Abstract
To study epigenetic regulation of the human β-globin locus during hematopoiesis, we investigated patterns of histone modification and chromatin accessibility along this locus in hematopoietic progenitor cells (HPCs) derived from both humans and transgenic mice. We demonstrate that the developmentally related activation of human β-like globin genes in humans and transgenic mice HPCs is preceded by a wave of gene-specific histone H3 hyperacetylation and K4 dimethylation. In erythroid cells, expression of β-like globin genes is associated with histone hyperacetylation along these genes and, surprisingly, with local deacetylation at active promoters. We also show that endogenous mouse β major and human β-like genes are subject to different epigenetic control mechanisms in HPCs. This difference is likely due to intrinsic properties of the human β-globin locus since, in transgenic mice, this locus is epigenetically regulated in the same manner as in human HPCs. Our results suggest that a defined pattern of histone H3 acetylation/dimethylation is important for specific activation of human globin promoters during development in human and transgenic HPCs. We propose that this transient acetylation/dimethylation is involved in gene-specific potentiation in HPCs (ie, before extensive chromatin remodeling and transcription take place in erythroid cells).
Introduction
Regulation of the “on/off” state of transcription in eukaryotes plays a critical role in embryogenesis and cellular differentiation.1 This heritable process is linked to epigenetic states involving DNA methylation and changes in chromatin structure, which maintain transcriptional status throughout mitosis and DNA replication. Eukaryotic gene activation results from the interplay of trans-activators and/or repressors with nucleosome-modifying and/or -remodeling factors. Indeed, nucleosome organization is a key component of epigenetic regulation (Felsenfeld and Groudine2 and references therein). Observations made in recent years have led to the notion that a combination of histone modifications such as acetylation, phosphorylation, and methylation generates a histone code that regulates the use of information from the genetic code.3 These posttranslational modifications of histones are important determinants in nucleosome-nucleosome and nucleosome-DNA interactions4 and provide precise patterns recognized and bound by specific proteins.
Epigenetic regulation of transcription appears to play an important role during hematopoiesis. For example, abnormal patterns of DNA methylation and chromatin structure are common traits in hematologic malignancies,5 and chromosomal translocations that change the activity of histone acetyltransferases and histone deacetylases (HDACs) are associated with several forms of leukemia.6 Hematopoiesis is characterized by a gradual commitment of multipotent hematopoietic progenitors to become bipotent or unipotent progenitors and, eventually, mature blood cells. In hematopoietic progenitor cells (HPCs), lineage-specific genes are thought to be located in chromatin that is poised for activation,7,8 whereas the chromatin of nonexpressed genes would exist as or become increasingly restrictive. The mechanisms responsible for maintenance of an active chromatin state are poorly understood. In HPCs, basal expression of genes that eventually become highly transcribed in committed cells might play a role in transcriptional potentiation, although this may not always be a prerequisite.9,10
β-Like globin gene expression is tightly regulated during development and hematopoiesis. The human β-globin locus comprises 5 developmentally regulated genes (ϵ-Gγ-Aγ-δ-β) whose high-level expression depends on the locus control region (LCR) which, in turn, consists of 5 DNase I hypersensitive sites (HS). The LCR activates β-globin gene transcription through direct interaction with promoter regions11,12 and is a major determinant of chromatin structure at the locus.13 Mice transgenic for the human β-globin locus express the human genes in a developmentally regulated manner.14,15 The β-globin genes exhibit a basal level of transcription in hematopoietic progenitor cell lines16 and in HPCs of the aorta-gonad-mesonephros region,17 suggesting that in these cells the β-globin locus is characterized by an “open” chromatin structure. Low-level expression is maintained throughout erythropoiesis and only increases to full expression in differentiated erythroid cells. However, it is not known whether this basal level of expression corresponds to developmental-specific expression of the globin genes in erythroid cells.
The observation that the β-globin locus is uniformly hyperacetylated in chicken erythroid cells18 suggests that histone acetylation is important for the activation and/or the maintenance of an active globin locus. Additionally, active chicken globin genes are acetylated at lysine 9 of histone H3 (H3 K9), whereas histones across inactive genes are preferentially H3 K9–methylated.19 The mouse β major (βmaj) promoter is also acetylated in uninduced mouse erythroleukemia (MEL) cells.20 Furthermore, the murine β-globin locus is differentially acetylated during development, and active genes as well as the LCR are marked by H3 and H4 acetylation.21 In murine MEL cells containing a human chromosome 11,22 the human β-globin locus was found to be acetylated at histones H3 and H4 throughout, whereas peaks of H3 hyperacetylation were characteristic of transcriptionally active genes. However, MEL are proerythroblast-like transformed cells, and the role of histone modifications at the human β-globin locus in vivo at early hematopoietic stages remains to be elucidated.
We have investigated the epigenetic state of the human β-globin locus in human and transgenic HPCs and compared this with major epigenetic variations associated with globin activation in erythroid cells in vivo. We demonstrate that histone H3 is hyperacetylated and K4 dimethylated at the human β (huβ) promoter in HPCs and subsequently deacetylated in erythroid cells. The human γ (huγ) promoters are not acetylated in adult HPCs as is the case for 11.5 (days after coitus [dpc]) transgenic mouse–derived fetal liver HPCs. Our data therefore provide evidence for a transcriptional potentiation mechanism occurring at the human globin locus in HPCs.
Materials and methods
Bone marrow culture and in vitro colony assays
Mouse bone marrow was cultured in Dulbecco modified Eagle medium (DMEM) with 10% characterized fetal bovine serum (FBS; HyClone, Logan, UT), 3 U/mL erythropoietin (Epo), and 300 nM trichostatin A (TSA; Sigma, St Louis, MO) or equal volume of ethanol. Cells were incubated for 8 hours at 37°C in 5% CO2. For in vitro colony assays, cells were plated onto MethoCult M3434 medium (StemCell Technologies, Vancouver, BC, Canada). Single cultures contained 50, 100, or 200 c-Kit+/CD31high/Ly-6C- cells or 5 × 105 Ter119+ cells or 5 × 104 mouse bone marrow cells. Colony types were determined and scored at day 14 by microscopy and Wright-Giemsa staining.
Cell sorting
Staining with antibodies (PharMingen, San Diego, CA; or Caltag, Burlingame, CA) was carried out on ice for 30 minutes followed by one wash in phosphate-buffered saline (PBS) 5% heat-inactivated FBS. Cells were analyzed using high-speed fluorescence-activated cell sorter (FACS) Vantage with DIgitalized VAntage (DIVA) option (Becton Dickinson, San Jose, CA). For c-Kit+/CD31high/Ly-6C- sorting, cells were incubated with rat anti–Ly-6C antibodies (Abs) followed by goat antirat phycoerythrin (PE)–conjugated Abs, then biotinylated anti-CD31 Ab followed by streptavidin tricolor–conjugated Abs and anti–c-Kit fluorescein isothiocyanate (FITC)–conjugated Abs. Otherwise, cells were stained with rat anti-Ter119 Abs and goat antirat PE-conjugated Abs. Human mononuclear cells from leukapheresis of healthy donors were separated on Ficoll-Paque (Pharmacia, Uppsala, Sweden) and stained with mouse anti-CD36 Abs followed by rat antimouse PE-conjugated Abs and anti-CD14 FITC-conjugated Abs. Human CD34+ cells were stained with anti-CD34 Abs phycoerythrin-Cy5 (PC5) conjugated. The purity of the sorted populations was evaluated by postsorting analysis and Wright-Giemsa staining.
DNase I sensitivity assay
DNase I sensitivity assay was carried out as previously described.23 About 30 000 nuclei were digested with 0 or up to 0.35 U of DNase I (Roche, Indianapolis, IN) for 30 minutes on ice. Average molecular weight of DNase I–treated samples was determined by Southern blot.
Chromatin immunoprecipitation (ChIP) assay
Antibodies and ChIP kits were purchased from Upstate Biotechnology (Lake Placid, NY). Antibodies were raised against acetylated histones H3 (K9, K14), H4 (K5, K8, K12, K16), phosphorylated histone H3 (S10), dimethylated histone H3 (K4), and nonmodified histone H3. Cross-linked chromatin was reduced in size by sonication in order to obtain fragments of 500-bp average size. ChIP assays were carried out as per manufacturer's instructions.
Duplex PCR and single-cell RT-PCR analyses
Quantitative polymerase chain reaction (PCR) analysis on DNase I–treated and ChIP samples was performed as previously described.23 All primer sequences and amplicons, as well as their molecular weights, are available on the Blood website; see the Supplemental Materials link at the top of the online article.
For single-cell reverse transcriptase–PCR (RT-PCR), single cells were deposited into 96-well plates. RT-PCR was performed using Qiagen (Valencia, CA) one-step RT-PCR kit.
S1 nuclease protection assay
RNA samples were prepared using Trizol (Life Technologies, Gaithersburg, MD) and the assay was performed as previously described.23
Results
Purification of hematopoietic progenitor cells and erythroid cells
Murine HPCs were purified from adult bone marrow by sorting c-Kit+/CD31high/Ly-6C- cells. These cells comprise about 2% of the nucleated population and are early hematopoietic cells without mature or late-committed properties.24,25 Figure 1A shows a typical 3-color flow cytometric analysis. The sorted population is 97% pure and possesses a blastlike phenotype, as revealed by Wright-Giemsa staining (Figure 1B). The hematopoietic potentiality of these cells was ascertained by in vitro colony assays. On average, of 100 colonies 54 were CFU-GEMMs (granulocyte-erythrocyte-megakaryocyte-macrophage colony-forming units), 21 were CFU-GMs (granulocyte-macrophage colony-forming units), 21 were BFU-Es (erythroid burst-forming units), and none were CFU-Es (erythroid colony-forming units). The remaining 4% were large colonies with an undifferentiated morphology (Table 1). Thus, 79% of the HPC population is composed of progenitors with multilineage potential and only 20% of the progenitors already shows unilineage commitment. Relative to total bone marrow, the HPC population displays a 95-fold enrichment in colony-forming cell (CFC) activity. This cloning efficiency is influenced by cell mortality induced by the long procedure and high-speed sorting.
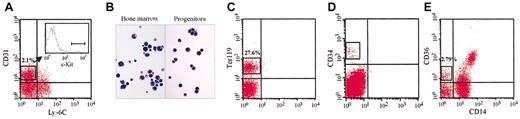
Flow cytometric analysis and Wright-Giemsa staining. Representative examples of sorting procedures. Log fluorescence distribution of mouse and human stained cells, showing the live gates used for flow cytometry. (A) Three-color flow cytometric analysis of adult mouse bone marrow cells stained with anti–c-Kit, CD31, and Ly-6C Abs. (B) Morphology of c-Kit+/CD31high/Ly-6C- cells (Progenitors) compared with total bone marrow cells (Bone marrow) stained with Wright-Giemsa (original magnification, × 40). (C) Flow cytometric analysis and sorting window of mouse bone marrow cells stained with Ter119 Abs. (D) Sorting profile of human CD34+ cells. (E) Two-color flow cytometric analysis and sorting of human mononuclear bone marrow cells tracked by CD36 and CD14 Abs.
Flow cytometric analysis and Wright-Giemsa staining. Representative examples of sorting procedures. Log fluorescence distribution of mouse and human stained cells, showing the live gates used for flow cytometry. (A) Three-color flow cytometric analysis of adult mouse bone marrow cells stained with anti–c-Kit, CD31, and Ly-6C Abs. (B) Morphology of c-Kit+/CD31high/Ly-6C- cells (Progenitors) compared with total bone marrow cells (Bone marrow) stained with Wright-Giemsa (original magnification, × 40). (C) Flow cytometric analysis and sorting window of mouse bone marrow cells stained with Ter119 Abs. (D) Sorting profile of human CD34+ cells. (E) Two-color flow cytometric analysis and sorting of human mononuclear bone marrow cells tracked by CD36 and CD14 Abs.
Clonogenic ability of c-Kit+/CD31high/Ly-6C- cells
. | Bone marrow cells, % . | c-Kit+/CD31high/Ly-6C-, % . |
|---|---|---|
| CFU-E | 2 | None |
| Mature BFU-E | 13 | None |
| Primitive BFU-E | 2 | 21 |
| CFU-GM | 73 | 2 |
| CFU-GM large | None | 19 |
| CFU-GEMM | 10 | 5 |
| CFU-GEMM large | None | 49 |
| Blastlike colonies | None | 4 |
| Total | 100 | 100 |
. | Bone marrow cells, % . | c-Kit+/CD31high/Ly-6C-, % . |
|---|---|---|
| CFU-E | 2 | None |
| Mature BFU-E | 13 | None |
| Primitive BFU-E | 2 | 21 |
| CFU-GM | 73 | 2 |
| CFU-GM large | None | 19 |
| CFU-GEMM | 10 | 5 |
| CFU-GEMM large | None | 49 |
| Blastlike colonies | None | 4 |
| Total | 100 | 100 |
Cells were seeded on methylcellulose and colonies were scored at day 14. Percentages shown are the results of 3 experiments.
CFU-E indicates erythroid colony-forming units; mature BFU-E, progenitors that give rise to colonies constituted by 3 up to 8 erythroblast clusters; primitive BFU-E, progenitors that give rise to 9 or more clusters of hemoglobinized erythroblasts; CFU-GM, granulocyte-macrophage colony-forming unit; and CFU-GEMM, granulocyte-erythrocyte-megakaryocyte-macrophage colony-forming unit.
Murine erythroid cells were isolated from bone marrow according to their Ter119+ phenotype and evaluated by colony assays. Ter119 Ab recognizes erythroid cells at different stages of maturation, from early proerythroblasts to mature erythrocytes.26 As expected,26 Ter119+ cells (Figure 1C) showed no CFC activity in colony assay. After sorting, more than 95% of the cells were Ter119+ (data not shown).
To enrich for human HPCs, CD34+ cells were purified from human leukapheresis samples (hereafter referred to as bone marrow cells). CD34+ cells represented 1% to 2% of total bone marrow27 (Figure 1D) and displayed 97% purity on average (data not shown).
Human erythroid cells were purified from bone marrow selecting for CD36+/CD14- cells. CD36 is a marker of erythroid commitment, which is present on CFU-Es, late BFU-Es,28 and erythroid cells.29 Since CD36 is also expressed on monocytes, these were purged using CD14 (Figure 1E), which is expressed at high levels exclusively on monocytes and macrophages. The CD36+/CD14- population displayed 97% purity on average (data not shown).
Single-cell pattern of globin gene expression in HPCs
It has been shown that mouse β globins are expressed at basal levels in HPCs16,17 and that in transgenic mice LacZ under the control of the huβ promoter is expressed before erythroid commitment.30 Nevertheless, it is unknown whether human β-like globin genes can be expressed in HPCs in vivo. To verify this, single-cell RT-PCR was performed on bone marrow HPCs of a transgenic mouse line carrying a 70-kb human β-globin locus (line 2,14 hereafter “ln2”) and on human bone marrow HPCs (Figure 2A-C). For RT-PCR of single-ln2 c-Kit+/CD31high/Ly-6C- cells or human CD34+ cells, we used primers that coamplify adult human (β and δ) and mouse (βmaj and βmin) globin transcripts31 or fetal human (γ) and mouse embryonic (βH1) transcripts.32 In ln2 HPCs, human and mouse transcripts were distinguished by restriction polymorphisms. In ln2 HPCs βmaj, transgenic huβ (tg-huβ), as well as huγ (tg-huγ) genes are expressed in about half of the cells subjected to RT-PCR. Interestingly, β and γ gene expression were also detected in about 50% of adult human CD34+ cells (Table 2). The level of huβ gene expression in human CD34+ cells was evaluated by S1 nuclease protection assay and shown to be at least 100-fold lower than in human total bone marrow (data not shown). Thus, the expression of β and γ genes in HPCs is not linked to their developmental stage–specific regulation.
Single-cell RT-PCR and S1 nuclease protection assay. Qualitative analysis of single-cell RT-PCR assays. (A) Huβ/huδ (266 bp) as well as βmaj/βmin (343 bp) transcripts were amplified and distinguished after EcoRI digestion.31 wt indicates wild-type mouse total bone marrow RNA; ln2, ln2 total bone marrow RNA; and -ctl, negative control. (B) Lanes 1-6 are representative examples of single-cell RT-PCR performed on ln2 c-Kit+/CD31high/Ly-6C- cells.(C) Lanes 1-9 are representative examples of single-cell RT-PCR performed on human CD34+ cells.
Single-cell RT-PCR and S1 nuclease protection assay. Qualitative analysis of single-cell RT-PCR assays. (A) Huβ/huδ (266 bp) as well as βmaj/βmin (343 bp) transcripts were amplified and distinguished after EcoRI digestion.31 wt indicates wild-type mouse total bone marrow RNA; ln2, ln2 total bone marrow RNA; and -ctl, negative control. (B) Lanes 1-6 are representative examples of single-cell RT-PCR performed on ln2 c-Kit+/CD31high/Ly-6C- cells.(C) Lanes 1-9 are representative examples of single-cell RT-PCR performed on human CD34+ cells.
Summary of one-step RT-PCR and S1 nuclease protection assays performed on In2 bone marrow and fetal liver c-Kit+/CD31high/Ly-6C— cells and human CD34+ cells
. | Single-cell RT-PCR analysis . | . | . | . | S1 assay . | |||
|---|---|---|---|---|---|---|---|---|
. | βmaj/βmin . | huβ/huδ . | huγ . | βH1 . | huβ . | |||
| Ln2 bone marrow c-Kit+/CD31high/Ly-6C— | 18/25 | 13/26 | 15/24 | 3/25 | ND | |||
| Ln2 fetal liver c-Kit+/CD31high/Ly-6C— | ND | ND | 13/26 | 3/25 | ND | |||
| Human bone marrow | — | ND | ND | — | 100 | |||
| Human CD34+ | — | 17/25 | 16/26 | — | < 1 | |||
. | Single-cell RT-PCR analysis . | . | . | . | S1 assay . | |||
|---|---|---|---|---|---|---|---|---|
. | βmaj/βmin . | huβ/huδ . | huγ . | βH1 . | huβ . | |||
| Ln2 bone marrow c-Kit+/CD31high/Ly-6C— | 18/25 | 13/26 | 15/24 | 3/25 | ND | |||
| Ln2 fetal liver c-Kit+/CD31high/Ly-6C— | ND | ND | 13/26 | 3/25 | ND | |||
| Human bone marrow | — | ND | ND | — | 100 | |||
| Human CD34+ | — | 17/25 | 16/26 | — | < 1 | |||
The level of human β-globin gene expression (determined by S1 nuclease protection assay) in CD34+ cells is relative to the expression level in human bone marrow cells.
ND indicates not done;—, not applicable.
Chromatin accessibility of the β-globin locus during hematopoiesis
Chromatin conformational changes at the mouse and human β-globin loci during hematopoiesis were investigated by sensitivity to DNase I digestion. For this purpose, we used ln2 and human bone marrows. Nuclei were treated with different concentrations of DNase I, and purified DNA was used as template for duplex PCR. One primer set was specific for either the mouse (Figure 3A) or the human (Figure 3B) locus. The second set was specific for genes transcriptionally inactive in hematopoietic cells, namely: ZFP37 (ZFP23 ) or amylase 2.1y (amy22 ) for mouse cells, and pax633 or necdin21,34 for human cells. PCR reactions were carried out within the linear range of amplification and all primer sets used in duplex PCR were tested to exclude any significant difference in amplification efficiency. Each data point depicted in Figure 3 represents the ratio of the 2 PCR products normalized to the input, DNase I–untreated sample. Curves showing a steep drop at the start indicate DNase I hypersensitivity, whereas the steepness of the curves at later points is a measure of general DNase I sensitivity. We observed that in ln2 HPCs, βmaj promoter is highly sensitive to DNase I (Figure 3C), whereas the tg-huβ promoter is weakly sensitive or insensitive (Figure 3D). To evaluate the degree of DNase I sensitivity at tg-huβ promoter, a third nonhematopoietic control (the kidney-specific Tamm-Horsfall [THP] gene35 ) was tested against ZFP. As shown in Figure 3E, DNase I sensitivity of THP and huβ promoters is comparable. This suggests that chromatin accessibility at the huβ promoter reflects the general situation for promoters of nonexpressed genes in hematopoietic cells. Mouse HS3 (mHS3) and the transgenic human HS3 (tgHS3) are both sensitive to DNase I in ln2 HPCs (Figure 3F-G). The same is also found for human HS2 (tgHS2) and HS5 (tgHS5) (Supplemental Materials). In ln2 erythroid (Ter119+) cells, DNase I sensitivity at the LCR and at globin promoters significantly increases, especially at tg-huβ and βmaj promoters (Figure 3C-G).
DNase I sensitivity assay of the β-globin locus. A map of mouse (A) and human (B) β-globin locus; genes are shown as ▪ and the location of LCR HS is indicated by ↓. Amplified regions used for PCR-based DNase I and ChIP assays are indicated by ▦. (C-I) PCR-based DNase I analysis. Each point of the curves represents samples of comparable molecular weight (ranging from 12 kb to 0.5 kb). For βmaj/ZFP and tg-huβ/ZFP, an additional DNase I–treated sample of 150-bp average size was included. The intensity of the PCR products was quantitated by Phosphorimager and plotted on graphs relative to the input (DNase I–untreated chromatin). Y-axis indicates relative amount of PCR products (globin relative to ZFP37 or pax6 products); X-axis, increasing DNase I concentration, ranging from 0 up to 0.35 U (see “Materials and methods”). Standard errors of mean (SEM) are indicated by vertical lines and are the results of at least 3 independent experiments. The P value was obtained using the unpaired Student t test. ZFP indicates ZFP37; βmaj, β major promoter; tg-huβ, human β promoter in transgenic ln2; mHS3, murine HS3; tgHS3, human HS3 in transgenic ln2; huβ, human β promoter in human bone marrow; and HS3, human HS3 in human bone marrow.
DNase I sensitivity assay of the β-globin locus. A map of mouse (A) and human (B) β-globin locus; genes are shown as ▪ and the location of LCR HS is indicated by ↓. Amplified regions used for PCR-based DNase I and ChIP assays are indicated by ▦. (C-I) PCR-based DNase I analysis. Each point of the curves represents samples of comparable molecular weight (ranging from 12 kb to 0.5 kb). For βmaj/ZFP and tg-huβ/ZFP, an additional DNase I–treated sample of 150-bp average size was included. The intensity of the PCR products was quantitated by Phosphorimager and plotted on graphs relative to the input (DNase I–untreated chromatin). Y-axis indicates relative amount of PCR products (globin relative to ZFP37 or pax6 products); X-axis, increasing DNase I concentration, ranging from 0 up to 0.35 U (see “Materials and methods”). Standard errors of mean (SEM) are indicated by vertical lines and are the results of at least 3 independent experiments. The P value was obtained using the unpaired Student t test. ZFP indicates ZFP37; βmaj, β major promoter; tg-huβ, human β promoter in transgenic ln2; mHS3, murine HS3; tgHS3, human HS3 in transgenic ln2; huβ, human β promoter in human bone marrow; and HS3, human HS3 in human bone marrow.
DNase I sensitivity was then analyzed in human CD34+ and CD36+/CD14- cells. As in ln2 HPCs, in human CD34+ cells the huβ promoter is not hypersensitive to DNase I digestion (Figure 3H). In contrast, HS3 is open in human HPCs just as was observed in ln2 (Figure 3I). Both huβ and HS3 acquire a more accessible chromatin conformation in human erythroid cells (Figure 3H-I).
Taken together, these results indicate that chromatin at the human β-globin LCR is moderately accessible in both human and ln2 HPCs, whereas the huβ promoter is not; hence, chromatin accessibility along the human LCR precedes that at the huβ promoter. On the other hand, unlike its human homolog, the βmaj promoter is already activated and accessible to DNase I in HPCs.
Histone covalent modifications at the β-globin locus during hematopoiesis
Patterns of histone covalent modification at the murine and human β-globin loci during hematopoiesis were assessed by ChIP assay36 using antibodies specific for acetylated histone H3 (AcH3), H4 (AcH4), dimethylated histone H3, or phosphorylated histone H3 (PhH3). ChIP material was used as a template for duplex PCR with one primer set specific for mouse or human globin locus and a second primer set specific for ZFP or amy (ln2 controls) and pax6 or necdin (human controls). PCR reactions were performed under conditions of linear amplification (Figure 4A). Ln2 as well as human controls showed no variation in histone acetylation (data not shown). To establish the relative enrichment or depletion of β-globin sequences for histone modifications, the ratio of the 2 PCR products (globin and control sequences) was determined in each immunoprecipitated sample and normalized to the input ratio. Both ZFP (Figure 4B) and amy (not shown, but same results) were used as controls for ln2 cells, whereas pax6 (Figure 4C) and necdin (not shown, but same results) were used as controls for human cells. All results were confirmed by 2 or 3 independent ChIP assays.
ChIP analysis of the human β-globin LCR in ln2 and human bone marrow cells. Immunoprecipitated and unbound (input) chromatin samples were subjected to duplex PCR analysis with one primer set specific for the human globin locus LCR and a second primer set specific for ZFP37 (ZFP) or pax6 gene. All PCR reactions were performed in parallel under conditions of linear amplification. Products were quantified by Phosphorimager. The level of enrichment of globin regions relative to the control and input samples is represented by bars with their corresponding SEM deviations. A value of 1 indicates that no enrichment was detected. (A) Duplex PCR ran in linear range of amplification. The same template DNA was subject to 29, 31, or 33 cycles of PCR amplification. Bars show the total intensity of the 2 PCR products and the line indicates the globin-control ratio. In the example, HS3 and pax6 primer sets were used. (B) ChIP performed with antiacetylated histone H3 (AcH3) and H4 (AcH4) Abs. Shown is the level of acetylation of HS3 (tgHS3) and HS4 (tgHS4) in ln2 HPCs and erythroid cells. Error bars indicate SEM deviations. (C) ChIP performed with anti-AcH3 and AcH4 Abs. Shown is the level of acetylation of HS3 and HS4 in human HPCs and erythroid cells. Error bars indicate SEM deviations.
ChIP analysis of the human β-globin LCR in ln2 and human bone marrow cells. Immunoprecipitated and unbound (input) chromatin samples were subjected to duplex PCR analysis with one primer set specific for the human globin locus LCR and a second primer set specific for ZFP37 (ZFP) or pax6 gene. All PCR reactions were performed in parallel under conditions of linear amplification. Products were quantified by Phosphorimager. The level of enrichment of globin regions relative to the control and input samples is represented by bars with their corresponding SEM deviations. A value of 1 indicates that no enrichment was detected. (A) Duplex PCR ran in linear range of amplification. The same template DNA was subject to 29, 31, or 33 cycles of PCR amplification. Bars show the total intensity of the 2 PCR products and the line indicates the globin-control ratio. In the example, HS3 and pax6 primer sets were used. (B) ChIP performed with antiacetylated histone H3 (AcH3) and H4 (AcH4) Abs. Shown is the level of acetylation of HS3 (tgHS3) and HS4 (tgHS4) in ln2 HPCs and erythroid cells. Error bars indicate SEM deviations. (C) ChIP performed with anti-AcH3 and AcH4 Abs. Shown is the level of acetylation of HS3 and HS4 in human HPCs and erythroid cells. Error bars indicate SEM deviations.
We first analyzed the level of AcH3 and AcH4 at human HS3 and HS4 in HPCs and erythroid cells derived from ln2 (tgHS3 and tgHS4) and human bone marrow (HS3 and HS4). In ln2 HPCs, AcH3 level is higher than the control at tgHS4 but not at tgHS3, whereas histone H4 is acetylated at both tgHS3 and tgHS4 (Figure 4B). In mature erythroid cells AcH3 and AcH4 level increases at both sites (Figure 4B). A similar pattern of acetylation is seen in human HPCs and erythroid cells (Figure 4C).
We then examined patterns of histone modification across the βmaj and huβ promoters. At the βmaj promoter only H4 is significantly acetylated in HPCs; in erythroid cells AcH3 increases substantially, whereas the increase in AcH4 is much smaller (Figure 5A). In contrast to the mouse βmaj promoter, the huβ promoter shows a high level of AcH3 in ln2 HPCs and human HPCs, which surprisingly decreases in the case of differentiated erythroid cells (Figure 5B-C). The level of AcH4 is very similar to that of the control in HPCs as well as erythroid cells. The rather unexpected H3 hyperacetylation at the huβ promoter in HPCs and its decrease in erythroid cells appears to be mediated by intrinsic properties of the human β-globin locus since the same patterns of histone acetylation are observed in human and ln2 mouse HPCs and erythroid cells.
ChIP analysis of mouse and human β-globin genes and promoters. Immunoprecipitated samples were subjected to duplex PCR analysis with one primer set specific for human or mouse β globin regions and a second primer set specific for ZFP37 (ZFP) or pax6 gene. The level of enrichment of globin regions relative to the control regions and input samples is represented by bars, with their corresponding SEM deviations. □ indicates progenitors; and ▪, erythroid cells. (A-B) ChIP analysis of ln2 HPCs and erythroid cells. Either βmaj or the transgenic huβ promoter was analyzed by PCR-based ChIP assay. X-axis indicates antibodies used for ChIP assays. AcH3 indicates antiacetylated H3; AcH4, antiacetylated H4; MeK4, antidimethylated H3; PhH3, antiphosphorylated H3; and H3, antinonmodified histone H3. (C) ChIP analysis of huβ promoter in human HPCs and erythroid cells. For ChIP analysis anti-AcH3 or AcH4 Abs were used. (D) Schematic representation of the huβ region; huβ exon 1 and 2 are indicated by gray boxes and amplicons are shown by dotted lines. (E) huβ5, huβ1 and (F) human Ψβ regions in ln2 (tg-huβ5, tg-huβ1, and tg-psβ) and human (huβ5, huβ1, and psβ) HPCs and erythroid cells were investigated by PCR-based ChIP assays performed with anti-AcH3 Abs.
ChIP analysis of mouse and human β-globin genes and promoters. Immunoprecipitated samples were subjected to duplex PCR analysis with one primer set specific for human or mouse β globin regions and a second primer set specific for ZFP37 (ZFP) or pax6 gene. The level of enrichment of globin regions relative to the control regions and input samples is represented by bars, with their corresponding SEM deviations. □ indicates progenitors; and ▪, erythroid cells. (A-B) ChIP analysis of ln2 HPCs and erythroid cells. Either βmaj or the transgenic huβ promoter was analyzed by PCR-based ChIP assay. X-axis indicates antibodies used for ChIP assays. AcH3 indicates antiacetylated H3; AcH4, antiacetylated H4; MeK4, antidimethylated H3; PhH3, antiphosphorylated H3; and H3, antinonmodified histone H3. (C) ChIP analysis of huβ promoter in human HPCs and erythroid cells. For ChIP analysis anti-AcH3 or AcH4 Abs were used. (D) Schematic representation of the huβ region; huβ exon 1 and 2 are indicated by gray boxes and amplicons are shown by dotted lines. (E) huβ5, huβ1 and (F) human Ψβ regions in ln2 (tg-huβ5, tg-huβ1, and tg-psβ) and human (huβ5, huβ1, and psβ) HPCs and erythroid cells were investigated by PCR-based ChIP assays performed with anti-AcH3 Abs.
To exclude the possibility that detectability of AcH3 is lost at the tg-huβ promoter in erythroid cells, we performed ChIP assays with anti-PhH3 and nonmodified histone H3 (H3) Ab. Indeed, it has been shown that the anti-AcH3 Ab used for ChIP assays may not recognize AcH3 when also phosphorylated at S10.37 As shown in Figure 5B, no significant enrichment for PhH3 was detected at the tg-huβ promoter in erythroid cells. Therefore, the weak acetylation of histone H3 is not the consequence of its phosphorylation. Moreover, ChIP performed with anti-H3 Ab showed that nonmodified histone H3 is underrepresented at the βmaj promoter in erythroid cells, confirming that histones are mainly acetylated (Figure 5A). Instead, the detection of nonmodified histone H3 at the tg-huβ promoter suggests that the promoter is not devoid of nucleosomes in erythroid cells (Figure 5B). Since histone H3 lysine 4 dimethylation (MeK4) has been shown to play an important role during gene activation,38 MeK4 was investigated at the tg-huβ promoter. Figure 5B shows that the tg-huβ promoter displays a significant level of MeK4 in HPCs, which undergoes a 3-fold enrichment in erythroid cells.
To better define the pattern of acetylation at the tg-huβ gene, AcH3 was investigated at 2 additional regions situated approximately 300 bp 5′ (huβ5) and approximately 400 bp 3′ (huβ1) of the huβ minimal promoter (Figure 5D). In ln2 and human HPCs, no enrichment for AcH3 is detected at huβ5 (Figure 5E), whereas huβ1 is significantly acetylated. In highly expressing erythroid cells, huβ5 remains equally hypoacetylated as in HPCs; however, in ln2 and human erythroid cells the downstream huβ gene (huβ1) is significantly more acetylated than observed in HPCs (Figure 5E). As a control, we also analyzed histone acetylation at a nontranscribed region of the locus, the intergenic Ψβ region. In human and ln2 HPCs and erythroid cells, Ψβ is not AcH3 (Figure 5F).
Thus, the minimal huβ promoter is epigenetically marked by AcH3 and MeK4 in HPCs. In erythroid cells, AcH3 at the minimal promoter decreases, whereas AcH3 across the gene and MeK4 at the tg-huβ promoter further increase. These results suggest the following: (1) globin gene expression in erythroid cells does not depend on promoter acetylation, and (2) AcH3 and MeK4 epigenetically mark the huβ gene and promoter before the establishment of high-level transcription in erythroid cells.
Globin gene transcription: silencing and induction
An interesting aspect of the human β-globin locus is the switching process during development and the possibility of using a variety of drugs interfering with acetylation to modify this switch in β-thalassemic patients. We therefore compared histone acetylation at huβ and huγ promoters. The γ genes are expressed during embryonic and fetal stages in humans and in ln2 mice. In adult ln2 and human bone marrow cells, human γ gene expression becomes negligible.14,39 ChIP analyses revealed that, in contrast to huβ (Figure 5B-C), histone H3 at Gγ/Aγ promoters (Figure 6A) is not acetylated in HPCs purified from human or ln2 bone marrow, suggesting that the acetylation mark in HPCs would specifically occur at promoters of genes that will become active in erythroid cells. If true, this lends to the prediction that huγ promoters should exhibit H3 acetylation in HPCs at earlier stages of development when γ genes are expressed. We therefore purified HPCs (c-Kit+/CD31high/Ly-6C- cells) from ln2 11.5-dpc fetal livers for ChIP analysis. At this stage fetal liver HPCs will give rise to erythroid cells that express both human γ and β genes. It should be noted however, that embryonic γ-but not β-expressing cells are still in circulation and thus present in the fetal liver erythroid cells.14 In our hands, the HPCs represent 8% of the 11.5-dpc fetal liver and they possess the same morphology, potential in colony assay, and transcriptional activity as bone marrow–derived HPCs (Figure 2; data not shown). As shown in Figure 6B, in ln2 fetal liver HPCs, both tg-huγ and tg-huβ promoters are significantly H3 acetylated. Similar to what was observed for the huβ promoter in adult erythroid cells, AcH3 at tg-huγ promoters decreases in 11.5-dpc erythroid cells. Hence, H3 deacetylation at tg-huγ again correlates with gene expression. In effect, AcH3 is largely maintained at tg-huβ promoter in erythroid cells, part of which is embryonic non-β–expressing cells.
ChIP analysis of human γ and β promoters in bone marrow and 11.5-dpc fetal liver cells. Chromatin from ln2 and human bone marrow cells was immunoprecipitated with antiacetylated H3 (AcH3) or H4 (AcH4) Abs. Immunoprecipitated and input samples were subjected to duplex PCR analysis with one primer set specific for huγ or huβ promoters and another specific for ZFP37 (mouse) or pax6 (human) gene. (A) ChIP analysis of huγ promoters in ln2 and human bone marrow HPCs and erythroid cells. (B) ChIP analysis of huγ and huβ promoters (tg-huγ and tg-huβ) in ln2 11.5-dpc fetal liver HPCs and erythroid cells. (C) ChIP and S1 nuclease protection assays of tg-huβ and tg-huγ promoters in ln2 TSA-treated Ter119+ cells. ChIP assays were performed with anti-AcH3 Abs, and the level of acetylation of TSA-treated samples (TSA) relative to their respective ethanol-treated (ETOH) controls is represented by bars with their corresponding SEM deviations. Mouse β-actin transcript was used as internal control for S1 nuclease protection assay; tg-huβ and tg-huγ expression level in TSA-treated cells is relative to the ethanol treated controls.
ChIP analysis of human γ and β promoters in bone marrow and 11.5-dpc fetal liver cells. Chromatin from ln2 and human bone marrow cells was immunoprecipitated with antiacetylated H3 (AcH3) or H4 (AcH4) Abs. Immunoprecipitated and input samples were subjected to duplex PCR analysis with one primer set specific for huγ or huβ promoters and another specific for ZFP37 (mouse) or pax6 (human) gene. (A) ChIP analysis of huγ promoters in ln2 and human bone marrow HPCs and erythroid cells. (B) ChIP analysis of huγ and huβ promoters (tg-huγ and tg-huβ) in ln2 11.5-dpc fetal liver HPCs and erythroid cells. (C) ChIP and S1 nuclease protection assays of tg-huβ and tg-huγ promoters in ln2 TSA-treated Ter119+ cells. ChIP assays were performed with anti-AcH3 Abs, and the level of acetylation of TSA-treated samples (TSA) relative to their respective ethanol-treated (ETOH) controls is represented by bars with their corresponding SEM deviations. Mouse β-actin transcript was used as internal control for S1 nuclease protection assay; tg-huβ and tg-huγ expression level in TSA-treated cells is relative to the ethanol treated controls.
We then investigated whether TSA,40 a known histone deacetylase inhibitor, would influence the acetylation state of tg-huγ and/or tg-huβ promoters and change their expression levels. Ln2 bone marrow was treated with TSA or with ethanol (TSA solvent) and AcH3 level was evaluated at the tg-huγ and tg-huβ globin promoters in erythroid cells. A 2- to 3-fold enrichment in AcH3 (TSA-relative to ethanol-treated cells) was seen at both tg-huβ and tg-huγ promoters (Figure 6C). This increase does not significantly modify the level or the balance of expression of these genes, as observed by S1 nuclease assay on ethanol- and TSA-treated bone marrow (Figure 6C). Thus, a higher level of AcH3 at tg-huβ and tg-huγ promoters in erythroid cells does not modify globin gene transcription.
Discussion
The work presented here assesses, for the first time, variations of chromatin conformation and histone covalent modifications at the human β-globin locus during hematopoietic differentiation in vivo. Our results suggest that a defined pattern of histone H3 acetylation/dimethylation is important for the specific activation of human globin promoters before high-level expression in erythroid cells. We do not know when this gene-specific potentiation is set, but since multipotent progenitors represent the vast majority of the HPCs investigated (Table 1), it could be present before erythroid lineage commitment.
Chromatin modifications of the human globin LCR in HPCs and erythroid cells
It has been shown that chromatin within lineage-restricted regulatory regions,8,10,41 including the mouse β-globin locus,7,17 can be remodeled in HPCs. Using transgenic mice and human bone marrow, we demonstrate here that chromatin at the human β-globin LCR is acetylated and accessible (to DNase I) in HPCs in vivo. Previous in vitro investigations using uninduced MEL cells22 provided some evidence in this transformed erythroid-committed cell line that the human LCR displays characteristics of chromatin poised for transcriptional activation. We now provide evidence that the human β-globin LCR is activated in a population of HPCs purified from fresh bone marrow and composed mostly of uncommitted progenitors. Accessibility at HS2, HS3, and HS5 increases in mature erythroid cells when compared with HPCs, suggesting that the LCR is further remodeled during differentiation to allow high-level β-globin gene expression. The human LCR therefore follows an activation pattern similar to that of other hematopoietic loci.10,41 Thus, the LCR may already be activated to some extent by partial HS occupancy in HPCs, but complete occupancy of HS and extended chromatin remodeling would only be attained in erythroid cells upon additional binding of stage-specific transactivators. The analysis of histone modifications at different HSs within the LCR suggests that histone acetylation has already facilitated an activated structure at the human LCR, with the exception of HS3, in both human and ln2 bone marrow HPCs. In mature erythroid cells the acetylation is further increased at all sites including HS3. The exceptional behavior of HS3 is possibly related to its “chromatin opening” ability42 and/or to the particular chromatin organization of the HS3 core region.43
Chromatin at the human β and mouse β major genes is differently activated during erythropoiesis
The huβ and mouse βmaj promoters show different patterns of histone modifications and DNase I accessibility during differentiation from HPCs to erythroid cells. The mouse βmaj promoter is already largely accessible to DNase I digestion in HPCs with a low level of acetylated H3 and a moderate level of acetylated H4. During differentiation, accessibility is further increased (in particular, the appearance of the hypersensitive site in the promoter illustrated by the early points in the curve in Figure 3C), H3 acetylation is increased several fold, whereas H4 acetylation only doubles when compared with controls. In contrast, in human bone marrow the endogenous huβ promoter shows a very different pattern and, importantly, maintains this pattern when the locus is introduced in mice. It is not accessible to DNase I in HPCs and shows a low level of H4 acetylation but, curiously, a high level of H3 acetylation. Upon cell differentiation, the huβ promoter becomes more accessible to DNase I, H4 acetylation hardly changes, but H3 acetylation decreases significantly. The upstream part of the promoter shows a low level of H3 acetylation, which barely changes upon differentiation, whereas downstream in the gene histone H3 is already acetylated in HPCs and the level increases several folds upon differentiation. Using a cell culture–based system (transformed MEL cells), it has been reported that the huβ promoter is highly acetylated when active22 ; however, we cannot be certain whether our data agree or contradict those results as it is not clear whether the human locus in MEL cells was analyzed before (nonexpressing) or after induction of differentiation (expressing).
We conclude that the activation of the human and mouse β-globin promoters is different and that this difference is intrinsic to these loci since the huβ promoter maintains its own activation program when the human β-globin locus is introduced in the mouse. These differences may explain some of the controversies that have arisen with respect to activation of β-globin loci, where data from patients and mice show that expression and DNase I accessibility at the huβ globin gene are lost when the LCR is deleted (see Grosveld13 for review), whereas expression and sensitivity at the mouse βmaj gene is not lost when the LCR is deleted.44 Our data indicate that the mouse βmaj, but not the huβ globin gene, is already accessible to DNase I digestion in HPCs, suggesting that the LCR may indeed not be required to achieve accessibility across the mouse locus. Therefore, even though studies on the mouse β-globin locus have been invaluable tools to understand aspects of globin gene regulation, differences between the epigenetic regulation of the human and the mouse loci should be considered before comparing results obtained with these 2 β-globin loci.
Interestingly, histone hypoacetylation as we observed at the active huβ promoter is not unique. Indeed, it has been shown that several active genes are as hypoacetylated as the surrounding bulk chromatin.45,46 For example, histone acetylation, as exemplified at interferon β (IFN-β)47 and hormone receptor–dependent genes48 in mammals, or at PHO8 gene in Saccharomyces cerevisiae,49 is a transient signal that does not engender chromatin accessibility by itself but rather provides a mark that facilitates recruitment of remodeling complexes. The synergy between histone acetylation and the activity of switching/sucrose nonfermenting (SWI/SNF)–related chromatin-remodeling complex has been reported previously.50
Our results are consistent with a multistage activation model in which the huβ gene and promoter would first be epigenetically marked by histone H3 acetylation and K4 dimethylation in bone marrow HPCs before the onset of high-level transcription. This mark would subsequently be recognized by activators and/or remodeling complexes that in turn would guarantee high-level globin expression in erythroid cells, where indeed chromatin at the huβ promoter is dimethylated and remodeled but no longer acetylated.
The observed decrease of acetylation at the huβ promoter in erythroid cells can be either the consequence of an active deacetylation mechanism (carried out by HDACs) or of a passive process. In TSA-treated Ter119+ cells the tg-huβ promoter is significantly more acetylated than in nontreated cells, which suggests that the tg-huβ promoter is actively deacetylated by TSA-sensitive HDACs in erythroid cells. The transcription factor erythroid Krüppel-like factor (EKLF), which is involved in huβ promoter regulation, might play a causal role in huβ promoter deacetylation in erythroid cells since EKLF was found to interact with HDAC1.51 However, to date it is still unknown whether the EKLF-HDAC1 interaction at the huβ promoter occurs sometime during hematopoiesis.
In summary, we show that the huβ gene and promoter are epigenetically marked in adult HPCs, presumably poising the promoter for activation. The mark at the promoter occurs within a very defined region, spanning the promoter TATA, CCAAT, and CACCC boxes since no significant H3 acetylation at the upstream huβ5 and Ψβ intergenic regions was detected. During differentiation, the huβ promoter undergoes an extensive chromatin remodeling accompanied by histone H3 deacetylation, as revealed by hypersensitivity to DNase I digestion and histone H3 hypoacetylation in erythroid cells. Such a pattern of modification is not unlike what has been observed for the hepatocyte nuclear factor 4α (HNF 4α) gene in differentiating CaCo-2 (human, white, colon, adenocarcinoma) cells.52
Human β versus human γ gene epigenetic regulation during development
The fetal γ globin genes are silenced around birth in humans, but in ln2 the transgenic locus switches from γ to β gene expression in the early fetal liver stage, and only β is expressed in erythroid cells derived from adult bone marrow.14 We show that huγ promoters are not acetylated in ln2 and human bone marrow HPCs, whereas in HPCs derived from 11.5-dpc fetal livers these are H3 acetylated. The fact that the huβ promoter is acetylated in both 11.5-dpc and adult HPCs suggests that this acetylation is linked to an epigenetic and developmental gene–specific regulatory mechanism. Using somatic cell hybrids between fetal human or murine transgenic erythroblasts and MEL cells, it was shown that fetal erythroblasts could express the γ genes also in an adult MEL environment.53,54 This observation together with other cell fusion experiments55,56 invoked the suggestion that epigenetic changes taking place sometime during erythroid differentiation might be important for globin gene switching. Here, we have provided evidence that in freshly isolated human and murine HPCs, defined epigenetic mark(s) are linked to developmental-specific globin gene expression in mature erythroid cells.
Interestingly, TSA-induced H3 acetylation is not sufficient to reactivate tg-huγ gene expression in bone marrow erythroid cells. As previously shown, some HDAC inhibitors are able to induce γ globin gene reactivation in human erythroid cells.57-61 From these studies, it is unclear whether or not these HDAC inhibitors directly influence histone acetylation of the human β-globin locus. On the other hand, treatment of bone marrow cells with HDAC inhibitors is not sufficient to modify the pattern of globin gene expression in adult β–yeast artificial chromosome (β-YAC) transgenic mice62 carrying the whole human β-globin locus. Thus, epigenetic mechanisms other than histone acetylation might influence huγ gene regulation. As previously suggested,63 the difference in γ gene reactivation in human and transgenic mouse could also be explained by the absence of a “fetal-regulated” globin gene in mice. Then, in human cells, HDAC inhibitors could influence the expression of trans-acting factors, which in mice are either missing or not influenced by HDAC inhibitors.
It has been shown that mouse and human globin genes can be expressed at basal levels in adult HPCs but it is not known whether this transcription is involved in the maintenance of a local “potentiated” chromatin structure. Our results confirm that huγ and huβ genes are transcribed in bone marrow and fetal liver HPCs and show for the first time that this basal expression is neither linked to the level of promoter acetylation nor to the stage-specific activation of these genes in erythroid cells. Thus, basal globin gene expression in HPCs appears to be linked to the human β-globin locus potentiation during hematopoiesis but not to the developmental-specific regulation of globin genes.
Taken together, our results strongly suggest that the pattern of histone acetylation in HPCs is important for the transcriptional potentiation of globin genes and, more interestingly, for the developmentally regulated expression of these genes in erythroid cells. We show that the human γ and β globin genes and promoters are epigenetically marked by histone H3 acetylation/dimethylation in HPCs and we suggest that this mark can be recognized during differentiation by activators and/or remodeling complexes, such as EKLF coactivator-remodeling complex 1 (E-RC1),63 to allow proper expression in terminally differentiated erythroid cells.
Prepublished online as Blood First Edition Paper, August 14, 2003; DOI 10.1182/blood-2003-05-1540.
Supported by grants from the National Cancer Institute of Canada (Terry Fox Foundation) and from the Canadian Cancer Research Society (E.M.). S.B. was supported by the Guy-Bernier Immuno-Oncology fellowship and E.M. is a scholar of the Canadian Institutes of Health Research.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Silvana Lachance and Sophie Ouellette for technical assistance, Marella de Bruijn for useful comments, Denis-Claude Roy for providing us with leukapheresis samples, and Elliot Drobetsky and Marie Trudel for critical review of the manuscript.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal