Abstract
The nuclear receptor ligand all-trans retinoic acid (ATRA) causes dramatic terminal differentiation of acute promyelocytic leukemia (APL) cells in vitro and in patients, but it is less active in other malignancies. However, downstream mediators of the effects of ATRA are not well understood. We used a cDNA microarray to search for ATRA-regulated genes in the APL cell line NB4 and found that ATRA regulated several members of the tumor necrosis factor (TNF) pathway. Here we show that TNF can synergize with ATRA to induce differentiation, showing monocytic characteristics more typical of differentiation mediated by TNF than by ATRA. ATRA and TNF can also induce differentiation of the non-APL cell line U937. Underlying this response was an increase in TNF-induced nuclear factor-κB (NF-κB) DNA binding within 2 hours in the presence of ATRA and activation of NF-κB DNA binding and transcriptional activity in response to ATRA alone within 48 hours of ATRA treatment. Furthermore, we found a synergistic induction of the NF-κB target genes BCL-3, Dif-2, and TNF receptor 2 (TNFR2) in response to the combination of TNF and ATRA. These genes have been previously shown to play a role in TNF signaling, and amplification of such genes may represent a mechanism whereby TNF and ATRA can act synergistically. We propose that ATRA can prime cancer cells for differentiation triggered by TNF and suggest that targeting the TNF pathway in combination with ATRA may represent a novel route to treat leukemias. (Blood. 2003;102:237-245)
Introduction
All-trans retinoic acid (ATRA) is a naturally occurring vitamin A derivative that plays a critical role in diverse cellular processes, including proliferation, embryo morphogenesis, and differentiation. It has also been found effective in promoting differentiation of various types of tumor cells in vitro and in vivo. Because of its growth-inhibiting and differentiation-promoting properties, retinoic acid and its derivatives hold therapeutic promise in treating cancer, especially when used in combination with other chemotherapeutic agents. Most impressively, retinoic acid has been shown to effect complete remission in patients with acute promyelocytic leukemia (APL).
The biologic actions of ATRA are mediated through interaction with the nuclear retinoic acid receptors (RAR types α, β, γ) and retinoid X receptors (RXR types α, β, γ). When bound by ligand, these receptors release corepressor complexes and recruit coactivator complexes. Such action results in potent transcriptional activation of a number of target genes, but the specific genes responsible for ATRA-driven differentiation of normal and transformed cells remain undetermined. In the APL model of differentiation, cells of myeloid origin are blocked at the promyelocyte stage. The presence of the PML/RAR oncoprotein plays an integral role in this differentiation block. ATRA-induced degradation of the PML/RAR oncoprotein appears to be an important initial step in allowing APL cells to differentiate. However, there is evidence that the disappearance of the fusion protein may not be necessary for ATRA-driven differentiation.1 Furthermore, the removal of PML/RAR by ribozymes results in cell apoptosis,2 indicating that this action alone is insufficient for ATRA-mediated differentiation. There is evidence that members of the CEBP family of transcription factors3-5 and the cell-cycle inhibitor p216,7 may play a role, but undoubtedly other genes have an impact on the granulopoietic response to ATRA.
Finding biologic targets of ATRA is complicated by the fact that cells exist in a milieu containing a variety of hormones and cytokines. When combined with ATRA, these factors may exert different cellular responses than when these agents are used alone. This concept is supported by reports demonstrating that cytokines such as granulocyte–colony-stimulating factor (G-CSF), granulocyte macrophage–colony-stimulating factor (GM-CSF), and interferons can combine with ATRA in a synergistic manner to induce differentiation and gene expression.8-12 The clinical relevance of pathways that cross talk with ATRA has been demonstrated by some promising studies in the treatment of several malignancies.13-16 The molecular mechanisms of these synergistic effects on differentiation remain unclear. A possible explanation is that the transcription of some differentiation-promoting genes may be turned on only in the presence of both agents.11,17 Therefore, finding novel pathways that intersect with ATRA signaling is important for our understanding of the physiological functions of ATRA and in devising rational combination therapy regimens.
To find such pathways, we used cDNA microarray technology to search for genes modulated by ATRA in the APL cell line NB4. We reasoned that ATRA-mediated regulation of groups of genes belonging to a particular signaling pathway would indicate that this pathway is important for differentiation. We found several members of the tumor necrosis factor (TNF) pathway to be regulated by ATRA. TNF is a cytokine involved in numerous cellular processes, including apoptosis and differentiation.18-22 Once TNF binds its cognate receptors, TNF receptor 1 (TNFR1) and TNFR2, it relays downstream signals through at least 2 distinct pathways. The first signal activates apoptosis through a cascade beginning with caspase 8 and leading to DNA cleavage and cell death.23 The second pathway stimulates translocation of the nuclear factor-κB (NF-κB) family of transcription factors into the nucleus, where they activate numerous target genes. Activation of NF-κB inhibits TNF-induced apoptosis and mediates many of the nonapoptotic functions of the TNF pathway. The TNF stimulatory signal results in NF-κB transcription factors (p65, p52, p50, c-Rel, and RelB), binding their response elements as homodimers or heterodimers. The p65-p50 heterodimer is the most potent transactivator of NF-κB target genes.24,25
The idea of an interaction between the TNF and ATRA signaling pathways has been supported by several recent reports. In lung cancer cells, ATRA increased the levels of TNFR,26 and in melanoma cells there was a synergistic induction of interleukin-8 (IL-8) by TNF and ATRA.27 ATRA alone did not have an effect on NF-κB binding in either model, and novel downstream effectors of ATRA and TNF were not presented, but both reports demonstrated that the pretreatment of cells with ATRA increased the level of NF-κB binding induced by TNF.
In NB4 cells, it was found that ATRA could induce the expression of TNF-related apoptosis-inducing ligand (TRAIL).28 It was observed that TRAIL could induce apoptosis of cultured APL, ATRA-resistant APL, and patient-derived APL blast cells in vitro. Furthermore, ATRA-induced differentiation coincided with an increased expression of TRAIL and the antiapoptotic genes BCL2A1 and cIAP. These genes are known to be regulated by the NF-κB family of transcription factors, indicating a potential interaction between NF-κB and ATRA pathways in APL cells and suggesting that the NF-κB pathway may represent an additional target for future APL therapies.
We now report that ATRA and TNF can synergistically induce differentiation of the APL cell line NB4 and the monoblastic cell line U937 without a concomitant increase in levels of apoptosis. We report several pieces of evidence that ATRA is working directly on TNF-responsive genes whose transcriptional control is mediated in part by TNF. We found that ATRA-promoted TNF induced NF-κB DNA binding to its cognate DNA-binding sequence and transcription within 2 hours of treatment. In contrast to previous reports, we found that ATRA alone could induce NF-κB binding after 48 hours of treatment. Furthermore, we report novel target genes that are synergistically induced by TNF and ATRA.
Materials and methods
Materials
RPMI 1640 and fetal bovine serum (FBS) were purchased from Invitrogen (Burlington, ON, Canada). All-trans retinoic acid was obtained from Sigma (Oakville, ON, Canada). Recombinant human tumor necrosis factor was obtained from Peprotech (Princeton, NJ). Nitroblue tetrazolium dye was supplied by Sigma. 32 P adenosine triphosphate (ATP) types α and γ were from Perkin Elmer. Anti–c-Rel, p65/RelA, RelB, p50, and p52 monoclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The monoclonal anti-IκB antibody was a generous gift of Dr J. Hiscott.
Cell culture
NB4 and U937 cells were maintained in RPMI 1640 medium supplemented with 10% FBS. Cell growth was quantified using a standard hemocytometer technique with trypan blue exclusion assay. For cell growth assays, cells in logarithmic growth were seeded at a density of 104 cells/mL. Cells were treated with 10-6 M ATRA and 10 ng/mL TNF unless otherwise specified.
cDNA microarray analyses
Cells were harvested using the Invitrogen FAST Track RNA preparation kit, and 2 μg was used for labeling reactions. Labeling reactions and microarray analyses were performed as previously described.29
Luciferase assays
Cells (5 × 106) were electroporated (350 V; 950 μF) with 10 μg luciferase reporter construct containing 2 Igk κB motifs30 (generous gift of Dr A Beg) and 5 μg β-Gal reporter construct used to normalize luciferase assays. Assays were performed as recommended by the manufacturer (Promega) using a Lumat LB 9507 luminometer (EG&G Berthold).
Electrophoretic mobility shift assays
Nuclear extracts were used for all mobility shift reactions. Cells were washed with phosphate-buffered saline (PBS), resuspended in 400 μL ice-cold hypotonic buffer 1 (10 mM Tris-Cl pH 7.8, 5 mM MgCl2, 10 mM KCl, 0.1 mM EDTA [ethylenediaminetetraacetic acid], 300 mM sucrose, 5 mM β-glycerol phosphate, 0.5 mM dithiothreitol, plus protease inhibitors), and allowed to swell for 10 minutes. Cells were lysed by adding 5 μL 10% NP-40 and vortexing. Nuclei were pelleted (1 minute at 1000 rpm), and the supernatant was removed. The recovered pellet was then resuspended in 50 μL hypertonic buffer 2 (20 mM Tris-Cl pH 7.8, 5 mM MgCl2, 320 mM KCl, .2 mM EDTA, 25% glycerol, 0.5 mM dithiothreitol, 5 mM β-glycerol phosphate, plus protease inhibitors) and was incubated on ice for 15 minutes. Nuclear debris was removed through centrifugation at 13 000 rpm for 10 minutes. Analysis of DNA binding was performed using a 32 P-end–labeled probe corresponding to a palindromic κB motif31 : upper strand, 5′-GATCCAACGGCAGGGGAATTCCCCTCTCCTTA-3′. For electrophoretic mobility shift assay (EMSA), 12 μg extract was used for each binding reaction. Binding reactions were carried out for 30 minutes at room temperature in a 20-μL binding reaction mixture containing 20 mM HEPES (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid), 1 mM dithiothreitol, 1 μg poly(dI.dC), 100 mM NaCl, 5% glycerol, and 1 mM EDTA. Protein-DNA complexes were separated on a 5% polyacrylamide gel at 200 V in 0.5 × TBE.
In vitro kinase assay
Kinase assays (p65) were performed with 10 μg whole cell lysate and full-length glutathione S-transferase–p65 (GST-p65) (a gift from Dr J. Stavnezer) at 30°C for 30 minutes as previously described.32
Differentiation assays
Cells to be used in nitroblue tetrazolium (NBT) reduction assays and for fluorescence-activated cell sorter (FACS) analysis of differentiation markers were seeded at 3 × 105 cells per well in 6-well plates. NBT assays were performed as previously described.33 Briefly, approximately 2 × 105 cells were placed in RPMI 1640 medium and mixed with an equal volume of NBT (1.0 μg/mL). The mixture was then rotated at 37°C for 40 minutes. The fraction of NBT-positive cells was determined using a hemocytometer. Cell surface expression of CD14, CD18, and CD11b by flow-assisted cell sorting was performed according to the antibody manufacturer's specifications (PharMingen) using the FACScan cytometer (Becton Dickinson Labware).
TNFR1 and TNFR2 expression
Cells were seeded at 3 × 105 cells per well in 6-well plates and were treated for the indicated time periods. Approximately 3 × 105 cells were collected and washed with 1 × PBS containing 0.5% bovine serum albumin (BSA) and 0.02% sodium azide. TNFR1 and TNFR2 were detected using fluorescein isothiocyanate (FITC)–labeled antibodies according to the manufacturer's (R&D Systems) instructions
Western blot analysis
Whole cell protein was used for all Western blot analyses. Cells were washed in PBS, after which they were resuspended in lysis buffer (50 mM Tris-Cl, 150 mM NaCl, 0.02% sodium azide, 1% NP-40, 0.5% sodium deoxycholate, plus protease inhibitors) for 30 minutes. Debris was removed by centrifugation at 13 000 rpm for 15 minutes. Equal amounts of protein were separated on 10% polyacrylamide gels containing 0.1% sodium dodecyl sulfate (SDS) and were transferred to nitrocellulose membranes (Bio-Rad Laboratories, Mississauga, ON, Canada). Membranes were probed with anti–NF-κB subunit antibodies at a dilution of 1:500 and anti-IκB at a dilution of 1:1000. Antibody binding was detected with the enhanced chemiluminescence (ECL; Amersham Pharmacia) system.
mRNA analysis
Total RNA was isolated using guanidinium thiocyanate extraction, as previously described.34 For Northern blotting, 10 μg RNA was electrophoresed on a 1% formaldehyde agarose gel and blotted onto Zeta probe transfer membranes. cDNA probes were labeled by random priming (Amersham Pharmacia). Hybridization and autoradiography were performed as previously described.35 Dif-2 and BCL-3 full-length cDNAs were kind gifts of Dr G. Schmitz and Dr J. W. Lee, respectively.
Reverse transcription was performed on 2.5 μg total RNA, after heating at 70°C for 10 minutes, with random hexamer primers. The reaction was carried out at 42°C for 45 minutes in the presence of 160 U Moloney murine leukemia virus reverse transcriptase (Invitrogen). cDNA was amplified using an A20 gene-specific primer (sense, 5′-CTGGACGCACTTCGCAGC-3′; antisense, 5′-TTGCCCCGTTTCAGTTGTAT-3′). Polymerase chain reaction (PCR) was performed as follows: 94°C for 5 minutes; 30 cycles at 94°C for 30 seconds, 57°C for 30 seconds, 72°C for 2 minutes; and incubation at 72°C for 10 minutes. CD70 primers were as follows: sense, 5′ TACGTATCCATCGTGATG 3′; antisense, 5′ GTTGGTGCAGAGTGTGTC 3′. PCR products were resolved on 2% agarose gels. Amplification of β-actin gene transcripts was performed to assess RNA integrity.
Results
Synergistic induction of differentiation by ATRA and TNF-α in NB4 and U937 cells
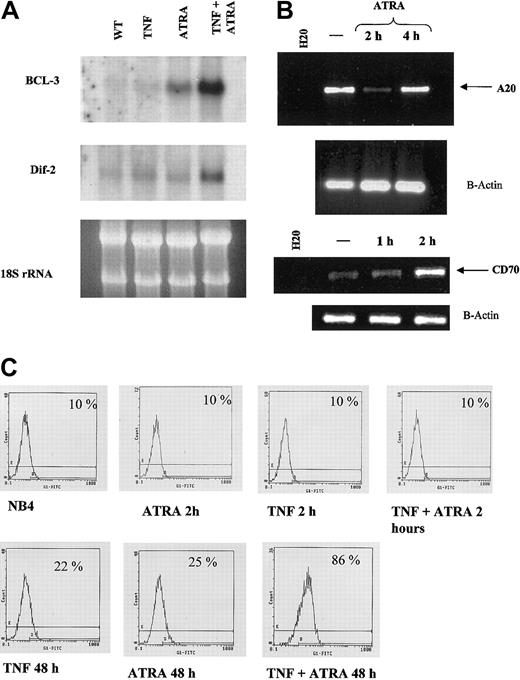
We used a cDNA microarray to find target genes of ATRA that may be regulated during NB4 cell differentiation (M.W. et al, manuscript in preparation). We found that ATRA increased mRNA expression of several genes reportedly involved in TNF signaling (Table 1). Only those genes having a 2.5-fold or greater change in mRNA levels on ATRA treatment are reported. Additionally, the expression of A20, a negative regulator of the TNF pathway, was inhibited by ATRA. Regulation by ATRA was confirmed for BCL-3, Dif-2, A20, and TNFR2 (Figure 6).
Synergistic induction of target genes by ATRA and TNF. (A) Northern blot of BCL-3 and Dif-2. Total RNA (10 μg) was loaded in each lane. Ribosomal RNA levels indicate equal loading. BCL-3 mRNA levels were seen to increase after 2 hours of RA treatment in NB4 cells, whereas neither RA nor TNF alone had a significant effect on Dif-2 mRNA levels. mRNA levels of both genes were strongly induced on combined treatment with TNF and RA. (B) RT-PCR analysis of A20 RNA levels in response to ATRA. RT-PCR confirmed array results in that A20 levels decreased after 2 hours of ATRA treatment. —indicates no treatment. (C) Cytofluorometric analysis of TNFR2 levels on NB4 cells in response to 72-hour treatment with ATRA and TNF. Synergism between ATRA and TNF could be observed at 48 and 72 (data not shown) hours after treatment, whereas little effect could be seen at 2 hours. Percentages represent the number of positive cells that show FITC-positive staining.
Synergistic induction of target genes by ATRA and TNF. (A) Northern blot of BCL-3 and Dif-2. Total RNA (10 μg) was loaded in each lane. Ribosomal RNA levels indicate equal loading. BCL-3 mRNA levels were seen to increase after 2 hours of RA treatment in NB4 cells, whereas neither RA nor TNF alone had a significant effect on Dif-2 mRNA levels. mRNA levels of both genes were strongly induced on combined treatment with TNF and RA. (B) RT-PCR analysis of A20 RNA levels in response to ATRA. RT-PCR confirmed array results in that A20 levels decreased after 2 hours of ATRA treatment. —indicates no treatment. (C) Cytofluorometric analysis of TNFR2 levels on NB4 cells in response to 72-hour treatment with ATRA and TNF. Synergism between ATRA and TNF could be observed at 48 and 72 (data not shown) hours after treatment, whereas little effect could be seen at 2 hours. Percentages represent the number of positive cells that show FITC-positive staining.
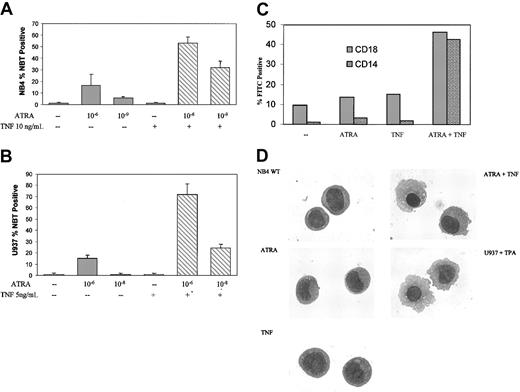
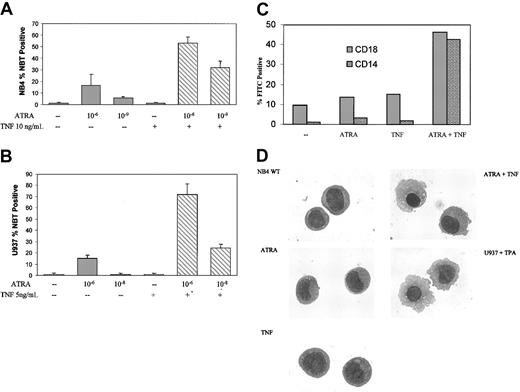
To investigate possible interactions between ATRA and TNF signaling in APL and other acute leukemias, we studied the maturation of NB4 and PML/RAR-negative U937 monoblastic cells in response to ATRA and TNF. ATRA induced a significant amount of terminal granulocytic differentiation of NB4 cells after 6-day treatment,36 as assessed by NBT reduction analyses, but this occurred to a lesser degree in U937 cells.37 Consistent with previous reports,36,37 only a moderate increase in terminal differentiation was observed in response to 10-6 M ATRA after 3 days (Figure 1A). TNF alone had little discernible effect on differentiation in either cell line. However, ATRA and TNF combined synergistically in both cell lines to induce NBT reduction within 3 days (Figure 1A-B). Synergism between ATRA and TNF could be observed at concentrations of ATRA as low as 10-9 M in NB4 cells and 10-8 M in U937 cells (Figure 1A-B). To extend the results from our NBT analyses, we looked at expression levels of cell surface markers associated with differentiation. CD18 and CD14, primarily markers of monocytic differentiation, were also significantly increased in response to TNF and ATRA (Figure 1C) after 3-day treatment.
ATRA- and TNF-induced differentiation. Differentiation of NB4 cells in response to 3-day exposure to ATRA and TNF. Results are representative of 3 experiments performed in triplicate. (A-B) Results of NBT reduction assay performed on NB4 and U937 cells treated with ATRA and TNF for 3 days. Synergism in the number of NBT-positive cells could be observed at concentrations of 10-9 M ATRA in NB4 and 10-8 M ATRA in U937 cells. Error bars represent SD. (C) Cytofluorometric analysis of surface marker expression. Percentages of cells expressing the monocyte-specific CD14 and the myeloid-specific CD18 cell surface markers of differentiation were determined using monoclonal FITC-labeled antibodies. Results are representative of 1 of 3 experiments performed in triplicate. Standard errors were 5% or less. —indicates no treatment. (D) Morphologic analysis of representative NB4 cells treated with TNF and ATRA for 3 days. Cells were stained with Giemsa-Wright and were viewed at × 100 magnification. U937 cells were treated with 25 nM TPA for 24 hours as a control for monocyte/macrophage morphologic features.
ATRA- and TNF-induced differentiation. Differentiation of NB4 cells in response to 3-day exposure to ATRA and TNF. Results are representative of 3 experiments performed in triplicate. (A-B) Results of NBT reduction assay performed on NB4 and U937 cells treated with ATRA and TNF for 3 days. Synergism in the number of NBT-positive cells could be observed at concentrations of 10-9 M ATRA in NB4 and 10-8 M ATRA in U937 cells. Error bars represent SD. (C) Cytofluorometric analysis of surface marker expression. Percentages of cells expressing the monocyte-specific CD14 and the myeloid-specific CD18 cell surface markers of differentiation were determined using monoclonal FITC-labeled antibodies. Results are representative of 1 of 3 experiments performed in triplicate. Standard errors were 5% or less. —indicates no treatment. (D) Morphologic analysis of representative NB4 cells treated with TNF and ATRA for 3 days. Cells were stained with Giemsa-Wright and were viewed at × 100 magnification. U937 cells were treated with 25 nM TPA for 24 hours as a control for monocyte/macrophage morphologic features.
Prior reports have shown that degradation of the PML/RAR oncoprotein correlates with ATRA-mediated differentiation of these cells. However, the combined effects of TNF and ATRA on the differentiation in NB4 or resistant subclone cells could not be explained because of an increased rate of PML/RAR degradation (data not shown). Previous work has demonstrated that NB4 cells have the potential to differentiate into granulocytes or monocytes.38 ATRA alone promotes the differentiation of NB4 cells along a granulocytic lineage. It has also been shown that ATRA has a modest effect on differentiation along a monocytic lineage in U937 cells,39 but it is less than the granulocytic response in APL. Increased levels of the monocyte-specific marker CD14 and morphologic analysis of NB4 cells treated with TNF and ATRA for 3 days suggested that the cells were maturing primarily into a monocytic phenotype (Figure 1C-D). As seen in Figure 1D, this treatment induced a morphology similar to that of U937 cells induced to differentiate along a monocyte/macrophage pathway, as previously described.40 TNF alone had little discernible effect on cell morphology (Figure 1D).
TNF and ATRA have proapoptotic properties in some cells. When fully differentiated, myeloid cells have a limited lifespan, with the cells terminated through apoptosis. Therefore, we investigated whether there was an increase in apoptosis associated with ATRA- and TNF-induced differentiation. We observed no increase in the number of apoptotic NB4 or U937 cells in response to the combination of 10 ng/mL TNF and 10-6 M ATRA at 24, 48, or 72 hours after treatment, compared with ATRA or TNF alone, as determined by annexin V staining (data not shown). This suggests that TNF activation of a proapoptotic caspase cascade in these cells did not play a role in the differentiation response.
These results demonstrate that the combination of TNF and ATRA induced cells to differentiate at lower concentrations of ATRA and modulated the differentiation program toward a monocytic phenotype without changing the propensity to undergo apoptosis. Interestingly, we also found that TNF and ATRA could overcome ATRA resistance in cells, with ATRA resistance acquired in vitro and patient-derived, ATRA-resistant APL cells (M.W. et al, manuscript in preparation).
Induction of NF-κB binding by ATRA
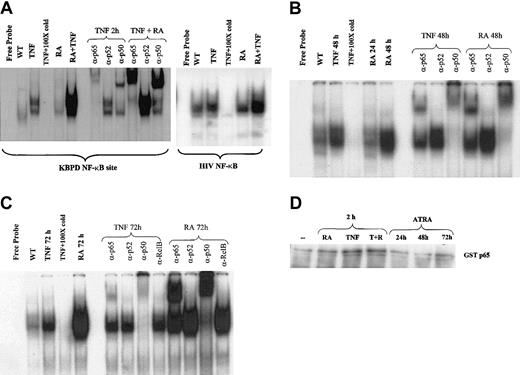
To further explore the mechanisms by which ATRA and TNF pathways might interact, we performed gel shift analyses to determine whether ATRA influenced the binding of NF-κB family members to their DNA response element. Recent data have indicated that the activation of NF-κB in response to proinflammatory stimuli results in differential gene expression of NF-κB target genes at early and late time points.41,42 We therefore examined the effects of ATRA on NF-κB binding at intervals from 2 hours to 3 days. Using nuclear extracts from NB4 cells, we found that ATRA enhanced the TNF-induced binding of NF-κB members to their response elements after 2 hours of cotreatment in NB4 cells (Figure 2A). ATRA alone could induce binding to the same response elements after 48 and 72 hours of treatment (Figure 2B-C). Supershift experiments revealed that ATRA-induced–bound complexes were composed primarily of p65-p50 heterodimers at all time points studied (Figure 2A-C). In contrast, extracts from cells treated with TNF showed a gradual shift from complexes composed primarily of p65-p50 heterodimers at 2 hours to complexes composed mainly of p50 homodimers at 72 hours after treatment (Figure 2A, C). This is consistent with a previous report demonstrating that stimulation of the NF-κB pathway resulted in a shift from cRel-p65 heterodimers to p50 homodimers at late time points.41 This may result in the differential expression of NF-κB target genes. Alternatively, because p50 homodimers are considered to have low transcriptional activity, this may be a mechanism whereby the NF-κB pathway can shut itself off. The continued detection of p65 in the binding complex induced by ATRA at 72 hours suggests that ATRA may prolong NF-κB–mediated transcription. Western blot analysis was performed to determine whether the increased level of NF-κB binding in response to ATRA resulted from increased levels of cellular NF-κB protein (Figure 3). As shown in Figure 3, levels of the NF-κB proteins p65, p50, and p52 were not significantly changed with ATRA treatment, but RelB levels were decreased at 72 hours. The NF-κB subunit c-Rel was not found in significant quantities in NB4 cells (data not shown).
ATRA regulation of NF-κB binding. EMSAs and supershift assays showing binding to a palindromic NF-κB response element using nuclear extracts from NB4 cells treated with ATRA and TNF for various times. (A) ATRA is seen to heighten TNF-induced NF-κB binding after 2 hours and to increase these complexes alone after 48 and 72 hours (B-C). Supershift assays indicated that complexes induced by ATRA were composed primarily of p65 and p50 proteins. (D) In vitro kinase assay using GST-p65 as a substrate. While a slight increase in p65 phosphorylation is observed in response to TNF, no change is seen in response to ATRA.
ATRA regulation of NF-κB binding. EMSAs and supershift assays showing binding to a palindromic NF-κB response element using nuclear extracts from NB4 cells treated with ATRA and TNF for various times. (A) ATRA is seen to heighten TNF-induced NF-κB binding after 2 hours and to increase these complexes alone after 48 and 72 hours (B-C). Supershift assays indicated that complexes induced by ATRA were composed primarily of p65 and p50 proteins. (D) In vitro kinase assay using GST-p65 as a substrate. While a slight increase in p65 phosphorylation is observed in response to TNF, no change is seen in response to ATRA.
NF-κB protein levels in NB4 cells. Western blot of NF-κB protein levels in NB4 cells in response to RA demonstrates that increased NF-κB binding in response to ATRA is not attributed to increased NF-κB family member levels. NB4 whole cell extract (30 μg) was analyzed for NF-κB subunit levels in response to ATRA for the time periods indicated. Antibodies were used at a dilution of 1:500.
NF-κB protein levels in NB4 cells. Western blot of NF-κB protein levels in NB4 cells in response to RA demonstrates that increased NF-κB binding in response to ATRA is not attributed to increased NF-κB family member levels. NB4 whole cell extract (30 μg) was analyzed for NF-κB subunit levels in response to ATRA for the time periods indicated. Antibodies were used at a dilution of 1:500.
Previous work has demonstrated that phosphorylation and acetylation of p65 can play a role in its transcriptional activity.32,43,44 Because ATRA is a known activator of several kinase cascades, we explored the possibility that the phosphorylation status of p65 may play a role in ATRA enhancement of TNF-induced NF-κB binding at 2 hours. In vitro kinase assays using recombinant GST-p65 (Figure 2D) demonstrated a small but consistent increase in p65 phosphorylation in response to TNF, but we could not conclude that ATRA enhanced this effect of TNF. The increased induction of NF-κB by 2 hours of treatment with ATRA and TNF may be explained by the stimulation of signaling pathways by cytokines that are early targets of ATRA (Figure 6B and “Discussion”). Other cytokines, such as interferon,45 have been reported to enhance TNF activation of NF-κB, and combinations of cytokines, such as interferon and interleukin, also activate NF-κB in a synergistic manner.46
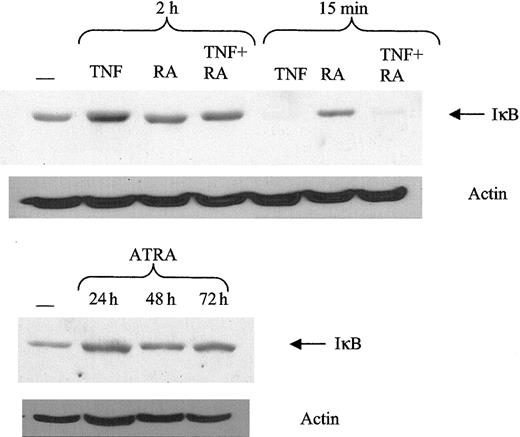
NF-κB subunits are normally found sequestered in the cytoplasm by the IκB complex.47 TNF can induce phosphorylation of IκB, making it a target of the ubiquitin proteasome pathway. Once IκB is degraded, NF-κB subunits are free to move into the nucleus to bind DNA. Therefore, it is possible that ATRA might mediate it effects on NF-κB by stimulating IκB degradation. Consistent with previous results, TNF can induce a significant decrease in IκB levels after 15 minutes (Figure 4). However, ATRA did not decrease IκB levels at any time point studied, nor did the addition of ATRA to TNF affect its response to TNF. These data indicate that the increase in NF-κB binding is not mediated by effects of ATRA on IκB.
Response of IκB to ATRA and TNF. ATRA does not promote IκB degradation. Protein levels of IκB were determined using Western blotting of 30 μg whole cell extract. Typical IκB degradation was observed in response to TNF at 15 minutes after treatment.
Response of IκB to ATRA and TNF. ATRA does not promote IκB degradation. Protein levels of IκB were determined using Western blotting of 30 μg whole cell extract. Typical IκB degradation was observed in response to TNF at 15 minutes after treatment.
Synergistic induction of transcription by ATRA and TNF
The increased binding of NF-κB in response to ATRA indicated that ATRA may be able to regulate transcription through NF-κB response elements. To further investigate this possibility, we transfected NB4 cells with a luciferase construct containing a NF-κB response element and measured the level of luciferase activity in response to RA and TNF. Although TNF had only a modest 1.8-fold induction of transcription in this model, we found that ATRA alone had a strong stimulatory effect on the level of luciferase activity after 48 hours (Figure 5). When combined with TNF, there was an increase in the induction of luciferase activity from 10.8- to 24-fold compared with untreated cells (Figure 5).
ATRA promotes transcription through NF-κB response element. NB4 cells were electroporated with Igk κB constructs and treated for 48 hours. The combination of TNF and ATRA gave a stronger induction of luciferase than did either agent alone. Results shown are representative of 3 experiments performed in triplicate. Standard errors are less than 2%. Numbers above the bars indicate fold induction.
ATRA promotes transcription through NF-κB response element. NB4 cells were electroporated with Igk κB constructs and treated for 48 hours. The combination of TNF and ATRA gave a stronger induction of luciferase than did either agent alone. Results shown are representative of 3 experiments performed in triplicate. Standard errors are less than 2%. Numbers above the bars indicate fold induction.
Our differentiation data showed that ATRA had a modest influence on differentiation as a single agent after 3 days, whereas TNF by itself had little effect. Therefore, we considered it possible that in addition to the observed enhancement of ATRA on the TNF pathway, TNF might act on the RAR signaling pathway. Using reverse transcription-PCR (RT-PCR), we found that ATRA could increase levels of RARβ and C/EBP epsilon in NB4 cells as previously reported. However, we did not observe any further increase in mRNA levels of RARβ or C/EBPϵ genes when TNF was combined with ATRA. Additionally, TNF had no effect on ATRA-driven transcription of a CAT construct containing DR5 RARE (data not shown). These data suggest that the interaction between ATRA and TNF is mediated primarily by ATRA acting on the TNF pathway as opposed to the converse.
Therefore, we studied the effects of ATRA on the transcription of endogenous genes containing NF-κB response elements. Our array data identified several genes that are regulated by ATRA at early and late time points. We used Northern blot analysis to investigate levels of BCL-3 and Dif-2 mRNA in response to TNF and RA in NB4 cells. BCL-3 is an IκB-like protein that enhances and represses transcription by NF-κB p50 and p52 homodimers.48-50 Dif-2 is a lipopolysaccharide-inducible gene isolated because of its regulation during monocyte differentiation.51 ATRA was seen to have a stronger stimulatory effect on BCL-3 mRNA levels after 2-hour treatment than on Dif-2 levels (Figure 6A). TNF increased Dif-2 levels slightly after 2 hours but had no effect on BCL-3 levels (Figure 6A). However, treatment of NB4 cells concurrently with TNF and ATRA for 2 hours substantially increased the expression of BCL-3 and Dif-2 mRNA (Figure 4A). RT-PCR analysis also confirmed our array data demonstrating that ATRA could increase CD70 levels at early time periods. The CD70 pathway is a known stimulator of NF-κB,52 and the interplay between this (and other) signaling pathways stimulated by RA contributes to the enhancement of TNF-induced NF-κB binding observed at 2 hours after treatment. Our arrays identified one gene as down-regulated. RT-PCR analysis confirmed that A20, a TNF-inducible gene that inhibits NF-κB activity,53 was significantly down-regulated by 2 hours of ATRA (Figure 6B) but returned to near basal levels by 4 hours of ATRA exposure. Thus, the regulation of A20 provides another potential means whereby ATRA can enhance NF-κB activity at early time periods.
Our EMSA and luciferase assay results indicated that the ATRA and TNF pathways may also interact on TNF-responsive promoters at late time points (Figures 2B-C, 5). Previous work has shown that TNFR1 and TNFR2 can be regulated by ATRA at late time points.26 Promoter analysis of the 2 receptors has shown that the type 2 receptor promoter contains an NF-κB response element. FACS analysis revealed that there was little regulation of TNFRI by either TNF or RA at 48 and 72 hours after treatment (data not shown). However, TNFR2 expression levels were increased at 48 and 72 hours after treatment (Figure 6C and data not shown). A combination of the 2 agents resulted in a synergistic induction of TNFR2 at both 48 and 72 hours but not at 2 hours (Figure 6C and data not shown).
Discussion
The ability of ATRA to induce the differentiation of APL cells is well documented. However, the target genes of ATRA that mediate differentiation remain unclear. Using a cDNA microarray, we found that a number of genes involved in the TNF signaling pathway are regulated by ATRA in NB4 cells. This suggested that modulation of the TNF pathway could be important for the prodifferentiation effects of ATRA. TNF and ATRA have been reported to inhibit clonal growth of patient-derived myeloid leukemia cells and to influence myeloid differentiation, but mechanistic data have been lacking.54 We found that costimulation with TNF and ATRA resulted in a more rapid and synergistic induction of differentiation than with either agent alone. Differentiation could be observed after a period of only 3 days at concentrations of RA as low as 10-9 M in NB4 and 10-8 M in the PML/RAR-negative cell line U937. These concentrations are substantially lower than those required to promote differentiation by ATRA alone after a period of 6 days.36
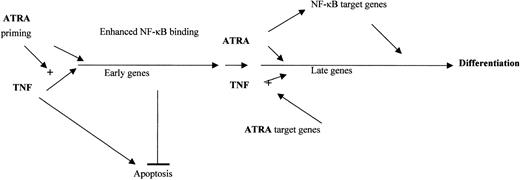
Synergistic effects between ATRA and various other agents on the differentiation of NB4 cells have been reported. These include interferons, GM-CSF, and cyclic adenosine monophosphate (cAMP).8,12,55 The mechanisms that underlie the synergy between these agents remain unclear. Our data show that the strong prodifferentiation effects of TNF and ATRA differ from those of ATRA alone. Although TNF is able to promote the differentiation of some myeloid cells along a monocytic pathway, TNF alone had little effect on the maturation of NB4 or U937 cells within 3 days. The dramatic increase in CD14 expression in response to both agents suggested that ATRA can activate the promonocytic actions of TNF, consistent with our findings that ATRA influenced the activity of the TNF pathway. However, we observed no effect of TNF on the RAR pathway. This supports the idea that in the presence of other differentiation regulatory factors, RA may play an important regulatory role56,57 and that induction of alternative differentiation programs may complement ATRA as a therapeutic agent. A hypothetical model of the interaction between TNF and ATRA is presented in Figure 7.
Model of ATRA and TNF interaction. ATRA primes NB4 cells for TNF by increasing the production of target genes involved in TNF signaling at early and late time points. ATRA and TNF may combine at early time points to promote the production of genes that have the dual role of inhibiting TNF-mediated apoptosis and promoting differentiation. New target genes are regulated at late time points and at the induction of NF-κB binding by ATRA. The interplay between ATRA-mediated enhanced TNF signaling and synergistic induction of common target genes by TNF and ATRA lead to NB4 differentiation.
Model of ATRA and TNF interaction. ATRA primes NB4 cells for TNF by increasing the production of target genes involved in TNF signaling at early and late time points. ATRA and TNF may combine at early time points to promote the production of genes that have the dual role of inhibiting TNF-mediated apoptosis and promoting differentiation. New target genes are regulated at late time points and at the induction of NF-κB binding by ATRA. The interplay between ATRA-mediated enhanced TNF signaling and synergistic induction of common target genes by TNF and ATRA lead to NB4 differentiation.
To explain the synergistic effects of ATRA with other agents on differentiation, previous studies have described a so-called priming model of ATRA action.58,59 This model proposes that ATRA makes cells competent for differentiation triggered by an agent activating a second pathway. Based on our data, we propose that RA has a priming effect on myeloid cells whereby it potentiates the differentiating properties of other agents, as has been previously reported.58,59 We present here novel data that begin to explain the mechanisms whereby ATRA primes the cells for TNF-driven differentiation.
First, ATRA induced the expression of genes involved in TNF signaling. In addition to its ability to regulate genes involved in TNF signaling by itself, ATRA could also act synergistically with TNF to regulate common target genes. At 2 hours after treatment, we found that the 2 agents could synergistically induce mRNA levels of the NF-κB target genes BCL-3 and Dif-2. BCL-3 can act as a coactivator of NF-κB transcription through its interaction with p50 or p52 homodimers bound to DNA.48-50 It seems plausible that the regulation of BCL-3 represents another level through which RA can modulate transcription through NF-κB sites. The functionality of the Dif-2 proteins is unclear. It is highly homologous to the IEX-1 protein that acts as a regulator of apoptosis.60,61 We are performing analyses of the Dif-2 and BCL-3 promoter regions to better understand how the TNF and ATRA pathways converge on a transcriptional level. It is conceivable that the increasing number of antiapoptotic genes found to be targets of ATRA62-64 act in concert to promote differentiation and to delay apoptosis. The observation that ATRA can promote TNF-induced NF-κB binding at 2 hours in vitro may also help explain some of the observed gene regulation by the 2 agents in vivo. Although we could not find an ATRA-mediated effect on p65 phosphorylation, it is possible that the early induction by ATRA of cytokines that activate NF-κB, such as CD70, could mediate synergistic DNA binding of NF-κB. However, other possibilities, such as p65 acetylation, must be investigated.
Our finding that ATRA down-regulates A20, a negative regulator of the antiapoptotic NF-κB signaling arm of the TNF pathway, is consistent with a role of ATRA in tipping the balance of signals in the cell toward differentiation as opposed to apoptosis. It is possible that at early time points, the antiapoptotic genes up-regulated by ATRA and the down-regulated proapoptotic genes are important in offsetting any proapoptotic properties of ATRA or other extracellular signals and may be important in allowing the cells to proceed down the path to differentiation, especially in the presence of other differentiation inducers.
Our results indicate that costimulation with TNF and ATRA results in the increased expression of common target genes at late and early time periods. At 48 and 72 hours after treatment, we found that ATRA or TNF alone could slightly increase TNFR2 expression. When combined, they produced a much greater increase in cellular levels of TNFR2 without an accompanying increase in TNFR1 levels. This is consistent with the suggestion that TNFR2 may be the major effector of the NF-κB pathway, whereas TNFR1 may play a more proapoptotic role.65,66 Therefore, the up-regulation of TNFR2 may be part of an autoregulatory loop to amplify the effects of TNF on NF-κB.
Second, at later time points, we found that ATRA alone could stimulate NF-κB binding. In contrast to TNF, which stimulated binding of complexes composed primarily of p50 homodimers at these time points, ATRA stimulated binding of the transcriptionally active p50-p65 heterodimer complex. This increase in NF-κB binding may be attributed to an ATRA-mediated increase in TNFR2 levels or an increased release of proinflammatory cytokines such as interferons and TNF-α. NF-κB is the downstream mediator of many of the cellular effects of TNF. One of the functions of NF-κB is to turn on the transcription of genes that counter the proapoptotic effects of TNF. This again supports the idea that some antiapoptotic genes may play a role in differentiation.28,63,64,67,68 It is possible that by stimulating the production of NF-κB target genes, RA may not only block the proapoptotic effects of TNF but may also promote TNF-induced monocytic differentiation. It is unclear whether NF-κB activation is vital for the differentiation effects of TNF and ATRA. Although inhibitors of NF-κB could be useful in uncovering its role, we have found NB4 cells to be acutely sensitive to apoptosis in response to NF-κB inhibition (data not shown).
In conclusion, we found that there is significant interaction between the ATRA and TNF signaling pathways leading to differentiation. This interaction involves increased NF-κB activity followed by increased levels of TNFR2 and a combinatorial stimulation of gene transcription that neither agent alone can significantly induce. Our data support the idea that one of the roles played by ATRA in differentiation may be to prime cells so that they are more susceptible to the prodifferentiation effects of other signaling molecules. Recently, recombinant TNF and vectors carrying the TNF-α gene have been re-examined as anticancer agents not only in vitro but also in clinical trials.69-74 The strong combined effects of ATRA and TNF in NB4, U937, and lung cancer cells26 suggest that targeting the TNF pathway with ATRA may be useful for treating a variety of cancers.
Prepublished online as Blood First Edition Paper, February 13, 2003; DOI 10.1182/blood-2002-09-2725.
Supported by funds from the Canadian Institutes of Health Research (CIHR) and the Leukemia Research Fund of Canada. M.W. is supported by a student grant from the CIHR, and C.R. is supported by a student grant from the US Army Breast Cancer Research Program. W.H.M. is an Investigator of the CIHR.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mariantonietta Ricci for excellent discussions and advice.