerCP)–conjugated anti-B220 antibody, and FITC-conjugated anti-IAq, anti–B7-2, CD11b, anti-F4/80 or Gr-1 antibodies and analyzed on a FACS-Calibur flow cytometer. Only CD45+ cells were evaluated for cell phenotype. Values are the average ± SE from 3 experiments. (B) HPCs were plated on fibroblasts in 96-well plate and incubated for 2 or 4 days with 20 ng/mL GM-CSF Cell proliferation was measured by [3H]-thymidine incorporation. (C-D) HPCs were cultured on fibroblasts in 24-well plate with GM-CSF (C) or M-CSF (D) for 7 or 10 days. In GM-CSF culture (C), 5 ng/mL TNF-α was added 48 hours before cell phenotype analysis. Cells were labeled with APC-conjugated anti-CD11c or anti–Gr-1 antibodies, PE-conjugated anti-CD45, and FITC-conjugated anti-IAq, anti–B7-2, CD11b, or anti-F4/80 antibodies and analyzed on a FACSCalibur flow cytometer. Only CD45+ cells were evaluated for cell phenotype. Values are the average ± SE from 3 experiments. Asterisk indicates statistically significant differences between cells treated with 3T3-MSCV and 3T3-Jagged-1 fibroblasts.
Accumulation of immature myeloid cells after activation of Notch signaling in HPCs. (A) HPCs were cultured on fibroblasts in 24-well plate with FL and supernatants from control splenocytes for 10 days as described in “Materials and methods.” Cells were then labeled with APC-conjugated anti-CD11c or anti–Gr-1 antibodies, PE-conjugated anti-CD45 antibody, peridinin chlorophyll protein (PerCP)–conjugated anti-B220 antibody, and FITC-conjugated anti-IAq, anti–B7-2, CD11b, anti-F4/80 or Gr-1 antibodies and analyzed on a FACS-Calibur flow cytometer. Only CD45+ cells were evaluated for cell phenotype. Values are the average ± SE from 3 experiments. (B) HPCs were plated on fibroblasts in 96-well plate and incubated for 2 or 4 days with 20 ng/mL GM-CSF Cell proliferation was measured by [3H]-thymidine incorporation. (C-D) HPCs were cultured on fibroblasts in 24-well plate with GM-CSF (C) or M-CSF (D) for 7 or 10 days. In GM-CSF culture (C), 5 ng/mL TNF-α was added 48 hours before cell phenotype analysis. Cells were labeled with APC-conjugated anti-CD11c or anti–Gr-1 antibodies, PE-conjugated anti-CD45, and FITC-conjugated anti-IAq, anti–B7-2, CD11b, or anti-F4/80 antibodies and analyzed on a FACSCalibur flow cytometer. Only CD45+ cells were evaluated for cell phenotype. Values are the average ± SE from 3 experiments. Asterisk indicates statistically significant differences between cells treated with 3T3-MSCV and 3T3-Jagged-1 fibroblasts.
Withdrawal of Notch signaling permits differentiation of ImCs
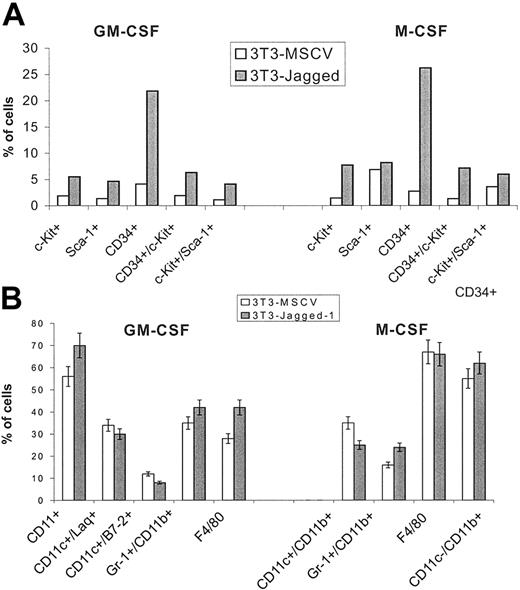
We analyzed the phenotype of ImCs accumulated as a result of Notch activation. HPCs were cultured for 7 days on control or Jagged-1 fibroblasts with GM-CSF or M-CSF. Gr-1+ cells were isolated using a magnetic bead separation technique. The resulting population had more than 93% Gr-1+CD11b+ cells. These cells were then labeled with antibodies against markers specific for HPCs and stem cells: CD34, c-kit ligand, and Sca-1. Gr-1+ cells generated on Jagged-1 fibroblasts had 3 to 5 times higher proportion of HPCs than Gr-1+ generated on control fibroblasts (Figure 6A). CD34+ cells represented almost 25% of all Gr-1+ cells generated on Jagged-1 fibroblasts, whereas their proportion in Gr-1+ cells generated on control fibroblasts was less than 5%.
Withdrawal of Notch signaling permitted Gr-1+/Mac1+ ImC differentiation. (A) HPCs were cultured on fibroblasts with either GM-CSF or M-CSF for 7 days. Gr-1+ cells were isolated and stained with antibodies against markers of hematopoietic progenitors: APC-conjugated c-Kit, FITC-conjugated Sca-1, and PE-conjugated CD34 and analyzed on a FACSCalibur flow cytometer. (B) HPCs were cultured on fibroblasts with GM-CSF for 5 days; CD45+ cells were isolated and further cultured without fibroblasts for additional 5 days with either GM-CSF or M-CSF. In the case of GM-CSF culture, 5 ng/mL TNF-α was added 48 hours before cell phenotype analysis. Cells were labeled with APC-conjugated anti-CD11c or anti–Gr-1 antibodies, PE-conjugated anti-CD11b, and FITC-conjugated anti-IAq, anti–B7-2, or anti-F4/80 antibodies and analyzed on a FACSCalibur flow cytometer. Values are the average ± SE from 3 experiments. Similar results were obtained when cells were cultured for an additional 5 days on control fibroblasts.
Withdrawal of Notch signaling permitted Gr-1+/Mac1+ ImC differentiation. (A) HPCs were cultured on fibroblasts with either GM-CSF or M-CSF for 7 days. Gr-1+ cells were isolated and stained with antibodies against markers of hematopoietic progenitors: APC-conjugated c-Kit, FITC-conjugated Sca-1, and PE-conjugated CD34 and analyzed on a FACSCalibur flow cytometer. (B) HPCs were cultured on fibroblasts with GM-CSF for 5 days; CD45+ cells were isolated and further cultured without fibroblasts for additional 5 days with either GM-CSF or M-CSF. In the case of GM-CSF culture, 5 ng/mL TNF-α was added 48 hours before cell phenotype analysis. Cells were labeled with APC-conjugated anti-CD11c or anti–Gr-1 antibodies, PE-conjugated anti-CD11b, and FITC-conjugated anti-IAq, anti–B7-2, or anti-F4/80 antibodies and analyzed on a FACSCalibur flow cytometer. Values are the average ± SE from 3 experiments. Similar results were obtained when cells were cultured for an additional 5 days on control fibroblasts.
Next, we asked whether Notch activation through Jagged-1 is necessary for maintaining the phenotype of immature cells. Enriched HPCs were cultured for 5 days with GM-CSF on control or Jagged-1 fibroblasts; CD45+ myeloid cells were isolated and transferred into new plates. These plates either contained control fibroblasts or were without any fibroblasts. Cells were cultured for additional 5 days with either GM-CSF or M-CSF. In culture with GM-CSF, 1 μg LPS was added 24 hours prior to cell analysis. In total, cells were cultured for 10 days, exactly the same time as in experiments shown in Figure 5B-C. However, the results of these experiments were dramatically different. The 5-day culture of the cells without Jagged-1 fibroblasts resulted in a significant decrease in the proportion of Gr-1+CD11b+ ImCs and accumulation of relatively mature DCs and macrophages (compare Figure 5B-C and Figure 6B). Previously observed differences between the cells grown on Jagged-1 and control fibroblasts (Figure 5B-C) now completely disappeared (Figure 6B). Taken together, these data demonstrate that Notch signaling mediated by Jagged-1 results in accumulation of HPCs and ImCs and prevents differentiation of mature DCs and macrophages. Withdrawal of this signal allows for terminal differentiation of DCs and macrophages.
Activation of Notch by Jagged-1 ligation together with IL-4 enhances DC differentiation from HPCs
IL-4 is known to provide strong signals supporting selective differentiation of DCs from HPCs. We evaluated Jagged-1 effects on DC differentiation in the presence of IL-4. HPCs were cultured on Jagged-1 or control fibroblasts for 7 days with GM-CSF and IL-4. After that time the number of CD45+ cells in these 2 groups was very similar (1.62 ± 0.25 × 106/well for control and 1.60 ± 0.26 × 106/well for 3T3-Jagged-1 fibroblasts, P > .1). No differences were also seen in cell proliferation assay (Figure 7A). However, HPCs cultured on Jagged-1 fibroblasts produced 2-fold more DCs than HPCs cultured on control fibroblasts (Figure 7B).
IL-4 enhances DC differentiation during Notch activation. (A) HPCs were plated on the fibroblasts in triplicate on 96-well plate with GM-CSF and IL-4 and cultured for 2 or 4 days. Cell proliferation was measured by [3H]-thymidine incorporation. (B) HPCs were cultured on control or Jagged-1 fibroblasts with GM-CSF and IL-4 for 7 days. Cells were labeled with APC-conjugated anti-CD11c, PE-conjugated anti-CD45, FITC-conjugated anti-IAq, anti–B7-2, Gr-1 or anti-CD11b antibodies, and PerCP-conjugated anti-B220 antibody and analyzed on a FACSCalibur flow cytometer. Only CD45+ cells were evaluated. Differences between the groups were statistically significant (P < .05) for all tested cell populations. (C) DCs were generated with GM-CSF and IL-4 from HPCs as described, irradiated at 150 Gy, and cultured with lymph node cells isolated from control allogeneic BALB/c mice at different ratios. Cell proliferation was measured in triplicates as described in “Materials and methods.” Values are the average ± SE from 2 performed experiments. Differences between the groups were statistically significant (P < .05) at all LN cell/DC ratios. (D) HPCs were cultured on control or Jagged-1 fibroblasts with FL and IL-4 for 7 days. Cells were analyzed as described in Figure 6B. Differences between the groups were statistically significant (P < .05) for all tested cell populations with the exception of CD11c+B7-2+ cells. (E) Bone marrow–enriched HPCs were cultured with GM-CSF or GM-CSF and IL-4 on control and Jagged-1 fibroblasts. On day 5, CD45+ cells were isolated and nuclear protein was used for CBF-1–binding activity as described in Figure 3. Lane 1, 50-fold excess of unlabeled “cold” probe; lane 2, mutant probe; lane 3, HPCs cultured with GM-CSF on 3T3-MSCV; lane 4, cells cultured with GM-CSF on 3T3-Jagged-1 fibroblasts; lane 5, HPCs cultures with GM-CSF and IL-4 on 3T3-MSCV fibroblasts; lane 6, cells cultured with GM-CSF nd IL-4 on 3T3-Jagged-1 fibroblasts.
IL-4 enhances DC differentiation during Notch activation. (A) HPCs were plated on the fibroblasts in triplicate on 96-well plate with GM-CSF and IL-4 and cultured for 2 or 4 days. Cell proliferation was measured by [3H]-thymidine incorporation. (B) HPCs were cultured on control or Jagged-1 fibroblasts with GM-CSF and IL-4 for 7 days. Cells were labeled with APC-conjugated anti-CD11c, PE-conjugated anti-CD45, FITC-conjugated anti-IAq, anti–B7-2, Gr-1 or anti-CD11b antibodies, and PerCP-conjugated anti-B220 antibody and analyzed on a FACSCalibur flow cytometer. Only CD45+ cells were evaluated. Differences between the groups were statistically significant (P < .05) for all tested cell populations. (C) DCs were generated with GM-CSF and IL-4 from HPCs as described, irradiated at 150 Gy, and cultured with lymph node cells isolated from control allogeneic BALB/c mice at different ratios. Cell proliferation was measured in triplicates as described in “Materials and methods.” Values are the average ± SE from 2 performed experiments. Differences between the groups were statistically significant (P < .05) at all LN cell/DC ratios. (D) HPCs were cultured on control or Jagged-1 fibroblasts with FL and IL-4 for 7 days. Cells were analyzed as described in Figure 6B. Differences between the groups were statistically significant (P < .05) for all tested cell populations with the exception of CD11c+B7-2+ cells. (E) Bone marrow–enriched HPCs were cultured with GM-CSF or GM-CSF and IL-4 on control and Jagged-1 fibroblasts. On day 5, CD45+ cells were isolated and nuclear protein was used for CBF-1–binding activity as described in Figure 3. Lane 1, 50-fold excess of unlabeled “cold” probe; lane 2, mutant probe; lane 3, HPCs cultured with GM-CSF on 3T3-MSCV; lane 4, cells cultured with GM-CSF on 3T3-Jagged-1 fibroblasts; lane 5, HPCs cultures with GM-CSF and IL-4 on 3T3-MSCV fibroblasts; lane 6, cells cultured with GM-CSF nd IL-4 on 3T3-Jagged-1 fibroblasts.
Cells generated on 3T3-Jagged-1 fibroblasts had a significantly higher ability to stimulate allogeneic T-cell proliferation in MLRs, which is indicative of higher proportion of DCs (Figure 7C). These data suggest that IL-4 may provide the necessary signal to promote the complete differentiation of immature cells accumulated as the result of the effect of Jagged-1. To test this hypothesis we used another system to generate DC from HPC using FL. The experimental system with FL provides conditions for generation of both lymphoid and myeloid DCs (Figure 7D). Incubation of HPCs on Jagged-1 fibroblasts did not affect the total number of cells (data not shown) but generated a significantly higher proportion of DCs (Figure 7D). This was especially evident in the population of myeloid-related DCs. A significant increase was also observed in the proportion of plasmacytoid DCs (from 0.8% to 4.7%, P < .05; Figure 7D).
IL-4 can interfere with Notch signaling and thus affect cell differentiation, or alternatively it can provide a strong differentiation signal independent on Notch. To test the direct effect of IL-4 on Notch activation HPCs were cultured with GM-CSF alone or with GM-CSF and IL-4 on control or Jagged-1 fibroblasts. On day 5, CD45+ cells were isolated and nuclear protein was used for CBF-1–binding activity. The presence of IL-4 did not affect activation of Notch by Jagged-1 (Figure 7E), which suggested that IL-4 did not affect Notch activation during differentiation of DCs.
Notch activation does not affect cytokine receptors and does not activate DCs
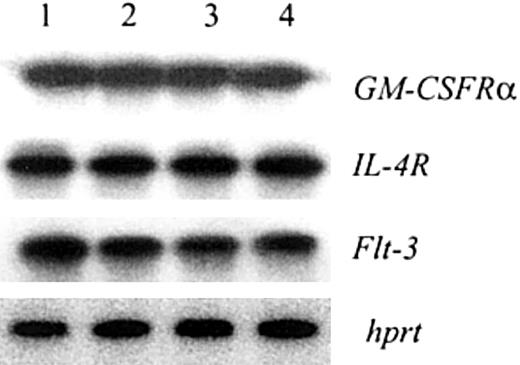
Notch activation in HPCs could result in up-regulation of the receptors for cytokines critically important for myelopoiesis. This could affect accumulation of ImCs or DCs. To address this possibility we evaluated the expression of the genes encoding receptors for GM-CSF, FL, and IL-4. Notch activation in HPCs did not affect the expression of any of the tested genes (Figure 8).
Jagged-1 ligation had no effect on the expression of GM-CSFRα, IL-4R, or Flt-3 in HPCs. Bone marrow–enriched HPCs were cultured with GM-CSF on control or Jagged-1 fibroblasts. After 48 hours or 72 hours of incubation CD45+ cells were isolated; RNA was extracted and used in reverse transcription-PCR followed by Southern blotting as described in “Materials and methods.” Lanes 1 and 2, HPCs cultured on 3T3-MSCV (1) or on 3T3-Jagged-1 (2) fibroblasts for 48 hours; lanes 3 and 4, HPCs cultured on 3T3-MSCV (3) or on 3T3-Jagged-1 (4) fibroblasts for 72 hours.
Jagged-1 ligation had no effect on the expression of GM-CSFRα, IL-4R, or Flt-3 in HPCs. Bone marrow–enriched HPCs were cultured with GM-CSF on control or Jagged-1 fibroblasts. After 48 hours or 72 hours of incubation CD45+ cells were isolated; RNA was extracted and used in reverse transcription-PCR followed by Southern blotting as described in “Materials and methods.” Lanes 1 and 2, HPCs cultured on 3T3-MSCV (1) or on 3T3-Jagged-1 (2) fibroblasts for 48 hours; lanes 3 and 4, HPCs cultured on 3T3-MSCV (3) or on 3T3-Jagged-1 (4) fibroblasts for 72 hours.
To investigate whether proinflammatory signals can provide effects similar to IL-4 on DC differentiation, enriched HPCs were incubated on control or Jagged-1 fibroblasts for 5 days with GM-CSF and with or without LPS (1 μg/mL). HPCs incubated on Jagged-1 fibroblasts generated a lower proportion of DCs than the cells incubated on control fibroblasts. LPS did not change this effect (data not shown). Similar results were obtained when cytosine-phosphorothiolated guanine-containing oligonucleotide (CpG) was used instead of LPS. It also did not change the effect of Notch-1 on DC differentiation.
Recently, it has been reported that a Jagged-1–soluble peptide can mature human monocyte-derived DCs.18 We asked whether cell-bound Jagged-1 could have the same effect. Immature DCs were generated from HPCs with GM-CSF and IL-4 for 5 days without the presence of fibroblasts. On day 5, cells were transferred onto Jagged-1 or control fibroblasts and cultured for 48 hours prior to DC phenotype analysis. No differences were found between these 2 groups of cells in the expression of CD11c, CD11b, MHC class II, B7-2, or CD40. The ability of DCs to stimulate allogeneic T cells also remained the same (data not shown). Addition of TNF-α or LPS resulted in activation of DCs, which was manifested in up-regulation of MHC class II, B7-2, and CD40. However, no differences between cells incubated on control or Jagged-1 fibroblasts were found. These data indicated that under these experimental conditions cell-bound Jagged-1 did not activate DCs.
Discussion
The data presented in this report indicate that Notch signaling is necessary but not sufficient for the differentiation of DCs. Lack of Notch-1 in ES cells significantly impaired their differentiation into DCs. We have previously found that Notch is required to maintain normal levels of nuclear factor κB (NF-κB) subunits in HPCs.19 It has been reported that RelB, one of the subunits of NF-κB, is critically important for differentiation of one particular type of DCs—myeloid-related DCs.27 Several subsets of DCs in mice have been described: myeloid-related, lymphoid-related, and plasmacytoid cells. However, specific markers for lymphoid and myeloid DCs are not well defined. CD8α, previously used as a marker of lymphoid DCs, was later found to be expressed on both committed lymphoid and myeloid progenitors. Another approach to separate these 2 populations is staining with anti-CD11c and anti-CD11b antibodies. CD11c+CD11b+ cells were characterized as myeloidrelated DCs and CD11c+CD11b- cells as lymphoid-related DCs (for a review, see Shortman and Liu4 ). Although these criteria are not absolute, they nevertheless allowed for more precise characterization of DC populations. We asked whether Notch signaling might be specifically important for only one subset of DCs. To address this question we generated all subtypes of DCs from HPCs using FL. Highly enriched HPCs obtained by negative selection of lineage-specific cells have been used. Previous studies have demonstrated that these cells are adequate for the analysis of DC differentiation in vitro. HPCs isolated from Notch-1-AS-Tg mice produced the same level of myeloid-related (CD11c+CD11b+), lymphoid-related (CD11c+CD11b-), or plasmacytoid (CD11c+ B220+Gr-1+) DCs as HPCs isolated from control mice, which was consistent with previous observations in Notch-1 conditionally knockout mice. However, when we analyzed the phenotype and function of DCs within each population we found that DCs generated from Notch-1–deficient HPCs had significantly lower levels of MHC class II and costimulatory molecules and a lower ability to stimulate allogeneic MLRs than control HPCs. These data confirmed the observations made in ES cell system and clearly indicated that Notch-1 is critically important for the differentiation of DCs. The differences in the magnitude of the effect between Notch-1 knockout ES cells and Notch-1–deficient HPCs could be explained by the fact that HPCs from Notch-1-AS-Tg mice still have about half of the control level of Notch-1 protein,19 whereas Notch-1 knockout ES cells were completely devoid of Notch-1.
Using bone marrow chimeric mice with conditionally knockout Notch-1, Radtke and colleagues found that neither thymic nor peripheral DCs were affected by Notch-1 deficiency.11 However, direct evaluation of DC phenotype and function in mice is very difficult, because of the very low proportion of these cells in tissues. Therefore, DC fractions in that study were enriched using gradient centrifugation.11 However, although this procedure is adequate for obtaining enriched DCs, it is much less suitable for the evaluation of the total populations of DCs due to considerable loss of cells. In addition, no expression of costimulatory molecules was analyzed nor were functional studies with isolated DCs performed. As we demonstrated here cells generated from Notch-1–deficient HPCs may have normal expression of CD11c, but significantly decreased B7-2 expression and functional activity. Probably the most accurate evaluation of DC populations in these mice can be achieved after in vivo stimulation with FL and direct assessment of DC phenotype and function.
Is Notch signaling sufficient to promote DC differentiation? We activated Notch in HPCs using a cell-bound ligand, which most closely reflects actual interaction between BMS cells and HPCs in the bone marrow. Notch ligand Jagged-1 was selected because it is expressed on BMS.28 Among members of Notch family, Notch-1 and Notch-2 are expressed in multiple lineages of hematopoietic cells. Notch-1 in particular appears to be regulated during myeloid differentiation.29
In the presence of all tested growth factors activation of Notch resulted in decreased production of mature DCs or macrophages. This was associated with accumulation of ImCs with Gr- 1+CD11b+ phenotype. These data were consistent with the results obtained previously on 32D myeloid cells14 and very recently on HPCs with activation of Notch via Delta-1.30 ImCs were enriched for HPCs and differentiated into mature DCs or macrophages once Jagged-1–expressing stroma was removed. It indicates that uninterrupted Notch signaling is required to keep cells in relatively undifferentiated state. Our data demonstrate that Notch activation in HPCs does not affect the expression of the receptors for main growth factors and cytokines involved in DC differentiation: GM-CSF, FL, or IL-4. It suggests that Notch effects are not mediated by increased sensitivity of HPCs to these cytokines. At this time we cannot exclude a possibility that expression of Jagged-1 on fibroblasts may affect expression of other molecules able to influence DC differentiation. This will require further testing.
IL-4 induced differentiation of DCs in the presence of Notch signaling. This phenomenon can be explained by the fact that activation of Notch leads to accumulation of the DC precursors. IL-4 apparently is able to override Notch-induced inhibition of DC differentiation, which eventually results in accumulation of mature DCs. Our data indicate that IL-4 does not interfere with Notch activation and apparently affects DC differentiation via other mechanisms currently under investigation.
Activation of Notch via cell-bound Jagged-1 ligand did not activate already differentiated DCs. This was in apparent contrast to recent data from Weijzen and coauthors, who showed that a Jagged-1–derived peptide was able to induce DC activation.18 It is possible that the level or duration of Notch activation is different between soluble and cell-bound ligands. If the soluble peptide is metabolized in culture, it would result in a transient Notch signal, which is then extinguished, whereas ligand expressed on the surface of feeder cells generates a continuous signal. In this case, exposure to Jagged peptide may mimic Notch stimulation followed by withdrawal of the signal, and trigger maturation.
Thus, our data suggest a model of DC differentiation involving Notch. In bone marrow, differentiation of DCs is controlled by cytokines and by direct cell-cell contact of HPCs with BMS cells. Activation of Notch signaling results in the accumulation of precursors of DCs and macrophages but prevents their terminal differentiation. As soon as the influence of BMS is terminated (when precursors leave bone marrow) those cells undergo rapid differentiation into macrophages or DCs. Such spatial control of cell differentiation may be necessary for keeping functionally competent mature DCs away from the bone marrow, thus maintaining homeostasis. Bacterial products such as LPS or CpG were not able to promote final steps of DC differentiation in the presence of Notch signaling. Apparently, their role is limited to activation of DCs on periphery. Our data also indicate that strong selection signals such as IL-4 may override that spatial control.
Prepublished online as Blood First Edition Paper, August 7, 2003; DOI 10.1182/blood-2003-04-1034.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr N. Luetteke and Dr M. Dikov for their technical assistance.

![Figure 5. Accumulation of immature myeloid cells after activation of Notch signaling in HPCs. (A) HPCs were cultured on fibroblasts in 24-well plate with FL and supernatants from control splenocytes for 10 days as described in “Materials and methods.” Cells were then labeled with APC-conjugated anti-CD11c or anti–Gr-1 antibodies, PE-conjugated anti-CD45 antibody, peridinin chlorophyll protein (PerCP)–conjugated anti-B220 antibody, and FITC-conjugated anti-IAq, anti–B7-2, CD11b, anti-F4/80 or Gr-1 antibodies and analyzed on a FACS-Calibur flow cytometer. Only CD45+ cells were evaluated for cell phenotype. Values are the average ± SE from 3 experiments. (B) HPCs were plated on fibroblasts in 96-well plate and incubated for 2 or 4 days with 20 ng/mL GM-CSF Cell proliferation was measured by [3H]-thymidine incorporation. (C-D) HPCs were cultured on fibroblasts in 24-well plate with GM-CSF (C) or M-CSF (D) for 7 or 10 days. In GM-CSF culture (C), 5 ng/mL TNF-α was added 48 hours before cell phenotype analysis. Cells were labeled with APC-conjugated anti-CD11c or anti–Gr-1 antibodies, PE-conjugated anti-CD45, and FITC-conjugated anti-IAq, anti–B7-2, CD11b, or anti-F4/80 antibodies and analyzed on a FACSCalibur flow cytometer. Only CD45+ cells were evaluated for cell phenotype. Values are the average ± SE from 3 experiments. Asterisk indicates statistically significant differences between cells treated with 3T3-MSCV and 3T3-Jagged-1 fibroblasts.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/12/10.1182_blood-2003-04-1034/6/m_h82335286005.jpeg?Expires=1766310611&Signature=xCHpdMpxYLkBT66CCW4IqKUnYILrLsnWfFlg7i89PYHABYEKzfy-WDB6mlrP1gdJ1obur7gY~w7LdKPRFl1Na9SFzFbF7DW2Rs6wWHv7DXePcAfOdNFG4uBo2lDP-EwIqBNe8N4a1E97ORViZ~vNTtcBlQWUkJW694~UBZZ3JurUkbKFL4sta~F1jGpywjwpdrzmsTK3QgaGHe-rfiJUDaAGWshQoxyKAKCQWNvpZSEv5ZT2YX4ZIepgcn3EXVkFUrzSXwkq2WRcvpJ6QKSaogcDl7IwHOYa3bqCWf7IOOllqf2G1I6Q59rHlkz4Ssx3B6BwLLA9C6D5cukHijCYjA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. IL-4 enhances DC differentiation during Notch activation. (A) HPCs were plated on the fibroblasts in triplicate on 96-well plate with GM-CSF and IL-4 and cultured for 2 or 4 days. Cell proliferation was measured by [3H]-thymidine incorporation. (B) HPCs were cultured on control or Jagged-1 fibroblasts with GM-CSF and IL-4 for 7 days. Cells were labeled with APC-conjugated anti-CD11c, PE-conjugated anti-CD45, FITC-conjugated anti-IAq, anti–B7-2, Gr-1 or anti-CD11b antibodies, and PerCP-conjugated anti-B220 antibody and analyzed on a FACSCalibur flow cytometer. Only CD45+ cells were evaluated. Differences between the groups were statistically significant (P < .05) for all tested cell populations. (C) DCs were generated with GM-CSF and IL-4 from HPCs as described, irradiated at 150 Gy, and cultured with lymph node cells isolated from control allogeneic BALB/c mice at different ratios. Cell proliferation was measured in triplicates as described in “Materials and methods.” Values are the average ± SE from 2 performed experiments. Differences between the groups were statistically significant (P < .05) at all LN cell/DC ratios. (D) HPCs were cultured on control or Jagged-1 fibroblasts with FL and IL-4 for 7 days. Cells were analyzed as described in Figure 6B. Differences between the groups were statistically significant (P < .05) for all tested cell populations with the exception of CD11c+B7-2+ cells. (E) Bone marrow–enriched HPCs were cultured with GM-CSF or GM-CSF and IL-4 on control and Jagged-1 fibroblasts. On day 5, CD45+ cells were isolated and nuclear protein was used for CBF-1–binding activity as described in Figure 3. Lane 1, 50-fold excess of unlabeled “cold” probe; lane 2, mutant probe; lane 3, HPCs cultured with GM-CSF on 3T3-MSCV; lane 4, cells cultured with GM-CSF on 3T3-Jagged-1 fibroblasts; lane 5, HPCs cultures with GM-CSF and IL-4 on 3T3-MSCV fibroblasts; lane 6, cells cultured with GM-CSF nd IL-4 on 3T3-Jagged-1 fibroblasts.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/12/10.1182_blood-2003-04-1034/6/m_h82335286007.jpeg?Expires=1766310611&Signature=4Q6AspjgpJqKLxScYFA9iro2Shhd4BT-QsAU1yQGpRGySZensl3kRW~GNFjO2xv~OE2LHX~XZkUfKyHt~RER14Qy0ULH2W2H2ROd6e~7Dow9q64EKrFoQ4Hhs1EopKVliYUMUBTLC6KaRtAS2NiWVuPHTx-pGSQBbeU1q-T9FGUQdrgfMfWhlk1IMtpm7myplG6hJumQ6a3Eceea1Y2-aJ7NrG7mqaeaD9EX1T~TIXBohLKhKPgA03vAxWWtvg94unLJGsxweYovPgpDotrG0M4IeTTbwVwNjlbHzAOR4ckFwbr5xRu2tam9nnX5w6fkwJo4fCv2~0C1v6sPvXZNVg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)