Abstract
CD18 (β2 leukocyte integrin) is transcriptionally regulated in myeloid cells, but the mechanisms that increase its expression in response to retinoic acid (RA) have not been defined. The CD18 promoter was activated by RA treatment in stably transfected U937 myeloid cells. We identified a retinoic acid response element (RARE) that lies nearly 900 nucleotides upstream of the CD18 transcriptional start site that was bound by the RA receptors, retinoic acid receptor (RAR) and retinoic X receptor (RXR). This RARE accounted for one half of the RA responsiveness of CD18. However, unexpectedly, one half of the dynamic response to RA was mediated by the 96-nucleotide CD18 minimal promoter, which lacks a recognizable RARE. Binding sites for the ets transcription factor, GA-binding protein (GABP), and Sp1 were required for full RA responsiveness of both the CD18 minimal promoter and the full-length promoter. The ets sites conferred RA responsiveness on an otherwise unresponsive heterologous promoter, and RA responsiveness was directly related to the number of ets sites. The transcriptional coactivator p300/CBP physically interacted with GABP in vivo, and p300 increased the responsiveness of the CD18 promoter to RA. These studies demonstrate a novel role for non-RAR transcription factors in mediating RA activation in myeloid cells. They support the concept that transcription factors other than RARs are required for RA-activated gene expression. We hypothesize that a multiprotein complex—an enhanceosome—that includes GABP, other transcription factors, and coactivators, dynamically regulates CD18 expression in myeloid cells.
Introduction
Granulocytes and monocytes, which are collectively known as myeloid cells, are phagocytes and antigen-presenting cells that are crucial for proper immune system function. Myeloid cells are derived from pluripotent, self-renewing stem cells, and gene transcription is tightly controlled during the differentiation of myeloid cells from stem cells. The central role of gene transcription in myeloid cells is demonstrated by the many forms of myeloid leukemia that are caused by aberrant expression of transcription factors.1
Retinoic acid (RA) plays an important role in both normal and abnormal myeloid cell development. RA acts as a ligand for retinoic acid receptors (RARs)—a family of nuclear proteins that bind to cognate DNA sequences and thereby regulate gene transcription. RARs bind to DNA sequences that contain repeats of a core consensus sequence, AGGTCA.2 RA induces granulocytic morphology, function, and gene expression of the myeloid cell line HL-60.3 A point mutation in the ligand-binding domain of retinoic acid receptor–α (RARα) blocks HL-60 differentiation, and this effect can be rescued by wild-type RARα.4 Chromosomal rearrangements that translocate RARα to unrelated partner proteins, such as promyelocytic leukemia (PML) and promyelocytic leukemia zinc finger (PLZF), cause acute promyelocytic leukemia (APL),5and RA treatment can induce remissions in the majority of affected patients.6 Thus, RA plays critical roles in both normal and malignant myeloid differentiation.
Myeloid genes are regulated by the combinatorial actions of transcription factors.7 Many myeloid gene promoters are regulated by ets transcription factors, which possess related DNA-binding domains that contain characteristic tryptophan repeats.8 Several etsfactors, including ets-2,9 PU.1,10and GA-binding protein (GABP),11 regulate gene expression during myeloid differentiation. Ets factors regulate myeloid gene promoters in combination with both lineage-restricted factors and more widely expressed transcription factors, including Sp1.7
CD18 is the β chain of the leukocyte integrins. It mediates cell-cell and cell-matrix interactions of leukocytes by noncovalently associating with CD11a, CD11b, and CD11c to form the antigens known as lymphocyte function associated antigen–1 (LFA-1), Mac-1 (Mo-1), and p150/95, respectively.12 Aberrant expression of CD18 results in leukocyte adhesion deficiency (LAD), a congenital immunodeficiency that causes recurrent infections and early death.13,14 CD18 is expressed exclusively by lymphocytes and myeloid cells, and it is transcriptionally regulated in myeloid cells.15 The CD18 promoter retains the leukocyte-specific and myeloid-inducible activity of the endogenous CD18 gene.16 A 96-nucleotide (nt) fragment of the CD18 promoter, whichis sufficient to direct myeloid gene expression, is bound by the transcription activator Sp1,17 and by theets factors GABP11 and PU.1.16These transcription factors act synergistically to activate CD18 expression in myeloid cells.
In this report, we demonstrate that RA-induced transcription of CD18 in myeloid cells is mediated by a novel mechanism. Transcriptional activation of CD18 by RA requires 2 distinct regions of the CD18 promoter: (1) a functional retinoic acid response element (RARE) that lies approximately 900 nt upstream of the transcriptional start site, and (2) ets and Sp1 sites in the proximal promoter. We propose that cooperative interactions between RARs and other transcription factors, including GABP and Sp1, are required for full RA responsiveness of myeloid gene expression. We identified a physical interaction between GABP and the transcriptional coactivator p300/CBP, and showed that transfected p300 further increased the responsiveness of the CD18 promoter to RA. These findings support the emerging view that responsiveness to retinoids and other hormonal agents depends on complex interactions between RARs and other transcription factors, and suggests that a multiprotein complex mediates the RA responsiveness of CD18.
Materials and methods
Cell culture
U937 cells (ATCC no. CRL 1593) (Rockville, MD) were passaged twice weekly in RPMI 1640 (GIBCO BRL, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal calf serum (FCS) (ICN, Costa Mesa, CA), l-glutamine, and penicillin/streptomycin (complete RPMI medium) in an atmosphere of 5% CO2. Human embryonic kidney cells (293 cells) were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FCS, l-glutamine, and penicillin/streptomycin. Where indicated, cells were treated with all-trans retinoic acid (RA; Sigma, St Louis, MO) at concentrations ranging from 10−12 M to 10−4 M in dimethyl sulfoxide (DMSO), or with DMSO vehicle alone as a negative control.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as previously described.11 CD18 RARE corresponds to CD18 promoter sequence −903 to −862 relative to the primary transcriptional start site, as previously described.15 EMSA probe was prepared by radiolabeling the following oligonucleotide primer pairs (Operon Technologies, Alameda, CA) with 32P deoxycytidine triphosphate (dCTP) and polynucleotide kinase (New England Biolabs, Beverly, MA):
CD18 RARE, top: 5′-GCGGGATCCTTGCAGTGAGCTGAGATCACGCCACTGCACTCCAGCTGGGTGAAGCTTGC G-3′
CD18 RARE, bottom 5′-CGCAAGCTTCACCCAGCTGGAGTGCAGTGGCGTGATCTCAGCTCACTGCAAGGATCCCGC-3′
Where indicated, 0.5 μg purified human RAR (Santa Cruz Biotechnology, CA) and/or 0.2 μg human retinoic X receptor (RXR) (Affinity Bioreagents, Golden, CO) were added to the binding reaction. EMSA was performed with 0.1 μg poly–deoxyinosine deoxycytidine (poly dIdC) (Amersham Bioscience, Piscataway, NJ) as nonspecific competitor, and binding was competed with 100 × molar excess of either unlabeled homologous probe or unlabeled irrelevant probe, which corresponds to CD18(−89) to CD18(−76):
Top: 5′-TCGAGTGCAACCCACCACA-3′
Bottom: 5′-AGCTTGTGGTGGGTTGCAC-3′.
Stable transfection
About 2 × 107 log phase U937 cells were electroporated (960 μF, 300 V), as previously described,15 with the following linearized plasmids: 5 μg pNeoNut (which confers resistance to G418) and 5 μg of the indicated luciferase reporter construct. Transfected cells were grown for 48 hours in complete RPMI medium, and then plated at a density of 3 × 105 cells per milliliter in complete RPMI medium supplemented with 1 mg/mL G418 (Sigma) and 0.9% methyl cellulose (Dow, Coral Gables, FL). Individual colonies were isolated 10 to 14 days later and expanded in complete RPMI medium supplemented with 1 mg/mL G418. Activation of the luciferase reporter gene was expressed as relative light units, and fold induction by RA was defined as luciferase activity in the presence of RA, divided by the activity of the paired DMSO control. For all DNA constructs, at least 3 independent clones were isolated and characterized. Where indicated, stably transfected cells were transiently transfected with 1 to 10 μg pCMVβp300 expression vector.18
DNA constructs
The RARE from the RAR promoter19 was cloned upstream of pTK81/luc.20 CD18 promoterets and Sp1 site mutations, which were previously described,11,16 17 were cloned into the CD18(−96)/luc promoter by polymerase chain reaction (PCR).
CD18 ets and Sp1 site mutations were cloned into the CD18(−918)/luc by PCR amplification of the region from −918 to −96, by PCR with CD18(−918)/luc as template and the following oligonucleotides:
5′-GCGGAGCTCCTTCACCCCTGCCCC-3′
5′-GCGAGATCTCCCGGGAGGCAGAGG-3′.
The resultant PCR product was cleaved with SacI andBglII and ligated upstream of CD18 proximal promoter mutant constructs, as described above.
The pTK/ets cluster constructs were prepared with the use of complementary pairs of oligonucleotides containing the CD18 sequence from −79 to −37 with flanking restriction sites, and cloned upstream of pTK81/luc.20 The sense strand of each oligonucleotide pair is as follows:
Ets-1: 5′-GCGGGATCCCCACTTCCTCCAAGGAGGAGCTGAGAGGAACAGGAAGTGTCAGCGGCCGCAAGCTTGCG-3′
Ets-3: 5′-GCGAAGCTTCCATGGCCACTTCCTCCAAGGAGGAGCTGAGAGGAACAGGAAGTGTCAGCTCGAGGCG-3′
Ets-4: 5′-GCGGCGGCCGCCCACTTCCTCCAAGGAGGAGCTGAGAGGAACAGGAAGTGTCAGCCATGGGCG-3′
RARE/TK was prepared with the use of complementary oligonucleotides containing CD18 sequence from −903 to −862, and cloned upstream of pTK81/luc. The sense strand of the oligonucleotide pair is as follows:
5′-GCGGGATCCTTGCAGTGAGCTGAGATCACGCCACTGCACTCCAGCTGGGTGAAGCTTGCG-3′.
RARE2/TK was prepared by PCR with the use of CD18(−918)/luc as template and the following oligonucleotides, and cloned upstream of pTK81/luc:
Top: 5′-GCGGGTACCCTGCAGAAGCTTTTGCAGTGAGCTGAGATC-3′
Bottom: 5′-GCGGGTACCCCATGGCACCCAGCTGGAGTGCAG-3′.
Immunoprecipitation
Whole-cell extracts were prepared from U937 cells, and protein concentration was determined by bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL). Then, 0.75 mg extract was incubated with antisera to p300 (Upstate Biotechnology, Lake Placid, NY) at 4°C for 1 hour, followed by incubation with protein-G sepharose (Amersham Bioscience) at 4°C for 1 hour. The immunoprecipitated material was boiled in 2 × loading buffer, electrophoresed in an 8% polyacrylamide gel, and transferred to nitrocellulose (Schleicher and Schuell, Keene, NH). In an adjacent lane, 50 μg whole-cell extract was electrophoresed. The blot was immunodetected with polyclonal antiserum against GABPα (Strategic Biosolutions, Newark, DE) or p300 (Santa Cruz Biotechnology), and horseradish peroxidase–conjugated goat antirabbit secondary antiserum, and detected by enhanced chemiluminescence (ECL) (Amersham Bioscience).
Results
RAR and RXR bind to the CD18 promoter
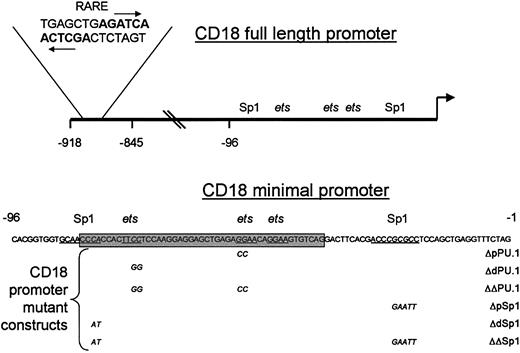
CD18 is transcriptionally regulated by RA in myeloid cells.15 For many genes, RA responsiveness is mediated by binding of RARs to retinoic acid response elements (RAREs). In general, RAREs consist of 2 or more DNA repeats that resemble the consensus sequence AGGTCA.2 We identified a potential RARE that lies nearly 900 nucleotides upstream of the CD18 transcriptional start site (Figure 1). Each half-site in this RARE matches the AGGTCA consensus sequence at 5 of 6 nucleotides, and the half-sites form an inverted repeat with a spacing of 1 nucleotide.
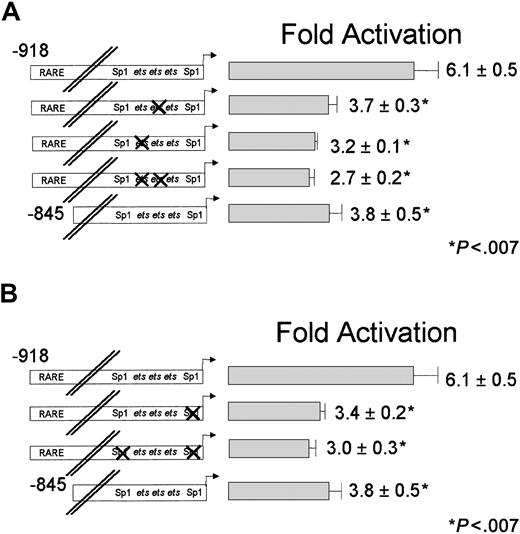
Diagrammatic illustration of the CD18 promoter and binding site mutants.
Diagram of the CD18 promoter with the sequence of the RARE shown in bold; arrows indicate the location and orientation of the half-sites. The positions of the Sp1 and ets family–binding sites within the CD18(−96) minimal promoter are underlined, and theets cluster is shaded. The sequences of the CD18 minimal promoter and relevant mutations are presented.
Diagrammatic illustration of the CD18 promoter and binding site mutants.
Diagram of the CD18 promoter with the sequence of the RARE shown in bold; arrows indicate the location and orientation of the half-sites. The positions of the Sp1 and ets family–binding sites within the CD18(−96) minimal promoter are underlined, and theets cluster is shaded. The sequences of the CD18 minimal promoter and relevant mutations are presented.
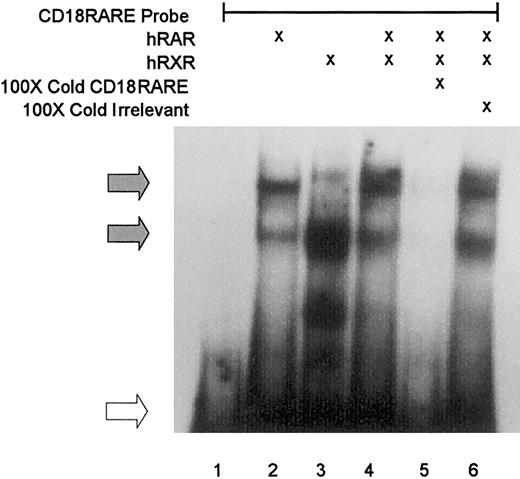
We prepared a radiolabeled, double-stranded DNA probe that includes the potential CD18 RARE for use in EMSA. Purified RAR bound to this DNA probe as a monomer and as a dimer (Figure2, lane 2, filled arrows). Similarly, RXR bound this probe as a monomer and as a dimer (lane 3, filled arrows). Together, these proteins formed a lower-mobility RAR/RXR heterodimeric complex (lane 4). Binding by the heterodimeric complex was abrogated by competition with unlabeled homologous probe (lane 5) or a known RARE (from the RARβ promoter; data not shown), but not by an irrelevant sequence (an unrelated region of the CD18 promoter, lane 6). Thus, RARα and RXRα bound to this region of the CD18 promoter in a sequence-specific manner.
CD18 RARE binding of RAR and RXR.
EMSA was performed following incubation of32P-radiolabeled CD18 RARE probe with human RAR (hRAR) and/or no added protein (lane 1), hRXR (lanes 2-6), and competition with 100-fold molar excess of unlabeled homologous probe (lane 5) or irrelevant probe (lane 6). Filled arrows indicate RAR/RXR–binding species, and the open arrow indicates unbound probe.
CD18 RARE binding of RAR and RXR.
EMSA was performed following incubation of32P-radiolabeled CD18 RARE probe with human RAR (hRAR) and/or no added protein (lane 1), hRXR (lanes 2-6), and competition with 100-fold molar excess of unlabeled homologous probe (lane 5) or irrelevant probe (lane 6). Filled arrows indicate RAR/RXR–binding species, and the open arrow indicates unbound probe.
The CD18 promoter is transcriptionally activated by retinoic acid
We previously showed that a 918-nucleotide fragment of the CD18 promoter, which includes the region of DNA that was bound by RAR/RXR (Figure 1), is transcriptionally active in myeloid cells. Because the endogenous CD18 gene is transcriptionally activated by RA,15 we sought to determine if the potential CD18 RARE mediates the responsiveness of the CD18 promoter to RA.
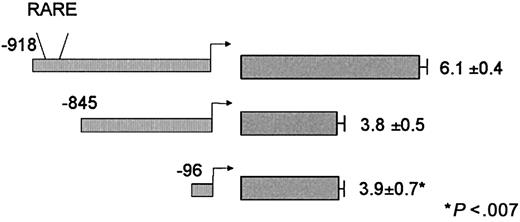
We stably cotransfected U937 cells with a plasmid that confers resistance to the antibiotic G418, along with one of the following CD18 promoter constructs (Figure 3): CD18(−918)/luc (which includes the region bound by RAR/RXR), CD18(−845)/luc (in which that region was deleted), or CD18(−96)/luc (the CD18 minimal promoter). Transfected cells were plated in methylcellulose, and individual colonies were isolated. Clones of cells were divided into 2 equal aliquots, and RA responsiveness was determined by comparing the luciferase activity of the cells treated with 10−5 M RA relative to the paired DMSO-treated control cells. Because the site of integration into chromatin may affect gene expression, at least 3 independent clones were examined for each DNA construct.
Responsiveness of CD18 to RA following integration into chromatin.
U937 cells were stably transfected with the indicated CD18 promoter constructs. Individual colonies were isolated, and equal aliquots of cells were treated with 10−5 M RA or DMSO (0.1%). Luciferase activity was measured 24 hours later; fold induction represents the relative activity of the RA-treated sample divided by the paired DMSO control sample. All data represent the mean and standard error of at least 3 independent clones, each tested in at least triplicate.
Responsiveness of CD18 to RA following integration into chromatin.
U937 cells were stably transfected with the indicated CD18 promoter constructs. Individual colonies were isolated, and equal aliquots of cells were treated with 10−5 M RA or DMSO (0.1%). Luciferase activity was measured 24 hours later; fold induction represents the relative activity of the RA-treated sample divided by the paired DMSO control sample. All data represent the mean and standard error of at least 3 independent clones, each tested in at least triplicate.
U937 cells that stably integrated CD18(−918)/luc (which includes the RARE) were transcriptionally activated 6.1-fold ± 0.4-fold by 10−5 M RA (Figure 3). RA responsiveness was dose dependent, and RA responsiveness was observed at concentrations as low as 10−11 M (data not shown). The CD18 promoter was activated 2-fold within 8 hours of treatment with 10−5 M RA, but promoter activity was maximal at 24 to 48 hours and declined thereafter (data not shown). For all subsequent experiments, cells were treated 10−5 M RA for 24 hours.
Unexpectedly, CD18(−845)/luc, which lacks the distal RARE, retained significant RA responsiveness (Figure 3). However, the responsiveness of CD18(−845)/luc to RA (3.8-fold ± 0.5-fold) was significantly lower than that of the full-length promoter (P < .007). Furthermore, the CD18 minimal promoter, CD18(−96)/luc, which lacks any recognizable RARE, exhibited the same RA responsiveness (3.9-fold ± 0.7-fold) as CD18(−845)/luc. These findings indicate that the distal element in the CD18 promoter is an authentic RARE, but that the proximal CD18 promoter also retains substantial RA responsiveness.
The CD18 minimal promoter requires functional Sp1- andets-binding sites for maximal RA responsiveness
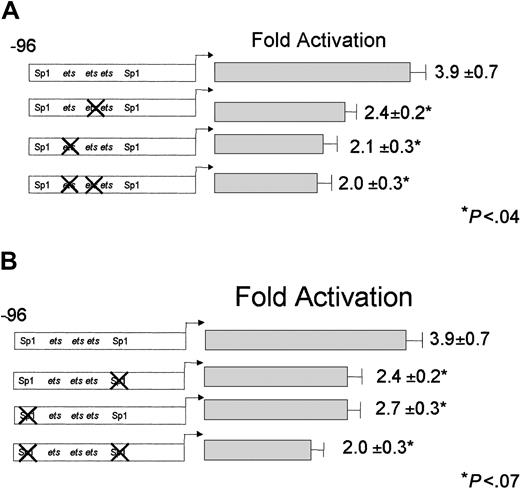
The CD18 minimal promoter, CD18(−96)/luc, was activated nearly 4-fold by RA, despite the absence of any recognizable RARE in this region of the gene. RAR and RXR, which bound avidly to the distal RARE (Figure 2), did not bind to this region (data not shown). We sought to identify the DNA sequences that are responsible for the RA responsiveness of the CD18 proximal promoter. We previously showed that the CD18 minimal promoter is bound by the ets factors GABP and PU.1, and by transcriptional activator Sp1. We stably transfected U937 cells with CD18 minimal promoter constructs that contain mutations that disrupt binding by ets factors or Sp1 (Figure 1). We examined the RA responsiveness of each promoter construct with at least 3 independent clones of cells.
Disruption of the individual ets sites reduced RA responsiveness of the CD18 minimal promoter by 30% to 40%; these effects were statistically significant (Figure4A). Mutation of both etssites reduced RA responsiveness by one half compared with the wild-type CD18(−96)/luc minimal promoter. Thus, disruption of the etssites dramatically reduced RA responsiveness of the CD18 promoter.
Effect of disrupting ets and Sp1 transcription factor–binding sites on RA induction of the minimal promoter.
RA induction of the minimal promoter is dependent on functionalets and Sp1 transcription factor–binding sites. CD18(−96)/luc or the corresponding DNA constructs withets-site mutations (indicated by X; panel A) or Sp1-site mutations (panel B) were stably transfected into U937 cells, and responsiveness to RA was measured, as described in the legend to Figure3. All data represent the mean and standard error of at least 3 independent clones, each tested in at least triplicate.
Effect of disrupting ets and Sp1 transcription factor–binding sites on RA induction of the minimal promoter.
RA induction of the minimal promoter is dependent on functionalets and Sp1 transcription factor–binding sites. CD18(−96)/luc or the corresponding DNA constructs withets-site mutations (indicated by X; panel A) or Sp1-site mutations (panel B) were stably transfected into U937 cells, and responsiveness to RA was measured, as described in the legend to Figure3. All data represent the mean and standard error of at least 3 independent clones, each tested in at least triplicate.
Similarly, we stably transfected U937 cells with promoter constructs that contained mutations of the Sp1 sites (Figure 4B). Mutation of either of the Sp1-binding sites reduced RA responsiveness by about 30%, while mutations in both Sp1 sites reduced RA responsiveness to approximately one half (P < .02). We conclude that the CD18 minimal promoter mediates RA induction through both the etssites and Sp1-binding sites.
The CD18 RARE and the CD18 minimal promoter function additively to mediate maximal RA responsiveness
We sought to determine if the RA responsiveness of the distal CD18 RARE requires the integrity of the ets and Sp1 sites in the CD18 minimal promoter. We introduced into the full-length CD18 promoter the same ets- and Sp1-site mutations that reduced activity of the CD18 minimal promoter. Mutation of the individualets sites in the minimal promoter reduced RA responsiveness of the full-length promoter (CD18(−918)/luc) by approximately 40% (Figure 5A). Mutation of bothets sites reduced its responsiveness by more than 50%. Similarly, mutation of both Sp1 sites reduced promoter activity by more than 50% (Figure 5B). Thus, maximal RA responsiveness of CD18 requires integrity of both the distal RARE and the ets and Sp1 sites in the minimal promoter. Mutation of the ets or Sp1 sites reduced, but did not fully abrogate, RA responsiveness of the full-length CD18 promoter. This indicates that the distal elements and the proximal promoter elements function additively.
Effect of disrupting ets and Sp1 transcription factor–binding sites and an intact RARE on RA induction of the full-length promoter.
RA induction of the full-length promoter is dependent on both functional ets and Sp1 transcription factor–binding sites and an intact RARE. CD18(−918)/luc or the corresponding DNA constructs with ets-site mutations (indicated by X; panel A) or Sp1-site mutations (indicated by X; panel B) were stably transfected into U937 cells, and responsiveness to RA was measured, as described in the legend to Figure 3. All data represent the mean and standard error of at least 3 independent clones, each tested in at least triplicate.
Effect of disrupting ets and Sp1 transcription factor–binding sites and an intact RARE on RA induction of the full-length promoter.
RA induction of the full-length promoter is dependent on both functional ets and Sp1 transcription factor–binding sites and an intact RARE. CD18(−918)/luc or the corresponding DNA constructs with ets-site mutations (indicated by X; panel A) or Sp1-site mutations (indicated by X; panel B) were stably transfected into U937 cells, and responsiveness to RA was measured, as described in the legend to Figure 3. All data represent the mean and standard error of at least 3 independent clones, each tested in at least triplicate.
The RARE and CD18 proximal promoter act independently to mediate RA induction
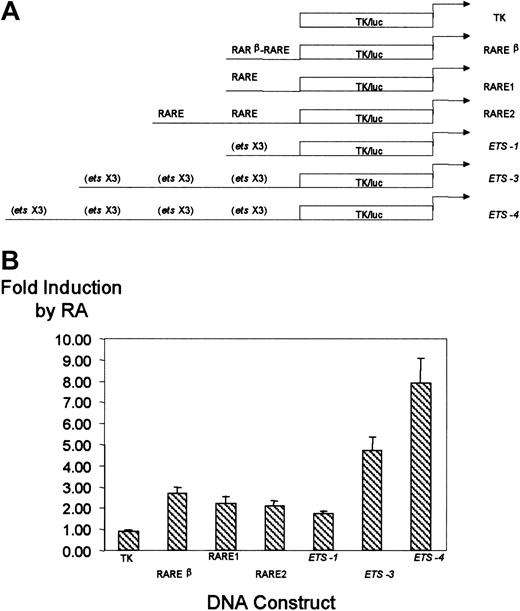
We sought to determine if the RA-responsive elements of the CD18 promoter could confer RA responsiveness on a heterologous promoter. We cloned CD18 promoter elements upstream of the Herpes simplex virus. Thymidine kinase minimal promoter linked to the luciferase gene (TK/luc), which, itself, is unresponsive to RA. A 50-nt fragment of the CD18 promoter that includes the distal RARE was cloned either as a monomer or as a tandem repeat (Figure6A). Similarly, a fragment of the CD18 minimal promoter that includes the cluster of 3 ets sites was cloned as a monomer, trimer, or tetramer upstream of TK/luc. U937 cells were stably transfected with these CD18 constructs; with the negative control, pTK81/luc; or with the RA-responsive plasmid, RARβ RARE-TK/luc. Each DNA construct was used to prepare stable lines, and 3 independent clones of each construct were examined.
Effect of CD18 RARE and proximal promoterets sites on RA responsiveness of a heterologous promoter.
The CD18 RARE and proximal promoter ets sites function independently to confer RA responsiveness to a heterologous promoter. (A) Diagrammatic representation of the TK/luc promoter, RARβ-RARE-TK/luc, RARE1-TK/luc, RARE2-TK/luc,ets-1–TK/luc, ets-3–TK/luc, andets-4–TK/luc. (B) The indicated promoter constructs were stably transfected into U937 cells, and responsiveness to RA was measured, as described in the legend to Figure 3. All data represent the mean and standard error of at least 3 independent clones, each tested in at least triplicate.
Effect of CD18 RARE and proximal promoterets sites on RA responsiveness of a heterologous promoter.
The CD18 RARE and proximal promoter ets sites function independently to confer RA responsiveness to a heterologous promoter. (A) Diagrammatic representation of the TK/luc promoter, RARβ-RARE-TK/luc, RARE1-TK/luc, RARE2-TK/luc,ets-1–TK/luc, ets-3–TK/luc, andets-4–TK/luc. (B) The indicated promoter constructs were stably transfected into U937 cells, and responsiveness to RA was measured, as described in the legend to Figure 3. All data represent the mean and standard error of at least 3 independent clones, each tested in at least triplicate.
As expected, the pTK/luc negative control was not responsive to RA (Figure 6B). The positive control, RARβ RARE-TK/luc, was activated 2.7-fold ± 0.3-fold in this stable transfection assay. The RARE1 and RARE2 constructs, which included 1 or 2 copies of the upstream CD18 RARE, respectively, were activated approximately 2-fold. Similarly, the CD18 proximal promoter ets cluster conferred RA responsiveness on pTK/luc. RA responsiveness was directly related to the number of copies of the ets cluster and ranged from 1.8- to 7.9-fold induction. Thus, both the CD18 distal RARE and the CD18 proximal promoter independently conferred responsiveness on the heterologous TK promoter. Importantly, this confirmed that the CD18 minimal promoter, which includes ets and Sp1 sites but which lacks any conventional RARE, mediates RA responsiveness.
Transcriptional coactivator p300 physically interacts with GABPα
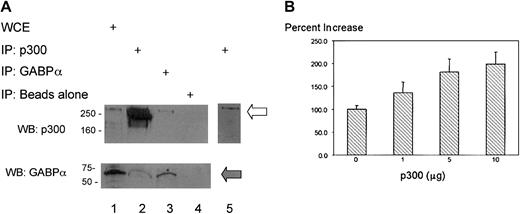
The nuclear coactivator p300/CBP physically contacts and functionally interacts with numerous transcription factors, includingets factors and retinoid receptors. We sought to determine if physical interactions between p300 and GABP might account for the RA responsiveness of the CD18 promoter. We immunoprecipitated whole-cell extracts from U937 cells with antiserum against p300 or against GABPα, and the precipitated products were immunoblotted with antisera against p300 or GABPα. Antiserum against p300 immunoprecipitated both p300 (Figure 7A; upper panel, lane 2) and GABPα (lower panel, lane 2). Similarly, antiserum against GABPα immunoprecipitated both p300 (upper panel, lane 3) and GABPα (lower panel, lane 3). Protein G beads alone did not precipitate either protein (lane 4). Thus, p300 and GABPα physically interact in U937 myeloid cells.
Physical interaction of GABP and p300 in myeloid cells and effect of p300 on transcriptional coactivation of the CD18 promoter.
Transcriptional coactivator p300 physically contacts GABP and increases responsiveness of the CD18 promoter to RA. (A) A 0.75-mg extract from U937 cells treated with 10−5 M RA for 48 hours was incubated with antibodies to p300 (lane 2), GABPα (lane 3), or no antibody (lane 4), followed by precipitation with protein-G sepharose. Immunoprecipitated products and 50 μg whole-cell extract (lane 1) were immunoblotted with antibodies to p300 and GABPα, as indicated. Open arrow indicates p300, and filled arrow indicates GABPα. Lane 5 represents a brief exposure of lane 2. WCE indicates whole cell extract; IP, immunoprecipitation; WB, Western blot. (B) U937 cells that were stably transfected with CD18(−918)/luc were transiently transfected with the indicated quantities of the transcriptional coactivator p300. Transfected cells were divided into 2 equal aliquots, which were treated with 10−5 M RA or DMSO (0.1%), and RA activation was measured 14 hours later. All data represent the mean and standard error of at least 3 independent transient transfections.
Physical interaction of GABP and p300 in myeloid cells and effect of p300 on transcriptional coactivation of the CD18 promoter.
Transcriptional coactivator p300 physically contacts GABP and increases responsiveness of the CD18 promoter to RA. (A) A 0.75-mg extract from U937 cells treated with 10−5 M RA for 48 hours was incubated with antibodies to p300 (lane 2), GABPα (lane 3), or no antibody (lane 4), followed by precipitation with protein-G sepharose. Immunoprecipitated products and 50 μg whole-cell extract (lane 1) were immunoblotted with antibodies to p300 and GABPα, as indicated. Open arrow indicates p300, and filled arrow indicates GABPα. Lane 5 represents a brief exposure of lane 2. WCE indicates whole cell extract; IP, immunoprecipitation; WB, Western blot. (B) U937 cells that were stably transfected with CD18(−918)/luc were transiently transfected with the indicated quantities of the transcriptional coactivator p300. Transfected cells were divided into 2 equal aliquots, which were treated with 10−5 M RA or DMSO (0.1%), and RA activation was measured 14 hours later. All data represent the mean and standard error of at least 3 independent transient transfections.
RA responsiveness of the CD18 promoter increased by p300
We transiently transfected the p300 expression plasmid into U937 cells that were stably transfected with CD18(−918)/luc. The responsiveness of the CD18 promoter to RA was doubled by transfection with p300 and was directly dependent on the amount of cotransfected p300 (Figure 7B). Thus, p300 acts as a transcriptional coactivator and increases the activity of the chromatin-integrated CD18 promoter in response to RA treatment. We conclude that p300/CBP physically interacts with GABPα and functionally cooperates with it to increase the transcriptional response of the CD18 promoter to RA.
Discussion
RA plays important roles in both normal and malignant (leukemic) myeloid differentiation. RA induces granulocytic differentiation of myeloid cell lines3; whereas mutant RAR blocks HL-60 granulocytic differentiation, expression of wild-type RAR rescues this defect.4 Rearrangements of RAR, such as PML-RAR and PLZF-RAR, are causally associated with APL,5 and treatment with RA can induce remissions in the majority of APL patients.6 Expression in mice of these chimeric RAR proteins generates syndromes that resemble APL.21 Thus, defining the mechanisms by which RA induces myeloid differentiation has important implications for normal myeloid biology and for the origins of myeloid leukemia.
CD18 is transcriptionally regulated by RA,15 but the mechanism by which RA activates CD18 expression was not previously defined. We have now identified a novel mechanism by which RA transcriptionally activates CD18. We identified a region of CD18 that resembles a conventional RARE that is bound by RARα and RXRα (Figure 2). This element resembled conventional RAREs that were previously identified in other myeloid genes, such as CCAAT/enhancer–binding protein–ε (C/EBPε)22and E3.23 This element was required for full transcriptional activation of CD18, but it accounted for only one half of the CD18 response to RA (Figure 3). Unexpectedly, we found that binding sites for GABP and Sp1 in the CD18 minimal promoter were required for full RA responsiveness of CD18 (Figures 4 and 5), even though this region was not bound by nuclear hormone receptors. These sites conferred RA responsiveness on the otherwise unresponsive TK promoter, and this effect was dependent on the number of etssites (Figure 6). Together, the distal RARE and the CD18 proximal promoter functioned cooperatively to mediate the full transcriptional response to RA. These findings indicate a novel mechanism of transcriptional regulation by RA in myeloid cells that uses nonsteroid receptors to activate gene expression. These data add to a growing body of literature that describes complex interactions between nuclear hormone receptors, ets factors, and Sp1 in the activation of gene expression in response to retinoids and other hormones.
Ets factors and Sp1 cooperatively interact to transcriptionally activate several myeloid genes. We previously showed that GABP and Sp1 are critical regulators of CD18 transcription and that they functionally interact to regulate its expression.17 Similarly, Sp1 and ets factors regulate a distal enhancer of the neutrophil elastase gene.24 PU.1 and Sp1 activate the promoters of the CD18 integrin partner proteins CD11c25 and CD11b,26 and other myeloid genes, such as c-fes,27p47phox,28 and the macrophage mannose receptor.29Ets factors and Sp1 cooperate to mediate the response of the HIV long terminal repeat (LTR) to lipopolysaccharide in macrophages.30 Thus, binding by ets factors and Sp1 is necessary for expression of several myeloid genes.
There is increasing evidence that hormone responsiveness is not mediated by steroid receptors alone. Sp1 and RARs cooperate to transcriptionally activate thrombomodulin31 and retinol-binding protein.32 Sp1 also functionally interacts with RARs to activate p21 and NGF1-A,33,34 and it physically interacts with RARs on the urokinase promoter35and the interleukin 1B (IL-1B) promoter.36 These studies indicate that hormone and vitamin responsiveness is mediated by the combinatorial action of Sp1 and ets factors, and that protein-protein interactions between RARs and other transcription factors regulate the response of numerous genes to RA.
How might transcription factors such as GABP and Sp1 mediate RA responsiveness of CD18? One possible mechanism is that RA might alter the expression or activity of these transcription factors. GABP is an obligate multimeric transcription factor that consists of 2 distinct proteins.37,38 GABPα is an ets-related transcription factor that binds to DNA in a sequence-specific manner. GABPα recruits GABPβ, which has a glutamine-rich transcriptional activation domain in its carboxy terminus. Together, GABPα and GABPβ form a transcriptionally active complex.39 40
GABPβ undergoes alternative splicing to generate 4 distinct isoforms that have different transcriptional activation and multimerization properties.41,42 For example, GABPβ isoforms differ in their ability to interact with the transcription factor E2F1.43 Similarly, GABPβ isoforms differentially interact with the recently described transcriptional corepressor, YEAF-1.44 RA-induced granulocytic differentiation of HL-60 myeloid cells causes substantial changes in the expression pattern of GABPβ isoforms. Expression of GABPα and Sp1 does not change during granulocytic differentiation (A.G.R., J. Shang, M. Luo, and A. Hebert, manuscript in preparation). Thus, changes in the expression of GABPβ isoforms during granulocytic differentiation might partially account for the RA-induced increase in CD18 transcription.
An alternative mechanism by which GABP and Sp1 might mediate RA responsiveness of the CD18 proximal promoter is through protein-protein interactions. The transcriptional coactivators p300 and CBP appear to integrate intracellular signaling by interacting with several classes of transcription factors, including ets factors and nuclear hormone receptors.45 We showed that p300 amplified the responsiveness of CD18 to RA (Figure 7B). Glutathione-S-transferase (GST)–GABPα fusion protein was previously shown to physically interact with p300 in vitro (Bannert et al46 and our unpublished data, September 2001). We have now shown that in U937 cells GABPα and p300 physically interact in vivo (Figure 7A). These data suggest that GABP mediates the RA responsiveness of CD18 by binding to p300 and thereby recruiting RARs to the CD18 proximal promoter.
In myeloid cells, p300 might serve as a platform for the assembly of a multiprotein complex, or enhanceosome.47 Such a multiprotein complex might recruit RARs bound to the distal RARE to the vicinity of the CD18 proximal promoter. Interestingly, we found that CD18 is responsive to RA in stable transfection assays (Figures 3-6), but that it is not responsive to RA in transient transfection (data not shown). Thus, chromatin assembly may play a role in the higher-order protein complex formation by which RA mediates responsiveness of CD18. We conclude that RA responsiveness of CD18 is critically dependent on both GABP and Sp1 in the proximal promoter, and RARs on the distal RARE. These 2 regulatory regions may cooperate via protein-protein interactions with common transcriptional coactivators to mediate RA responsiveness of CD18 in myeloid cells.
We thank Jonathan Licht for the pCMVβp300 expression plasmid.
Suppported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant R29 DK 44728 (A.G.R.); American Cancer Society grant RPG-92-002-04-DHP (A.G.R.); and the Herbert W. Savit '49 Endowed Research Fund (A.G.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alan G. Rosmarin, The Miriam Hospital, Research 215, 164 Summit Ave, Providence, RI 02906; e-mail:rosmarin@brown.edu.