Abstract
Retroviral vectors are the most efficient means of stable gene delivery to hematopoietic stem cells (HSCs). However, transgene expression from retroviral vectors is frequently subject to the negative influence of chromosomal sequences flanking the site of integration. Toward the development of autonomous transgene expression cassettes, we inserted the human interferon-β scaffold attachment region (IFN-SAR) and the chicken β-globin 5′ DNase I hypersensitive site 4 (5′HS4) insulator both separately and together into a series of self-inactivating (SIN) lentiviral vector backbones. Transduced cells of the human CD34+ hematopoietic progenitor line KG1a—pooled populations as well as individual clones harboring single integrants—were analyzed for reporter expression during culture periods of up to 4 months. Vectors carrying both the 5′HS4 insulator and the IFN-SAR consistently outperformed control vectors without inserts as well as vectors carrying either element alone. The performance of a set of vectors containing the murine stem cell virus long terminal repeat as an internal promoter was subsequently assessed during in vitro monocytic differentiation of transduced primary human CD34+ cord blood cells. Similar to what was observed in the KG1a hematopoietic progenitor cell model, optimal reporter expression in primary monocytes was obtained with the vector bearing both regulatory elements. These findings indicate that the 5′HS4/IFN-SAR combination is particularly effective at maintaining open chromatin domains permissive for high-level transgene expression at early and late stages of hematopoietic development, and thus could be of utility in HSC-directed retroviral vector–mediated gene transfer applications.
Introduction
Sustained expression of clinically relevant transgenes in the progeny of hematopoietic stem cells (HSCs) would provide novel treatments for a wide range of inherited blood diseases.1,2 Replication-defective recombinant oncoretroviral vectors derived from Moloney murine leukemia virus have been the most widely used vehicles for HSC gene transfer because of their capacity to introduce and stably express their genomes in mammalian cells.3 A limitation of oncoretroviral vectors, however, is that cell division is required for proviral integration into the host genome.4 By comparison, lentiviruses such as human immunodeficiency virus type 1 (HIV-1) have evolved a nuclear import machinery that allows them to infect nondividing as well as dividing cells.5 Thus, the recent development of HIV-1–based lentiviral vectors3,6 has provided the opportunity to efficiently deliver transgenes into minimally cultured HSCs, better preserving the long-term repopulating potential of the graft.7-13 In addition, as a consequence of the self-inactivating (SIN) lentiviral vector design, promoter sequences that are broadly or lineage-specifically expressed within the hematopoietic compartment can be readily incorporated into the vector backbones.13-18
Regardless of these advances, transgenes delivered by lentiviral vectors are still susceptible to chromosomal position effects that result in variable expression and frequently lead to transcriptional silencing.13,16,19,20 In this regard, recent work of others has demonstrated that insulator sequences can increase the probability of transgene expression when inserted into oncoretroviral vectors.21-23 Insulators are genetic elements near chromatin domain boundaries that function as barriers against repressive effects of neighboring inactive chromatin or to prevent inappropriate activation of a promoter by nearby heterologous enhancers.24 The best studied vertebrate element is a 1.2-kilobase (kb) fragment containing the chicken β-globin 5′ DNase I hypersensitive site 4 (5′HS4) that is constitutively present in all tissues.25-30 Extensive characterization by Felsenfeld and colleagues has shown that the 5′HS4 element can protect against position effects and also provide enhancer blocking function—satisfying both criteria of an insulator—and that these are separable properties.26,29,30 It is noteworthy that although tandem flanking copies of the 5′HS4 element are required for full insulating activity.25 The 5′HS4 element has also been reported to be incapable of preventing transcriptional silencing of oncoretroviral vectors in murine embryonic stem cells.21 Since there was no evidence of DNase I hypersensitivity, the 5′HS4 element did not appear to be acting as a domain boundary at the silenced integration sites in this instance. These observations indicate that the 5′HS4 element does not exhibit dominant chromatin barrier activity under all circumstances.
Other elements exist in the eukaryotic genome that are believed to contribute toward delimiting the topologic borders between chromatin domains.31 Termed scaffold or matrix attachment regions (S/MARs), these elements have been proposed to anchor chromatin to a skeleton of protein cross-ties called the nuclear scaffold (in metaphase) or nuclear matrix (in interphase), forming chromosomal loops.32-40 It has been proposed that S/MARs may function by bringing enhancers and other distal regulatory elements in close proximity to their corresponding promoter sequences. While insulator function has been attributed to some S/MARs,35 this does not appear to be a general property of these elements. Conversely, the 1.2-kb 5′HS4 element does not have S/MAR activity.26 Whatever the mechanism of action, 2 groups have shown that configuration-dependent insertion of the SAR believed to define the upstream border of the human interferon-β locus (IFN-SAR) into oncoretroviral vectors resulted in improved transgene expression.36-39
Given the ability of the 5′HS4 insulator and the IFN-SAR to separately reduce the sensitivity of oncoretroviral vectors to position effects and enhance transgene expression, we decided to comparatively examine the effect of these elements in the context of lentiviral vectors. Furthermore, in view of their complementary properties and structural features—that is, the core element of the 5′HS4 insulator is GC-rich with a high density of cytidineguanosine (CpG) dinucleotides (reminiscent of a CpG island), whereas S/MARs are AT-rich sequences26,34 —plus the fact that lentiviral vectors can readily accommodate large DNA inserts, we were interested in evaluating the utility of combining these elements. Our findings in the human KG1a hematopoietic progenitor line and in primary human CD34+ cord blood cells demonstrated that position-effect protection of lentiviral vector–mediated transgene expression provided by combinatorial association of the 5′HS4 insulator and the IFN-SAR was superior to that conferred by either element alone.
Materials and methods
Construction of lentiviral vectors
The SIN lentiviral transfer vectors used in this study were derived from the SIN-MU3-GW, SIN-EF-G, and SIN-CAG-G vectors described previously14 and contain, respectively, the U3 promoter region of the murine stem cell virus long terminal repeat (MSCV LTR; abbreviated MU3), the human elongation factor 1α (EF1α) promoter, and the composite CAG promoter (consisting of the human cytomegalovirus immediate early enhancer linked to chicken β-actin promoter sequences). The SIN-MU3-GW and SIN-EF-G vectors were further modified to contain the central polypurine tract (cPPT) and central termination sequence of HIV-1, which specify the creation of a plus strand overlap during reverse transcription referred to as the central DNA flap.41 A 178–base pair (bp) DNA fragment encompassing these sequences was amplified by polymerase chain reaction (PCR) from the pCMVΔR8.91 plasmid (provided by D. Trono, University of Geneva, Geneva, Switzerland). The PCR product was digested with NarI (provided by the forward primer, 5′-AGTCATGGCGCCACAAATGGCAGTATTCATCC-3′) and ClaI (provided by the reverse primer, 5′-ATTTATATCGATCCAAAGTGGATCTCTGCTGTC-3′) and subcloned into the vectors at unique ClaI sites upstream of the respective promoters in SIN-MU3-GW and SIN-EF-G, giving rise to the SINF-MU3-GW and SINF-EF-G vectors (F denotes central DNA flap sequences).
The 5′HS4-containing vectors were constructed by inserting a 1.2-kb SacI-SspI fragment containing the chicken β-globin 5′HS4 insulator in direct orientation into the 3′ LTRs of the vectors. The 5′HS4 insulator was obtained from plasmid pJC13-1, which contains 4 copies of the element25,26 (provided by G. Felsenfeld, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD). Following digestion with XbaI, the 1.2-kb 5′HS4-containing fragment was isolated from pJC13-1, blunted, and subcloned into the SmaI site in plasmid pBSP-3′LTR. The SmaI site was generated by insertion of a polylinker (upper strand: 5′-ATCCCGGGTCGCGA-3, lower strand: 5′-TAGGGCCCAGCGCT-3′) into the 3′ HIV-1 LTR of plasmid pBSP-3′LTR, replacing the EcoRV-PvuII fragment of the U3 region. The 3′ SIN LTR fragment containing the 5′HS4 element was then excised by XhoI-XbaI digestion and used to replace the corresponding sequences in SINF-MU3-GW, SINF-EF-G, and SIN-CAG-G, generating the SINF-MU3-GW-I, SINF-EF-G-I, and SIN-CAG-G-I vectors, respectively (I denotes the 5′HS4 insulator).
The IFN-SAR–containing vectors were constructed by inserting a 0.8-kb fragment of the human IFN-SAR from plasmid pCL36 (provided by J. Bode, Gesellschaft für Biotechnologische Forschung mbH, Braunschweig, Germany) in reverse orientation into the 3′ untranslated region of the enhanced green fluorescent protein (GFP) reporter, immediately upstream of the 3′ LTR. Following HindIII and BamHI digestion, the 0.8-kb IFN-SAR–containing fragment was isolated from pCL, and subcloned into the corresponding sites in the polylinker of the plasmid MINV.42 The IFN-SAR was then removed as an XhoI-SalI fragment and inserted into SINF-MU3-GW, SINF-EF-G, and SIN-CAG-G at unique XhoI sites to generate the SINF-MU3-GW-S, SINF-EF-G-S, and SIN-CAG-G-S vectors, respectively (S denotes the IFN-SAR).
The 0.8-kb XhoI-SalI IFN-SAR fragment was also subcloned into the plasmid pBSP-3′LTR-I at an XhoI site upstream of the 3′ LTR to generate pBSP-S-3′LTR-I. This plasmid was subsequently digested with XhoI and XbaI to obtain the S-3′LTR-I–containing fragment, which was used to replace the XhoI-XbaI fragment of SINF-EF-G, SINF-MU3-GW, and SIN-CAG-G, generating the SINF-EF-G-SI, SINF-MU3-GW-SI, and SIN-CAG-G-SI vectors, respectively (SI denotes the IFN-SAR/5′HS4 insulator combination).
Generation of lentiviral vector particles
Human embryonic kidney 293T cells43 (obtained from M. Eiden, National Institute of Mental Health, National Institutes of Health, Bethesda, MD) and human fibrosarcoma HT1080 cells44 (American Type Culture Collection [ATCC], CCL-121) were propagated in Dulbecco modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; BioWhittaker, Walkersville, MD), l-glutamine (2 mM; Invitrogen), penicillin (50 IU/mL; Invitrogen), and streptomycin (50 μg/mL; Invitrogen) at 37°C in a humidified atmosphere containing 5% CO2. Vesicular stomatitis virus (VSV)–G glycoprotein–pseudotyped lentiviral vector particles were produced by transient transfection of 293T cells, and high-titer vector stocks were prepared by ultracentrifugation (45 000g, 90 minutes), titered on HT1080 cells, and assayed for the presence of replication-competent virus as described previously.14,45
Transduction and fluorescence-activated cell sorting (FACS) analysis of KG1a cells
The human CD34+ hematopoietic progenitor line KG1a46 (ATCC, CCL-246.1) was cultured in Iscoves modified Dulbecco medium (IMDM; Invitrogen) plus 10% heat-inactivated FBS, l-glutamine (2 mM), penicillin (50 IU/mL), and streptomycin (50 μg/mL) at 37°C in a humidified atmosphere containing 5% CO2. KG1a cells were stably transduced by spinoculation at a multiplicity of infection (MOI) of 1 as described.14,45 Control (mock-transduced) and vector-transduced KG1a cells were pelleted, washed, and resuspended in phosphate-buffered saline plus 2% FBS. GFP+ cells were sorted as bulk populations or as single cells deposited into 96-well plates using a FACSVantage SE (BD Biosciences, San Jose, CA) equipped with CloneCyt Plus and an Innova 70C-Spectrum mixed argon-krypton ion laser (Coherent, Santa Clara, CA) tuned to 488 nm. Viable cells were gated by a combination of forward and orthogonal light scatter. The GFP fluorescent signal was detected in the FL1 channel with a 530/30 bandpass filter. Subsequent FACS analyses were performed on a BD LSR analyzer (BD Biosciences) utilizing similar excitation and detection strategies.14,42 Homogeneity of GFP transgene expression was indicated by the value of the percent coefficient of variation (CV) of the fluorescent signal, which is the standard deviation divided by the mean channel number of the maximum signal multiplied by 100.
Transduction and in vitro monocytic differentiation of CD34+ cord blood cells
Human umbilical cord blood samples were obtained after informed consent in conformity with an institutionally approved protocol. Mononuclear cells were isolated by density gradient centrifugation on Ficoll-Paque (Amersham Pharmacia Biotech, Piscataway, NJ). CD34+ cells were purified from the mononuclear cells by super paramagnetic microbead selection using a Miltenyi Biotec varioMACS CD34 progenitor cell isolation kit (Auburn, CA), according to the manufacturer's instructions. The CD34+ cells were cultured in 12-well tissue culture plates coated with CH-296 recombinant fibronectin fragment (2 μg/cm2; Takara Shuzo, Shiga, Japan) at a density of 1 × 106 cells per well. The cells were prestimulated for 48 hours in IMDM containing 10% FBS, 100 μM β-mercaptoethanol (Sigma, St Louis, MO) plus stem cell factor (100 ng/mL), Flt-3 ligand (100 ng/mL), and thrombopoietin (20 ng/mL) (all cytokines were purchased from Peprotech). The cells were transduced for 24 hours with lentiviral vector particles (2 × 106 transducing units [TU]/mL; MOI, 2) in the presence of protamine sulfate (4 μg/mL). Fresh medium was added and the cells were cultured for an additional 72 hours. The cells were then harvested with cell dissociation buffer (Invitrogen), washed, and resuspended in phosphate-buffered saline plus 2% FBS, and CD34+GFP+ cells were sorted. To evaluate and compare the levels of GFP transgene expression in CD14+ monocytes derived from transduced CD34+ cells, the cells were cultured for 7 weeks in IMDM medium containing 10% heat-inactivated FBS, L-glutamine (2 mM), and penicillin (50 IU/mL), plus interleukin-3 (IL-3; 20 ng/mL), IL-6 (20 ng/mL), and granulocyte-macrophage colony-stimulating factor (GM-CSF; 20 ng/mL). All cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2. Staining with anti-CD34 and anti-CD14 monoclonal antibodies conjugated to allophycocyanin (purchased from BD Biosciences Pharmingen, San Diego, CA) and FACS analysis for GFP expression were performed as described.14,42
Southern blot analysis
High-molecular-weight genomic DNAs (10 μg) were digested with BamHI (for pools of stably transduced cells) or EcoRI (for individual clones) and used for Southern blot analysis. A 0.8-kb fragment of pEGFP was used as a GFP-specific probe and a 1.3-kb EcoRI-AccI fragment of pCΔj-989 was used to detect the endogenous human bcl-2 gene as described.14
Results
Construction of lentiviral vectors containing the 5′HS4 insulator and/or the IFN-SAR
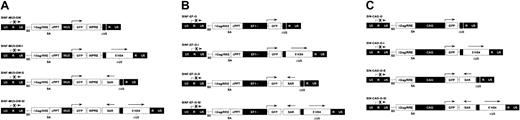
The series of lentiviral vectors used in this study were based on SIN lentiviral vector backbones developed previously that contain internal promoters capable of directing high levels of transgene expression in human hematopoietic cells.14 The promoters included a 302-bp fragment of the U3 region and part of the R region of the MSCV LTR minus a “negative control region” coincident with a binding site for the transcription factor YY1 (MU3) (Figure 1A);47 a 1.2-kb fragment of the human EF1α gene promoter containing a 943-bp intron (Figure 1B); and a 1.8-kb fragment of the CAG promoter containing a 908-bp intron (Figure 1C). Because the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) was previously shown to enhance transgene expression from the MU3 promoter (but not the intron-containing EF1α or CAG promoters),14 it was inserted into the MU3 promoter–containing constructs. The sets of vectors carrying the MU3 and EF1α promoters were further modified to include a 178-bp DNA fragment (cPPT) that functions as a cis-acting facilitator of HIV-1 DNA nuclear import.41
HIV-1–based SIN lentiviral vectors. There were 3 sets of vectors constructed containing (A) the MU3, (B) the EF1α, or (C) the CAG promoter driving expression of the enhanced GFP gene as reporter. Each set of vectors includes a control vector, a 5′HS4 element–containing vector, an IFN-SAR–containing vector, and a 5′HS4/IFN-SAR–containing vector. A 1.2-kb fragment containing the 5′HS4 insulator was inserted in direct orientation into the deleted U3 region (ΔU3) of the 3′ LTR, which is copied to the 5′ LTR following reverse transcription. The 0.8-kb IFN-SAR was introduced in reverse orientation into the 3′ untranslated region of the GFP gene, immediately upstream of the 3′ LTR. The arrows above the 5′ LTR and the internal promoters depict sites and direction of transcription. WPRE indicates woodchuck hepatitis virus posttranscriptional regulatory element; SAR, IFN-SAR; SD, splice donor; SA, splice acceptor; ΔGag, deleted Gag region; and RRE, Rev-responsive element.
HIV-1–based SIN lentiviral vectors. There were 3 sets of vectors constructed containing (A) the MU3, (B) the EF1α, or (C) the CAG promoter driving expression of the enhanced GFP gene as reporter. Each set of vectors includes a control vector, a 5′HS4 element–containing vector, an IFN-SAR–containing vector, and a 5′HS4/IFN-SAR–containing vector. A 1.2-kb fragment containing the 5′HS4 insulator was inserted in direct orientation into the deleted U3 region (ΔU3) of the 3′ LTR, which is copied to the 5′ LTR following reverse transcription. The 0.8-kb IFN-SAR was introduced in reverse orientation into the 3′ untranslated region of the GFP gene, immediately upstream of the 3′ LTR. The arrows above the 5′ LTR and the internal promoters depict sites and direction of transcription. WPRE indicates woodchuck hepatitis virus posttranscriptional regulatory element; SAR, IFN-SAR; SD, splice donor; SA, splice acceptor; ΔGag, deleted Gag region; and RRE, Rev-responsive element.
Each lentiviral vector set tested consisted of a vector carrying the 5′HS4 insulator alone, a vector carrying the IFN-SAR alone, a vector incorporating both elements, and a control vector without either element. In the 5′HS4 element–containing vectors, a 1.2-kb fragment of the 5′HS4 insulator was inserted into the deleted U3 region of the 3′ LTR in direct orientation. This configuration allowed it to be copied to the 5′ LTR during the reverse transcription and thus flank the vector sequences upon integration.21,22 To generate IFN-SAR–containing vectors, a 0.8-kb fragment of the human IFN-SAR was inserted immediately upstream of the 3′ LTR in reverse orientation, based on the findings of Plavec and colleagues that this location and orientation yielded optimal expression from oncoretroviral vector backbones.37-39
VSV-G–pseudotyped SIN lentiviral vectors were produced by transient transfection of 293T cells and titers determined by assaying on HT1080 cells. Titers of 1.7 × 106 transducing units (TU)/mL on average were obtained with control vectors lacking either the 5′HS4 or the IFN-SAR before concentration by ultracentifugation. Vectors containing the IFN-SAR (1.5 × 106 TU/mL), the 5′HS4 insulator (6 × 105 TU/mL), or both elements (4 × 105 TU/mL) had lower titers. The reduction in vector titers roughly correlated with the size of the inserts48 ; insert-specific and/or context-dependent effects on titers were not investigated. High-titer vector stocks were readily obtained by ultracentrifugation concentration of vector-containing supernatants as required.
Incorporation of the IFN-SAR together with the 5′HS4 insulator into SIN lentiviral vectors provides significant protection against repressive position effects
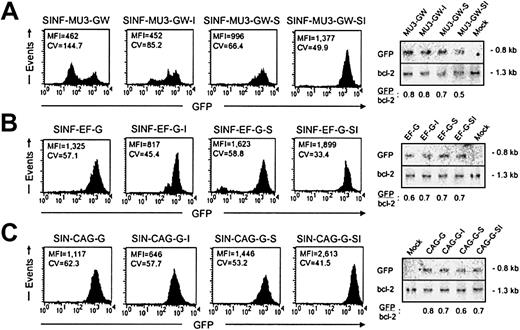
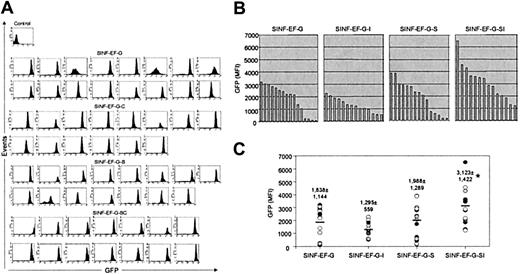
We initially investigated the effects of the 5′HS4 and IFN-SAR elements on lentiviral vector–mediated GFP transgene expression in the human CD34+ hematopoietic progenitor line KG1a, which models a precursor cell arrested prior to the myeloblast stage of differentiation.46 Pools of KG1a cells were transduced at low MOI with the MU3 promoter–containing set of vectors (4.2 ± 4.8% GFP+ cells before sorting) and analyzed for GFP transgene expression levels by FACS analysis during 4 months in culture (Figure 2A; Tables 1,2). When compared with the control vector SINF-MU3-GW (mean fluorescence intensity [MFI] = 462) at the 4-month time point, addition of the 5′HS4 insulator alone was found to provide no improvement in the level of GFP expression (MFI = 452), whereas inclusion of the IFN-SAR (MFI = 996) or both the IFN-SAR and the 5′HS4 insulator (MFI = 1377) resulted in maintenance of significantly higher GFP expression levels. A positive influence of the 5′HS4 and IFN-SAR elements on position effects was apparent from the profiles of GFP fluorescence intensity as reflected by the CV value of the signals, which is a relative indicator of homogeneity of expression. While the control vector displayed a broad pattern of GFP expression (CV = 144.7), the vector containing the IFN-SAR element maintained a relatively uniform pattern of GFP expression (CV = 66.4). A bimodal distribution of GFP expression was also observed for the population of cells transduced with the 5′HS4-containing SINF-MU3-GW-I vector (CV = 85.2). Notably, the SINF-MU3-GW-SI vector containing both elements had the most homogeneous profile of GFP expression (CV = 49.9).
Expression of lentiviral vector sets in pooled populations of KG1a cells. (A) FACS histograms of GFP fluorescence 4 months after transduction with the MU3 promoter–containing set of lentiviral vectors. (B) FACS histograms of GFP fluorescence 4 months after transduction with the EF1α promoter–containing set of lentiviral vectors. (C) FACS histograms of GFP fluorescence 4 months after transduction with the CAG promoter–containing set of lentiviral vectors. Shown in each case are the MFI values of fluorescence intensity and the CV values indicative of fluorescence homogeneity. To the right of each series of histograms are the Southern blot analyses of BamHI-digested genomic DNAs hybridized with a GFP probe (top panels) and a bcl-2 probe (bottom panels) showing comparable proviral vector copy numbers per cell.
Expression of lentiviral vector sets in pooled populations of KG1a cells. (A) FACS histograms of GFP fluorescence 4 months after transduction with the MU3 promoter–containing set of lentiviral vectors. (B) FACS histograms of GFP fluorescence 4 months after transduction with the EF1α promoter–containing set of lentiviral vectors. (C) FACS histograms of GFP fluorescence 4 months after transduction with the CAG promoter–containing set of lentiviral vectors. Shown in each case are the MFI values of fluorescence intensity and the CV values indicative of fluorescence homogeneity. To the right of each series of histograms are the Southern blot analyses of BamHI-digested genomic DNAs hybridized with a GFP probe (top panels) and a bcl-2 probe (bottom panels) showing comparable proviral vector copy numbers per cell.
To determine whether the changes in gene expression observed were specific to SIN lentiviral vectors containing the MU3 promoter, experiments were subsequently carried out in KG1a cells similarly transduced at low MOI with the EF1α promoter–containing set of vectors (5.2 ± 5.8% GFP+ cells before sorting). Although the effects were not as pronounced, when pools of transduced KG1a cells were analyzed after 4 months in culture (Figure 2B; Tables 1,2), the vector backbone with the 5′HS4/IFN-SAR combination was again found to maintain the highest level of GFP expression (MFI = 1899). The IFN-SAR–containing vector SINF-EF-G-S also maintained a comparably high level of GFP expression (MFI = 1623). In the case of SINF-EF-G-SI, the levels were approximately 1.4-fold higher than that achieved with the control vector SINF-EF-G (MFI = 1325). In contrast, the GFP expression level (MFI = 817) directed by the 5′HS4 insulator–containing vector SINF-EF-G-I was 1.5-fold reduced compared with that achieved with the control vector, suggesting the possibility of a negative influence on expression in this configuration. A more homogeneous pattern of GFP expression was again obtained in the case of the SINF-EF-G-SI vector containing both elements (CV = 33.4). The 5′HS4 insulator–containing SINF-EF-G-I vector (CV = 45.4) and the IFN-SAR–containing SINF-EF-G-S vector (CV = 58.8) also showed improved profiles of GFP fluorescence intensity compared with the pattern of GFP expression observed for the control SINF-EF-G vector (CV = 57.1).
When pools of KG1a cells transduced with the set of vectors containing the CAG promoter (0.2 ± 0.2% GFP+ cells before sorting) were likewise analyzed after a 4-month culture period (Figure 2C; Tables 1,2), the results obtained were similar to those observed for the EF1α promoter–containing set of vectors. By comparison with the control vector SIN-CAG-G (MFI = 1117), the SIN-CAG-G-SI vector carrying both elements exhibited a 2.3-fold higher level of GFP expression (MFI = 2613), while the IFN-SAR–containing vector SIN-CAG-G-S maintained a 1.3-fold higher level of GFP expression (MFI = 1446). On the contrary, the level of GFP expression directed by the 5′HS4 insulator–containing vector SIN-CAG-G-I was 1.7-fold lower (MFI = 646), again suggesting that the 5′HS4 insulator was exerting a negative influence on transgene expression. Moreover, as observed for the MU3 and EF1α promoter–containing sets of vectors, the most homogeneous GFP expression profile (CV = 41.5) was obtained with the CAG promoter–containing vector carrying both the 5′HS4 and the IFN-SAR. Both the IFN-SAR (CV = 53.2) and the 5′HS4 (CV = 57.7) carrying vectors also generated more homogeneous patterns of GFP expression than the control vector (CV = 62.3).
To confirm that the above differences in transgene expression levels were not due to different proviral vector copy numbers, genomic DNA was isolated from all of the pools of stably transduced KG1a cells and analyzed by Southern blotting using a GFP-specific probe. BamHI digestion of genomic DNAs revealed a 0.8-kb GFP-containing fragment in each case. To control for sample loading, the blots were stripped and rehybridized with a probe that detects endogenous bcl-2 gene sequences present on a 1.3-kb BamHI fragment. As shown in Figure 2, comparable proviral vector copy numbers were obtained for all 3 sets of vectors after normalization to the bcl-2 signal, indicating that the differing transgene expression levels were due to differential vector performance.
Clonal analysis of KG1a cells transduced with single copies of 5′HS4 insulator– and/or IFN-SAR–containing SIN lentiviral vectors
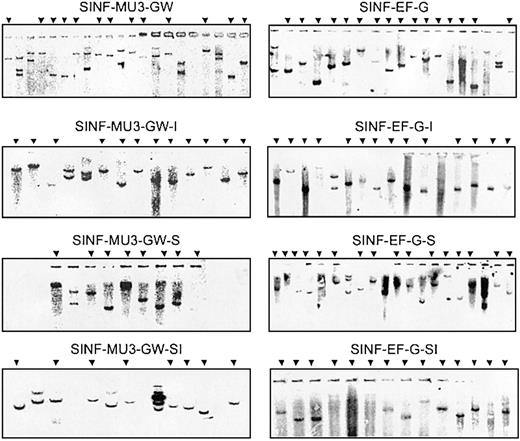
In order to further rule out vector copy number and/or cellular heterogeneity as a consequence of selective clonal outgrowth as potential explanations for the variations in transgene expression levels observed, we next investigated durability of transgene expression in KG1a clones harboring single vector integrants. KG1a cells were transduced with the MU3 and the EF1α promoter– containing sets of vectors at low MOI, and single GFP+ cells were sorted into 96-well plates. Genomic DNA was extracted from all clones that expanded and could be propagated in culture, and vector copy number was determined by Southern blot analysis with a GFP-specific probe following digestion with EcoRI (Figure 3). EcoRI cleaves once within all of the vectors to generate characteristic bands representative of unique integration sites. Southern blot analysis revealed distinct banding patterns, indicating that each clone represented an independent transduction event. Single-copy clones were readily identified at the 2-month time point. As can be seen in Figures 4A and 5A, a number of single-copy clones that had been transduced with either the MU3 promoter–containing control vector (4 of 13) or the EF1α promoter–containing control vector (3 of 16) developed heterogeneous GFP expression profiles. Bimodal, broad, or reduced levels of GFP expression were also observed in some instances in clones transduced with 5′HS4-containing or IFN-SAR–containing vectors. On the contrary, for both the MU3 promoter–containing set (8 of 8) and the EF1α promoter– containing set (14 of 14) all of the clones originating from transductions with vectors containing the 5′HS4/IFN-SAR combination exhibited unimodal patterns of GFP expression.
Copy number analysis of transduced KG1a clones. Genomic DNAs (10 μg) were digested with EcoRI, and Southern blot analysis was performed with a GFP hybridization probe. Arrows indicate single-copy clones transduced with the respective lentiviral vectors that were selected for FACS analysis of GFP expression.
Copy number analysis of transduced KG1a clones. Genomic DNAs (10 μg) were digested with EcoRI, and Southern blot analysis was performed with a GFP hybridization probe. Arrows indicate single-copy clones transduced with the respective lentiviral vectors that were selected for FACS analysis of GFP expression.
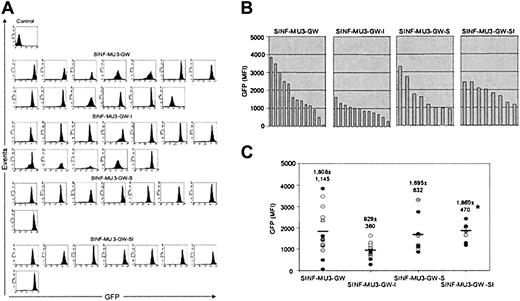
Expression of MU3 promoter–containing lentiviral vectors in KG1a clones. (A) FACS analysis of GFP expression of single-copy clones that were identified in Figure 3 after culturing for 2 months. (B-C) Comparison of MFI values of GFP fluorescence of the single-copy KG1a clones. *The MFI values of GFP fluorescence directed by the SINF-MU3-GW-SI vector were significantly higher than those directed by the SINF-MU3-GW-I vector (P < .05, Student t test). In addition, the CV values of GFP fluorescence homogeneity obtained with the SINF-MU3-GW-SI vector (CV = 46 ± 7) were significantly better than those obtained with the SINF-MU3-GW vector (CV = 88 ± 56) (P < .05, Student t test). In panel C, thick horizontal bars represent average MFI values of GFP fluorescence as indicated by the number above each column of data points, which are denoted by individual circles.
Expression of MU3 promoter–containing lentiviral vectors in KG1a clones. (A) FACS analysis of GFP expression of single-copy clones that were identified in Figure 3 after culturing for 2 months. (B-C) Comparison of MFI values of GFP fluorescence of the single-copy KG1a clones. *The MFI values of GFP fluorescence directed by the SINF-MU3-GW-SI vector were significantly higher than those directed by the SINF-MU3-GW-I vector (P < .05, Student t test). In addition, the CV values of GFP fluorescence homogeneity obtained with the SINF-MU3-GW-SI vector (CV = 46 ± 7) were significantly better than those obtained with the SINF-MU3-GW vector (CV = 88 ± 56) (P < .05, Student t test). In panel C, thick horizontal bars represent average MFI values of GFP fluorescence as indicated by the number above each column of data points, which are denoted by individual circles.
Expression of EF1α promoter–containing lentiviral vectors in KG1a clones. (A) FACS analysis of GFP expression of single-copy clones that were identified in Figure 3 after culturing for 2 months. (B-C) Comparison of MFI values of GFP fluorescence of the single-copy KG1a clones. *The MFI values of GFP fluorescence directed by the SINF-EF-G-SI vector were significantly higher than those directed by the SINF-EF-G-I and SINF-EF-G vectors (P < .05, Student t test). In addition, the CV values of GFP fluorescence homogeneity obtained with the SINF-EF-G-SI vector (CV = 41 ± 6) were significantly better than those obtained with the SINF-EF-G vector (CV = 63 ± 41) (P < .05, Student t test).
Expression of EF1α promoter–containing lentiviral vectors in KG1a clones. (A) FACS analysis of GFP expression of single-copy clones that were identified in Figure 3 after culturing for 2 months. (B-C) Comparison of MFI values of GFP fluorescence of the single-copy KG1a clones. *The MFI values of GFP fluorescence directed by the SINF-EF-G-SI vector were significantly higher than those directed by the SINF-EF-G-I and SINF-EF-G vectors (P < .05, Student t test). In addition, the CV values of GFP fluorescence homogeneity obtained with the SINF-EF-G-SI vector (CV = 41 ± 6) were significantly better than those obtained with the SINF-EF-G vector (CV = 63 ± 41) (P < .05, Student t test).
Comparative graphic representations of the MFI values of GFP expression levels for the 2 sets of vectors, presented in Figures 4B-C and 5B-C, allowed the position effects caused by random chromosomal integration and the potential benefits of the 5′HS4 and IFN-SAR elements on single-copy vector insertions to be better appreciated. For the MU3 promoter–containing set of vectors (Figure 4B-C), it was readily apparent that the control SINF-MU3-GW vector showed the most heterogeneous GFP expression pattern, ranging from extremely high levels in some clones to very low levels in others (MFI = 1808 ± 1145). This analysis also showed that clonal GFP expression levels were somewhat heterogeneous in the case of the IFN-SAR–containing SINF-MU3-GW-S vector, although to a lesser extent and with fewer instances of low level expressers observed (MFI = 1695 ± 832). While clones transduced with the 5′HS4-containing SINF-MU3-GW-I vector displayed the least interclonal variation, indicative of the ability of this element to overcome position effects to a certain degree, these clones exhibited the lowest levels of GFP expression on average (MFI = 929 ± 360). Notably, high-level homogeneous GFP expression was observed in the clones transduced with the 5′HS4/IFN-SAR–containing SINF-MU3-GW-SI vector (MFI = 1860 ± 470).
Similarly for the EF1α promoter–containing vector set (Figure 5B-C), this quantitative analysis revealed the ability of the 5′HS4 element to reduce position effects but at the expense of high level GFP expression (SINF-EF-G-I MFI = 1295 ± 559). As a single element, the IFN-SAR appeared to provide minimal benefit (SINF-EF-G-S MFI = 1988 ± 1289) compared with control SINF-EF-G–mediated GFP expression (MFI = 1838 ± 1144). The best overall performance was obtained with the SINF-EF-G-SI vector carrying the 5′HS4 element plus the IFN-SAR (MFI = 3123 ± 1422).
High-level lentiviral vector–mediated transgene expression in long-term myelomonocytic cultures of differentiated human CD34+ cord blood cells
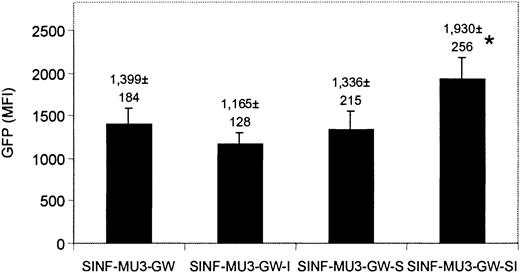
HSC-directed gene therapy holds promise for hereditary blood disorders such as type I Gaucher disease, a lysosomal storage disease that is due to a monogenic defect that results in pathologic accumulation of glucocerebroside in monocyte/macrophages.1 A previous study indicated that inclusion of the T-cell–specific CD2 locus control region could not block the decline in lentiviral vector–mediated transgene expression after extended culture of primary human T cells.49 To likewise study the effects of the 5′HS4 insulator and the IFN-SAR on transgene expression in primary human myelomonocytic cells, CD34+ progenitors were isolated from human cord blood and transduced at low MOI with the MU3 promoter–containing set of lentiviral vectors (3.9 ± 2.2% GFP+ cells before sorting). CD34+GFP+ cells were sorted and differentiated into monocytes during 7 weeks of culture in IL-3, IL-6, and GM-CSF. Shown in Figure 6 are the GFP expression levels averaged over weeks 4 to 7. All vectors, including the control SINF-MU3-GW vector, demonstrated high levels of GFP transgene expression in the myeloid progeny of transduced CD34+ cells throughout the observation period. These results are in marked contrast to our previous findings with oncoretroviral vectors in similar myelomonocytic differentiation cultures, in which a dramatic reduction in transgene expression was noted after 4 weeks.50 Similar to results obtained after prolonged culture of transduced KG1a cells, the SINF-MU3-GW-SI vector carrying both the 5′HS4 insulator and the IFN-SAR provided the best protection against position effects and/or directed the highest levels of transgene expression. At the 7-week time point in the experiment presented, when 30% to 50% of the nonadherent cells were CD14+ monocytes, the relative expression levels were SINF-MU3-GW-SI (MFI = 2619) more than SINF-MU3-GW (MFI = 1933) more than SINF-MU3-GW-S (MFI = 1927) more than SINF-MU3-GW-I (MFI = 1360).
Expression of MU3 promoter–containing lentiviral vectors in monocytes derived from CD34+cord blood cells. CD34+ cord blood cells were transduced with the MU3 promoter–containing set of lentiviral vectors, sorted, and cultured in the presence of IL-3, IL-6, and GM-CSF to promote monocytic differentiation. Shown is the comparison of MFI values of GFP fluorescence of nonadherent cells analyzed between 4 to 7 weeks of culture. Similar results were obtained in 2 independent experiments. *The MFI values of GFP fluorescence directed by the SINF-MU3-GW-SI vector were significantly higher than those directed by the other vectors (P < .05, Student t test). In addition, the CV values of GFP fluorescence homogeneity obtained with the SINF-MU3-GW-SI vector (CV = 76 ± 4) were significantly better than those obtained with the SINF-MU3-GW vector (CV = 89 ± 2) (P < .05, Student t test). Error bars represent SD.
Expression of MU3 promoter–containing lentiviral vectors in monocytes derived from CD34+cord blood cells. CD34+ cord blood cells were transduced with the MU3 promoter–containing set of lentiviral vectors, sorted, and cultured in the presence of IL-3, IL-6, and GM-CSF to promote monocytic differentiation. Shown is the comparison of MFI values of GFP fluorescence of nonadherent cells analyzed between 4 to 7 weeks of culture. Similar results were obtained in 2 independent experiments. *The MFI values of GFP fluorescence directed by the SINF-MU3-GW-SI vector were significantly higher than those directed by the other vectors (P < .05, Student t test). In addition, the CV values of GFP fluorescence homogeneity obtained with the SINF-MU3-GW-SI vector (CV = 76 ± 4) were significantly better than those obtained with the SINF-MU3-GW vector (CV = 89 ± 2) (P < .05, Student t test). Error bars represent SD.
Discussion
Gene delivery methods that use integrating vectors often result in poor expression levels because the vectors are negatively affected by dominant regulatory sequences and heterochromatin flanking the insertion sites.19 Others have demonstrated that when used separately insulators and S/MARs, which are believed to contribute to the organization of the eukaryotic genome into independent domains, can enhance oncoretroviral vector expression levels and provide a certain degree of protection against position effects.21,22,32,36-39 In this work we have comparatively analyzed the actions of the chicken β-globin 5′HS4 insulator and the human IFN-SAR on transgene expression levels in the context of several lentiviral vectors with different internal promoters. To achieve high efficiency transduction of HSCs and hematopoietic progenitors with VSV-G–pseudotyped lentiviral vectors, previous studies have used MOIs of up to 3000.7-13,15 As a result, the target cell genomes frequently contain multiple vector copies, precluding facile investigation of integration site–specific negative regulatory influences on transgene expression.10-13,15 Therefore, in the experiments reported here, we used 2 hematopoietic progenitor cell culture models—the KG1a cell line and primary CD34+ cord blood cells differentiated in vitro into monocytes—in which it was feasible to perform transductions at low MOIs to generate the requisite cells containing primarily single copies of lentiviral vector integrants. The principal finding of this investigation is that the combination of the 5′HS4 insulator and the IFN-SAR reproducibly resulted in higher and/or more homogeneous levels of lentiviral vector–mediated transgene expression in both model systems than was achieved with control vectors or vectors carrying either element alone.
To account for the possibility that some internal promoters might be more susceptible to position effects than others, we chose 3 promoters for this study on the basis of our previous results demonstrating that they all were capable of directing robust transgene expression in human hematopoietic cells: the U3 region of the MSCV LTR, the human EF1α promoter, and the composite CAG promoter.14 Interestingly, we found that regardless of the internal promoter, insertion of the 5′HS4 insulator into the U3 region of the 3′ LTR decreased transgene expression. A similar phenomenon was observed previously when the same 5′HS4 insulator fragment was incorporated into the U3 region of the 3′ LTR of an oncoretroviral vector.23 The authors proposed that the negative effect might be due to the enhancer blocking activity of the 5′HS4 element abrogating a positive influence of the LTR enhancer sequences on the internal promoter. As the LTR enhancer sequences were deleted from the SIN lentiviral vectors used in our experiments, this possibility can be ruled out. Rather, the 5′HS4 element may be preventing endogenous genomic enhancers from acting on the internal promoters. Because significantly higher levels of transgene expression were consistently obtained with corresponding lentiviral vectors carrying the IFN-SAR in addition to the 5′HS4 element, an alternative explanation is that the reduced expression is due to topological constraints that are alleviated by the IFN-SAR.36 A characteristic feature of S/MARs is that they exhibit an unusually high unwinding capability by stable base-unpairing, which may introduce and maintain a level of torsional stress conducive to active transcription by RNA polymerase II.33
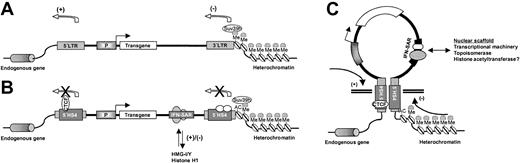
This hypothesis notwithstanding, the manner in which the 5′HS4 insulator and the IFN-SAR act in concert to reproducibly achieve improved transgene expression remains to be determined. Previous work examining whether the 5′HS4 insulator could protect oncoretroviral vectors from chromosomal position effects indicated that the element was capable of increasing the probability of transgene expression but was not sufficient to overcome silencing associated with methylation.21,22 In this regard, the IFN-SAR has been reported to prevent de novo methylation and to alleviate methylation-mediated transcriptional repression of oncoretroviral vector–directed transgene expression.39 However, the principal function of S/MARs is believed to be the attachment of chromosomal DNA to the nuclear scaffold/matrix, thereby forming topologically-independent chromatin loops favorable for gene expression.31,40 It is therefore tempting to speculate that the 5′HS4/IFN-SAR combination efficiently creates de facto chromatin domain boundaries (Figure 7).24,51,52
Potential mode of action of the 5′HS4/IFN-SAR combination. (A) Standard lentiviral vector showing negative (heterochromatin spreading) and positive (vector enhancer activation of endogenous gene expression) integration site effects. P indicates promoter. (B) SIN lentiviral vector containing the 5′HS4/IFN-SAR combination evaluated in this study. The enhancer-blocking function of the 5′HS4 element requires CTCF binding sites.27 It has been suggested that barrier activity depends on other signals within the 5′HS4 element that recruit histone acetyltransferases, which would acetylate (AC) histone H3 on lysine 9 and prevent encroachment of methylation (Me) by histone methyltransferases (Suv39h).28,29 The IFN-SAR could antagonize histone H1–mediated repression of gene expression by preferential interaction with high mobility group proteins (HMG-I/Y).53,54 X indicates block of function. (C) The IFN-SAR associates with the nuclear scaffold/matrix and, in conjunction with the pair of 5′HS4 elements, could facilitate formation of chromatin loops conducive to transgene expression.24,31,40 Arrows indicate direction in which the various elements exert their influence.
Potential mode of action of the 5′HS4/IFN-SAR combination. (A) Standard lentiviral vector showing negative (heterochromatin spreading) and positive (vector enhancer activation of endogenous gene expression) integration site effects. P indicates promoter. (B) SIN lentiviral vector containing the 5′HS4/IFN-SAR combination evaluated in this study. The enhancer-blocking function of the 5′HS4 element requires CTCF binding sites.27 It has been suggested that barrier activity depends on other signals within the 5′HS4 element that recruit histone acetyltransferases, which would acetylate (AC) histone H3 on lysine 9 and prevent encroachment of methylation (Me) by histone methyltransferases (Suv39h).28,29 The IFN-SAR could antagonize histone H1–mediated repression of gene expression by preferential interaction with high mobility group proteins (HMG-I/Y).53,54 X indicates block of function. (C) The IFN-SAR associates with the nuclear scaffold/matrix and, in conjunction with the pair of 5′HS4 elements, could facilitate formation of chromatin loops conducive to transgene expression.24,31,40 Arrows indicate direction in which the various elements exert their influence.
Accumulating data suggest that SIN lentiviral vectors based on HIV-1 may be a better platform for delivery and expression of transgenes in HSCs than oncoretroviral vectors.7-16 This is perhaps due to intrinsic structural or enzymatic properties of HIV-1 that are retained in current-generation vector systems that promote transcriptionally permissive chromatin domain reorganization and boundary remodeling.55 Alternatively, increased functionality may be the result of the unique integrative mechanism of HIV-1 that more efficiently targets certain sites in the genome that reside within open chromatin domains (eg, actively transcribed housekeeping genes). In accord with the latter mechanism, a recent report has demonstrated that in contrast to oncoretroviruses,56 HIV-1 preferentially integrates into transcriptionally active DNA.57 Nonetheless, as shown here and described previously,13,58 there is still clonal variability in transgene expression dependent on the lentiviral vector integration site. Moreover, it is also clear from this and other studies that unmodified HIV-1 vectors are not completely immune from progressive transcriptional down-regulation and silencing.16,20 Our observations indicate that the 5′HS4/IFN-SAR combination is particularly effective at reducing these repressive chromosomal position effects. The SIN HIV-1 vectors described herein carrying the chicken β-globin 5′HS4 insulator and the human IFN-SAR have thus been designated “HUMV” lentiviral vectors to reflect this property and their derivation from human immunodeficiency virus type 1.59
Further studies will be required to determine whether HUMV series lentiviral vectors are optimized for maintenance of high-level transgene expression in relevant large-animal models of HSC-directed gene therapy—an important prerequisite for future clinical trial consideration. In terms of safety issues though it seems plausible that the HUMV proviral form, which is devoid of HIV-1 U3 region transcriptional regulatory elements and flanked by the enhancer-blocking chicken β-globin insulator, might be less likely to activate cellular oncogenes by insertional mutagenesis than the conventional oncoretroviral vectors that are currently being used in human gene therapy applications (Figure 7).60,61
Prepublished online as Blood First Edition Paper, February 13, 2003; DOI 10.1182/blood-2002-09-2991.
Supported in part by National Institutes of Health grants HL65519 and HL66305 (R.G.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Gary Felsenfeld and Jurgen Bode for providing the 5′HS4 insulator and the IFN-SAR, respectively.