Abstract
Chronic myeloid leukemia (CML) is a clonal disease of hematopoietic stem cells caused by a reciprocal translocation of the long arms of chromosomes 9 and 22. In human leukocyte antigen A*0201+ (HLA-A*0201+) individuals, response after interferon-α (IFN-α) was shown to be associated with the emergence of CML-specific cytotoxic T cells that recognize PR-1, a myeloblastin (MBN)–derived nonapeptide. In contrast, imatinib potently induces remissions from CML by specific inhibition of the ABL tyrosine kinase. Here, we explored molecular regulations associated with CML responses under different treatment forms using cDNA-array. Expression of MBN was found to be down-regulated in remission under imatinib therapy (0 of 7MBN+ patients). In contrast, MBNtranscription was readily detectable in the peripheral blood in 8 of 8 tested IFN-α patients in complete remission (P = .0002). IFN-α–dependent MBNtranscription was confirmed in vitro by stimulation of peripheral blood mononuclear cells (PBMCs) with IFN-α and by IFN-α–mediated activation of the MBN promoter in reporter gene assays. Finally, with the use of HLA-A*0201–restricted,MBN-specific tetrameric complexes, it was demonstrated that all of 4 IFN-α–treated patients (100%), but only 2 of 11 imatinib patients (19%), in complete hematological or cytogenetic remission developed MBN-specific cytotoxic T cells (P = .011). Together, the induction of MBNexpression by IFN-α, but not imatinib, may contribute to the specific ability of IFN-α to induce an MBN-specific T-cell response in CML patients. This also implies that the character of remissions achieved with either drug may not be equivalent and therefore a therapy modality combining IFN-α and imatinib may be most effective.
Introduction
Chronic myeloid leukemia (CML) is a clonal disease of hematopoietic stem cells caused by a reciprocal gene translocation t(9;−22) (q34;q11), which creates the Philadelphia chromosome and results in the expression of a leukemia-specific oncoprotein,BCR-ABL.1,2BCR-ABL has been shown to activate various signal transduction pathways, such as theRAS-, STAT-, and PI3K-signaling pathways, leading to transformation, decreased apoptosis, loss of adhesive properties, and genetic instability.2,3 Even though there is solid evidence that BCR-ABL is the causative aberration in CML,4-6 other genetic events, such as decreased expression of the interferon-regulatory genesICSBP and IRF4, have also been shown to be implicated in the molecular pathogenesis of CML.7-10
Without allogeneic stem cell transplantation (SCT), CML inevitably progresses from a benign chronic phase to a fatal blast crisis.2 Among the established conventional CML-treatment regimens,11 interferon-α (IFN-α) has been demonstrated to significantly improve survival.3 This was especially apparent in patients where a cytogenetic remission could be achieved. The molecular mechanisms behind IFN-α responsiveness are not fully understood. However, the detection of minimal residual disease even in patients with cytogenetic remissions suggests that IFN-α allows control, rather than eradication, of CML.12A novel tyrosine kinase inhibitor, imatinib, has been found to induce high remission rates even in CML blast crisis and in IFN-α–resistant patients.13-16 Despite these very promising results, the long-term efficacy of imatinib cannot be predicted at this point, and among the available treatment options, currently only allogeneic SCT can cure CML.2 Cytotoxic lymphocytes (CTLs), which recognize distinct antigeneic peptides on CML cells, contribute greatly to the cure after SCT.17 Previously,BCR/ABL–specific peptides, and the PR1 nonapeptide, were found to elicit a CML-specific CTL response.18-22 PR1 is a peptide derived from myeloblastin (MBN), a 26-kDa serine protease, also known as proteinase 3.23MBN is abundantly expressed in azurophil granules of normal myeloid cells and is substantially overexpressed in certain immature myeloid leukemia cells,24 where it may be important for the maintenance of a leukemic phenotype.25 PR1-specific CTLs (PR1-CTLs) lysed and inhibited the proliferation of CML cells, but not of normal myeloid precursors, in a human leukocyte antigen A*0201 (HLA-A*0201)–restricted manner.18,19 Moreover, the association between the emergence of PR1-CTLs and a response to IFN-α in vivo strongly implied that IFN-α can induce remissions via induction of a CML-specific PR1-CTL response,26 although cause and effect of this correlation was not elucidated in this study.
Here, we investigated molecular and immunological mechanisms of a CML response under imatinib and IFN-α treatment.
Patients and methods
Patients and donors
Peripheral blood samples of healthy donors and patients treated with imatinib or IFN-α were obtained after informed consent was given. Imatinib patients were treated within 2 consecutive multicenter phase 2 protocols for CML patients in chronic phase. IFN-α–treated patients obtained a standard IFN-α–based treatment regimen.11 Nine of the 11 patients had been pretreated with IFN-α (2 months to 5 years) and were switched to imatinib because of (1) failure to achieve cytogenetic remissions (3 patients); (2) failure to maintain response (1 patient); or (3) IFN-α intolerance (5 patients). Treatment centers were the Mannheim or Marburg University clinic (Germany). High-resolution HLA testing was performed by the certified HLA-Laboratory at the University of Marburg. Only those patients with a confirmed HLA-A*0201 genotype were studied with the use of PR1-specific tetrameric complexes. The study protocol for the clinical study (STI 106) as well as the sample collection in our Mannheim and Marburg centers, have been approved by the local ethics committees.
Cell lines and in vitro stimulation
Cell lines U937, K562, Raji, and Jurkat were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) (Braunschweig, Germany). All cell lines were maintained at 5% CO2 in RPMI 1640 medium with 1% glutamine (Gibco/BRL, Eggenstein, Germany) supplemented with 10% FCS (Gibco/BRL), 1% penicillin/streptomycin (Biochrom, Berlin, Germany). Stimulation experiments were performed with 1000 to 1500 U/mL IFN-α (Biochrom) or 0.2 to 1 μM imatinib for up to 48 hours.
Hybridization by cDNA array
For hybridization of Atlas 1.2 human cDNA arrays (Clontech, Heidelberg, Germany), 33P-labeled cDNAs were prepared from 0.6 to 1.2 μg total RNA. RNA samples were mixed with 1 μL Atlas 1.2 CDS primer mix (Clontech) in a total volume of 10 μL and incubated at 70°C for 10 minutes. To 10 μL annealing reaction, 4 μL 5× Superscript reverse transcription buffer, 2 μL 10× deoxynucleoside triphosphate (dNTP) mix for deoxyadenosine triphosphate (dATP)–label, 3 μL α–33P-dATP, 2 μL 100 mM dithiothreitol (DTT), and 2 μL Superscript reverse transcriptase (50 U/μL) (GibcoBRL) were added, and samples were incubated at 42°C for 40 minutes. Samples were heated to 85°C for 3 minutes and, after the addition of 1 μL Superscript reverse transcriptase, were incubated again at 42°C for 30 minutes. Reactions were stopped with 2 μL termination mix (Clontech), and unincorporated nucleotides were removed by means of NucleoSpin Columns (Clontech) following the manufacturer's protocol. Hybridizations, phosphoimaging, and analysis of array hybridization data were done as previously described.27
Cell sorting and enrichment
Peripheral blood mononuclear cells (PBMCs) were separated from peripheral blood of IFN-α– or imatinib–treated patients by Ficoll density gradient centrifugation using Ficoll-Hypaque (1.077 g/dL Lymphoprep) (Nycomed Pharma, Oslo, Norway). CD15+ populations were enriched from peripheral blood after red cell lysis with the use of MiniMACS and directly labeled CD15 MicroBeads (Miltenyi Biotech, Bergisch Gladbach, Germany) as recommended by the manufacturer. The purity of the isolated CD15+ population ranged between 85% and 95%. CD3, CD14, and CD19 cell fractions were separated from peripheral blood of healthy volunteers and CML patients by means of a MoFlo cell sorter (Cytomation, Fort Collins, CO). The purity of the obtained fractions was verified to be 94% to 98%. Between 2 × 105 and 1 × 106 cells of each sorted fraction were subjected to RNA extraction.
RNA isolation and cDNA synthesis
RNA was extracted from whole peripheral blood; PBMCs; and MACS-enriched CD15 cells and CD3+-, CD14+-, and CD19+-sorted cell fractions from CML patients and healthy donors; stimulated PBMCs; and sorted cell fractions, respectively, with the use of the Qiagen RNA extraction kit (Hilden, Germany) as recommended by the manufacturer. Total RNA was then quantified, and equal amounts were reverse transcribed into cDNA as previously described.7-9
Polymerase chain reaction (PCR)
PCR was performed with the use of 1 μL (approximately 50 ng) single-stranded cDNA, essentially as previously described.7-9 The cycling conditions were 94°C for 2 minutes for denaturation; then 94°C for 1 minute, 55°C (for β-actin) or 61°C (for MBN) for 1 minute, 72°C for 1 minute for 21 (for actin) or 31 cycles (forMBN), followed by 90°C for 1 minute and 60°C for 10 minutes. The sequences of the primers are as follows: β-actin sense primer, 5-CCTTCCTGGGCATGGAGTCCT-3; β-actin reverse primer, 5-AATCTCATCTTGTTTTCTGCG-3, which results in a 407-bp PCR product; MBN sense primer, 5-CCTGCAGATGCGGGGGAACC-3; MBN reverse primer, 5-GTGAAAGCAGGGAGCGGCGTT-3, which results in a 452-bp PCR product. The products were electrophoresed on a 3% agarose gel. Gels were stained with ethidium bromide and photographed. BCR/ABL+minimal residual disease (MRD) was determined by quantitative real-time PCR as described.12
MBN reporter constructs
The upstream 677-bp fragment of the human MBNpromoter was PCR-amplified from genomic DNA extracted from peripheral blood of a healthy donor. Primers containing specific restriction sites on their 5′ ends were used to insert the fragment into the pGL3-Basic firefly luciferase reporter vector (Promega, Madison, WI): forward 5′-(KpnI)-TTC TCT GGG GCA GGC CCG TCC-3′; reverse 5′-(SacI)-TGG TGG GGT CCA GGG TGC ACC-3′. Products were sequenced on an automated sequencer (LI-COR) (MWG Biotech, Munich, Germany) to confirm sequence and orientation of the cloned product.
Reporter gene assays
MBN-promoter activation was measured by means of the dual luciferase assay (Promega) as described.28Briefly, 5 nM MBN-reporter construct, containing the firefly luciferase gene under control of the human MBN promoter, and the transfection control construct expressing the renilla luciferase gene, were transiently coexpressed by electroporation.7The control construct served as an internal reference for the efficiency of transfection and expression. At 24 hours after transfection, the medium was replaced by medium containing 1000 U/mL IFN-α or 0.2 to 1 μM imatinib. Luciferase activity was measured after 48 hours with an LB 96 P microlumat (EG&G Berthold, Bad Wildbad, Germany). MBN-specific promoter activation was quantified as a ratio of measured firefly light units (flus) relative to renilla luciferase units (rlus) and presented as fold stimulation to the unstimulated control. Each experiment was done at least 3 times.
PR1-CTL measurement
PR1-CTLs were measured by means of iTAg major histocompatibility complex (MHC) tetramers (Beckman Coulter, San Diego, CA). The iTAg MHC tetramers consisted of 4 HLA-A*0201 MHC class I molecules, each bound toMBN nonapeptide VLQELNVTV (PR1) and conjugated to phycoerythrin (PE), thus allowing the detection of CD8+ T cells specific for PR1. Detection of PR1-specific T cells was done according to the recommendations of the manufacturer. Briefly, 10 μL iTAg MHC tetramers and fluorescein isothiocyanate (FITC)–labeled CD8-specific monoclonal antibodies (Becton Dickinson, Heidelberg, Germany) were added per 100 μL whole blood, mixed, and incubated for 30 minutes at room temperature. Erythrocytes were then lysed with the use of red cell lysis buffer (OptiLyse; Beckman Coulter). Remaining cells were washed twice with cold phosphate-buffered saline (PBS), pelleted, resuspended, and measured on a FACScan (Becton Dickinson). The instrument was compensated with the isotype-matched control antibodies, and PBMCs of an HLA-A*0201+ patient (unidentified patient number 30 [UPN 30]) treated with IFN-α for more than 5 years served as positive control (Figure 5). Data were analyzed by means of CellQuest analysis software (Becton Dickinson).
Statistical analysis
Assessment of the statistical significance of the presence of PR1-CTLs and MBN expression in imatinib-treated versus IFN-α–treated patients was done by means of the Fisher exact test and an Apple Macintosh PowerBook with the use of StatView 4.5 (Abacus Concepts, Berkeley, CA). P < .05 was considered statistically significant.
Results
Differential regulation of myeloblastin expression under imatinib and IFN-α
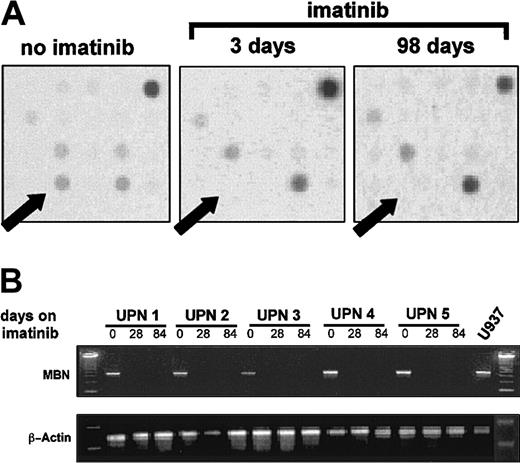
The gene-expression profile of 2 CML patients before, as well as 3 and 98 days after, initiation of imatinib treatment was analyzed by means of cDNA microarrays. MBN expression was found to be lost after initiation of imatinib therapy (Figure1A). This was confirmed by RT-PCR in 5 more patients in complete hematological remission (hCR) on imatinib therapy (Figure 1B).
MBN expression after initiation of imatinib therapy.
(A) A cDNA-array plot. A patient's mRNA before, as well as 3 and 98 days after, initiation of imatinib therapy was isolated, reverse transcribed, radiolabeled, and then used to probe Atlas 1.2 human cDNA arrays. Arrows indicate the dot location at which theMBN-specific DNA was deposited on the array membrane. The intensity of the signal dots corresponds with expression levels in the tested sample. Plots are representative of arrays on 2 individual CML patients treated with imatinib. A comparable loss of the MBN spot intensity was seen in both tested patients. (B) Regulation of MBN transcript levels under imatinib therapy as assessed by reverse transcriptase–PCR (RT-PCR). RNA was isolated from 5 patients in chronic phase of CML at diagnosis and at 4 and 12 weeks after start of imatinib therapy. At this time, at least a complete hematological remission was achieved. Lower panel: β-actin PCR was used to assess equal loading and integrity of RNA.
MBN expression after initiation of imatinib therapy.
(A) A cDNA-array plot. A patient's mRNA before, as well as 3 and 98 days after, initiation of imatinib therapy was isolated, reverse transcribed, radiolabeled, and then used to probe Atlas 1.2 human cDNA arrays. Arrows indicate the dot location at which theMBN-specific DNA was deposited on the array membrane. The intensity of the signal dots corresponds with expression levels in the tested sample. Plots are representative of arrays on 2 individual CML patients treated with imatinib. A comparable loss of the MBN spot intensity was seen in both tested patients. (B) Regulation of MBN transcript levels under imatinib therapy as assessed by reverse transcriptase–PCR (RT-PCR). RNA was isolated from 5 patients in chronic phase of CML at diagnosis and at 4 and 12 weeks after start of imatinib therapy. At this time, at least a complete hematological remission was achieved. Lower panel: β-actin PCR was used to assess equal loading and integrity of RNA.
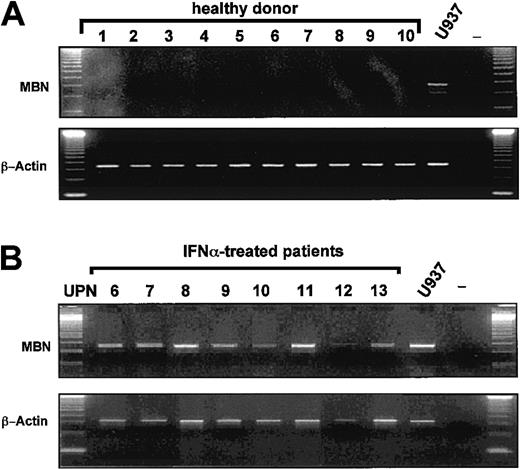
Because peripheral white blood cells of healthy donors (n = 10) wereMBN− by RT-PCR (Figure2A), we hypothesized that loss ofMBN expression in CML patients on imatinib was due to a clearance of MBN+ CML cells24from the peripheral blood. Unexpectedly, however, as compared with 0 of 7 imatinib-treated patients, 8 of 8 CML patients in major (MR) or complete cytogenetic remission (CR) under IFN-α wereMBN+ (Figure 2B) (P = .0002).
MBN transcription in healthy donors and IFN-α–treated patients as assessed by RT-PCR.
The lower blots in each panel display transcription levels of the housekeeping gene β-actin as reference gene for healthy donors and IFN-α patients. MBN+ U937 cells served as a positive control cell line. (A) Lack of MBNtranscription in the peripheral blood of healthy donors (1 to 10); positive control (U937), and PCR negative control. (B) MBNtranscript levels of IFN-α–treated CML patients in major or complete cytogenetic remission UPN6-13. Positive and negative controls, U937 and —.
MBN transcription in healthy donors and IFN-α–treated patients as assessed by RT-PCR.
The lower blots in each panel display transcription levels of the housekeeping gene β-actin as reference gene for healthy donors and IFN-α patients. MBN+ U937 cells served as a positive control cell line. (A) Lack of MBNtranscription in the peripheral blood of healthy donors (1 to 10); positive control (U937), and PCR negative control. (B) MBNtranscript levels of IFN-α–treated CML patients in major or complete cytogenetic remission UPN6-13. Positive and negative controls, U937 and —.
The possibility that differences in the MBN-expression levels of imatinib and IFN-α patients were due to variations in the blood differential or residual circulating immature CML cells could be ruled out, because both treatment groups were at least in hCR at the time of RT-PCR analysis and because quantitation of the BCR/ABL transcript levels from the peripheral blood revealed that MBN+ IFN-α patients (n = 6) had a significantly lower MRD than MBN−imatinib patients in hCR (n = 5) (P = .012) (Table1). Clinical characteristics of both patient groups are given in Table 1.
IFN-α treatment induces myeloblastin-transcription in CD14+ monocytes in vivo and in vitro
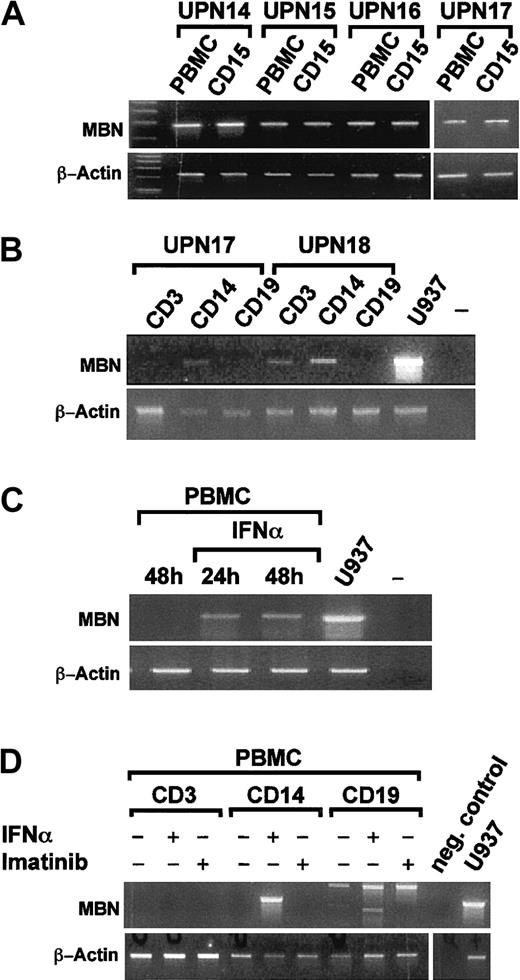
We next evaluated in which white blood cell compartmentMBN was induced by IFN-α. RT-PCR from Ficoll-enriched mononuclear cells (PBMCs) and CD15-enriched granulocytes of 4 CML patients in hCR under IFN-α revealed that both compartments wereMBN+ (Figure 3A). A more detailed analysis of sorted CD3, CD14, and CD19 PBMC subpopulations (n = 2) in hCR under IFN-α revealed that theMBN message could be detected in CD14+ monocytes and, in one patient, also in CD3+ T cells (Figure 3B). We then tested whether MBN transcription was also induced by IFN-α in vitro. PBMCs of healthy donors (n = 2) were stimulated with IFN-α for 24 and 48 hours (Figure 3C) or separated into CD3+, CD19+, and CD14+ cell fractions (n = 2) and then stimulated in vitro with IFN-α or imatinib. This resulted in the activation of MBNtranscription in PBMCs, specifically in CD14+ monocytes, by IFN-α but not imatinib (Figure 3D). Thus IFN-α, but not imatinib, induced MBN transcription in vivo and in vitro.
MBN transcription in sorted cell populations of IFN-α–treated patients and after in vitro stimulation of PBMCs by RT-PCR.
(A) PBMC- and MACS-enriched CD15+ cell fractions of 4 patients (UPNs 14-17) in complete hematological remission under IFN-α therapy. (B) Sorted CD14+ monocytic, CD3+T-lymphocytic, and CD19+ B-lymphocytic cell populations, as indicated, of 2 patients (UPN 17 and UPN 18) in complete hematological remission. (C) PBMCs of a healthy donor were cultured 48 hours without and for 24 hours or 48 hours in the presence of 1500 U/mL IFN-α. Also shown are unstimulated U937 positive control (U937) and a no-template control (–). (D) CD3, CD14, and CD19 cell populations were sorted on a Moflo cell sorter. Then, 0.2 to 0.3 × 106cells of each population were seeded into media supplemented with 1500 U/mL IFN-α, 1 μM imatinib, or no supplements as indicated. At 24 hours after treatment, cells were harvested and MBN was expression assessed by RT-PCR. Panels B and C are each representative for experiments performed with material from 2 distinct donors/patients. For reference, β-actin gene transcription was assessed as depicted.
MBN transcription in sorted cell populations of IFN-α–treated patients and after in vitro stimulation of PBMCs by RT-PCR.
(A) PBMC- and MACS-enriched CD15+ cell fractions of 4 patients (UPNs 14-17) in complete hematological remission under IFN-α therapy. (B) Sorted CD14+ monocytic, CD3+T-lymphocytic, and CD19+ B-lymphocytic cell populations, as indicated, of 2 patients (UPN 17 and UPN 18) in complete hematological remission. (C) PBMCs of a healthy donor were cultured 48 hours without and for 24 hours or 48 hours in the presence of 1500 U/mL IFN-α. Also shown are unstimulated U937 positive control (U937) and a no-template control (–). (D) CD3, CD14, and CD19 cell populations were sorted on a Moflo cell sorter. Then, 0.2 to 0.3 × 106cells of each population were seeded into media supplemented with 1500 U/mL IFN-α, 1 μM imatinib, or no supplements as indicated. At 24 hours after treatment, cells were harvested and MBN was expression assessed by RT-PCR. Panels B and C are each representative for experiments performed with material from 2 distinct donors/patients. For reference, β-actin gene transcription was assessed as depicted.
Myeloblastin promoter activation by IFN-α in U937 cells
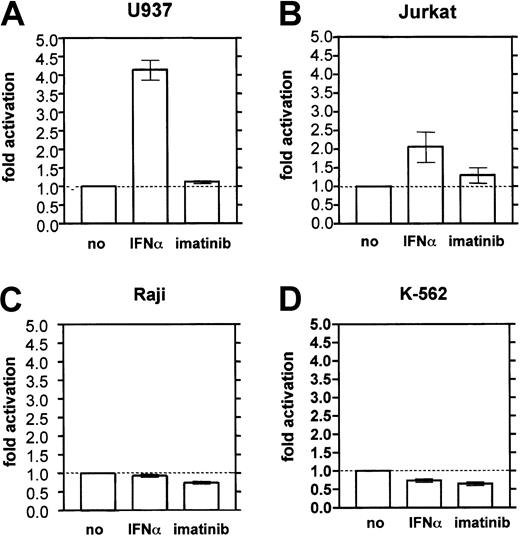
To assess whether IFN-α transactivates the MBNpromoter, MBN-reporter gene assays were performed. In the monocytic cell line U937, a 48-hour treatment with IFN-α resulted in a 4.2-fold increase of the MBN promotor activity relative to unstimulated control cells (Figure4A). IFN-α also moderately activated the MBN promoter by 2-fold above background in Jurkat T cells (Figure 4B), but had no effect in Raji B cells (Figure 4C) or in erythroleukemic K562 cells (Figure 4D). Imatinib did not significantly activate the MBN promoter in any of the tested cell systems (Figure 4).
MBN promoter reporter gene assay in hematopoietic cell lines.
Cells were stimulated with IFN-α (1000 U/mL) or imatinib (1 μM for U937, Raji, Jurkat; 0.2 μM for K562) for 48 hours and harvested for dual luciferase measurement. The results were calculated as the ratio of measured firefly light units (flus) relative to renilla luciferase units (rlus) and presented as fold stimulation compared with the unstimulated control. Each experiment was done at least 3 times. (A) U937 monocytes. (B) Jurkat T cells. (C) Raji B cells. (D) K562 erythroleukemic cells.
MBN promoter reporter gene assay in hematopoietic cell lines.
Cells were stimulated with IFN-α (1000 U/mL) or imatinib (1 μM for U937, Raji, Jurkat; 0.2 μM for K562) for 48 hours and harvested for dual luciferase measurement. The results were calculated as the ratio of measured firefly light units (flus) relative to renilla luciferase units (rlus) and presented as fold stimulation compared with the unstimulated control. Each experiment was done at least 3 times. (A) U937 monocytes. (B) Jurkat T cells. (C) Raji B cells. (D) K562 erythroleukemic cells.
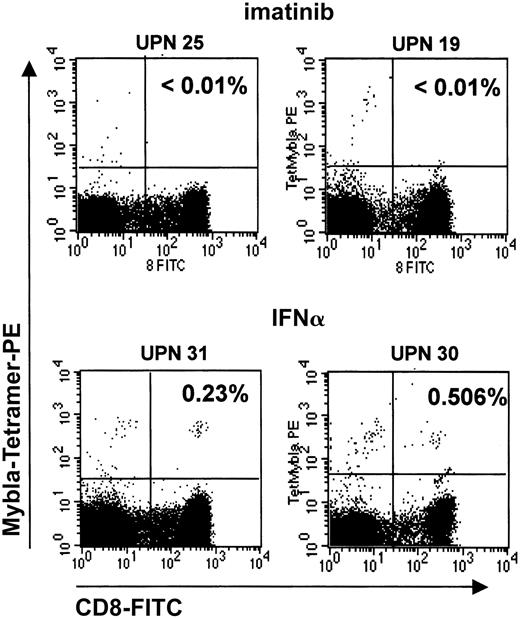
Remissions under imatinib are rarely associated with the emergence of myeloblastin-specific T cells
IFN-α induces a CML-specific CTL response, which is based on the recognition of an HLA-A*0201–presented MBN-derived peptide, PR1.18 19 We speculated that the induction ofMBN in CD14+ monocytes as weak antigen-presenting cells (APCs) and in other myeloid cells by IFN-α might promote the generation of an MBN-specific T-cell response. In turn, lack of MBN expression in imatinib-treated patients would imply a reduced likelihood of inducing MBN-specific CTL responses. To address this question, we compared the PR1-CTL frequencies in the peripheral blood of imatinib versus IFN-α patients in hCR or MR using PR1-specific tetramers. Indeed, imatinib treatment of HLA-A*0201+ CML patients was associated with the generation of a PR1-CTL response only in 2 of 11 (19%) eligible patients (Table2), as compared with IFN-α treatment, where 4 of 4 patients had developed PR1-CTLs. This difference was statistically significant (P = .011), and it is unlikely that it can be attributed to a short imatinib treatment period, because at the time of this analysis patients had on average been on imatinib for longer than 1 year (13.1 ± 4.6 months). Interestingly, 1 of the 2 patients who had developed PR1-CTLs under imatinib was in CR (Table 2). Figure5 shows representative dot plots of PR1-CTL+ and PR1-CTL− patients.
PR1-CTLs in peripheral blood of imatinib-treated CML patients.
PR1-CTLs are rarely found in peripheral blood of imatinib-treated CML patients. PR1-CTLs were measured by means of iTAg MHC tetramers (Beckman Coulter). PE-conjugated iTAg MHC tetramers and FITC-labeled CD8-specific monoclonal antibodies (Becton Dickinson) were added to 100 μL whole blood, mixed, and incubated for 30 minutes at room temperature. Erythrocytes were lysed, and the remaining cells were measured on a FACScan (Becton Dickinson). Data were analyzed by means of CellQuest analysis software. Percentages of cells that stain for both CD8 and PR1 are given in the upper right quadrants. The 2 upper plots depict representative PR1-CTL− patients (UPNs 25 and 19) in complete remission under imatinib treatment; 2 lower plots depict 2 PR1-CTL+, IFN-α–treated patients (UPNs 31 and 30) in complete hematological remission.
PR1-CTLs in peripheral blood of imatinib-treated CML patients.
PR1-CTLs are rarely found in peripheral blood of imatinib-treated CML patients. PR1-CTLs were measured by means of iTAg MHC tetramers (Beckman Coulter). PE-conjugated iTAg MHC tetramers and FITC-labeled CD8-specific monoclonal antibodies (Becton Dickinson) were added to 100 μL whole blood, mixed, and incubated for 30 minutes at room temperature. Erythrocytes were lysed, and the remaining cells were measured on a FACScan (Becton Dickinson). Data were analyzed by means of CellQuest analysis software. Percentages of cells that stain for both CD8 and PR1 are given in the upper right quadrants. The 2 upper plots depict representative PR1-CTL− patients (UPNs 25 and 19) in complete remission under imatinib treatment; 2 lower plots depict 2 PR1-CTL+, IFN-α–treated patients (UPNs 31 and 30) in complete hematological remission.
Discussion
Myeloblastin, also known as proteinase 3, plays a central role in the generation of a CML-specific CTL response.18,19 26Here, we demonstrate 3 key findings. First, remissions from CML under IFN-α and imatinib therapy are associated with a differential regulation of MBN expression. Whereas patients who responded to imatinib or healthy blood donors did not express detectable levels of MBN, IFN-α–treated patients readily expressedMBN in all peripheral white blood compartments. Second, it was found that MBN transcription is induced by the transactivating effects of IFN-α on the MBN promoter. Finally, it is demonstrated that IFN-α–treated patients regularly developed MBN-specific CTLs, whereas this was seen in fewer than 20% of the patients in remission under imatinib. This suggests that remissions from CML under imatinib and IFN-α are based on distinct effector mechanisms, which may be clinically relevant.
Using a cDNA array approach, we initially identified MBN as a down-regulated target gene under imatinib therapy (Figure 1). Loss ofMBN expression was conceivable, because imatinib eradicates immature, MBN+ CML cells24(Figure 2A). In contrast, IFN-α–treated patients in remission expressed MBN in the polymorphonuclear and in CD14+ PBMC fraction (Figure 3). This MBNpositivity was not due to potential BCR/ABL+residual disease, because the IFN-α patients had significantly less MRD compared with imatinib patients, who wereMBN− (P = .012) (Table 1). We concluded that IFN-α treatment activates MBN expression in vivo. This hypothesis could be confirmed in vitro by stimulation of unsorted PBMCs and sorted CD14+ monocytes of healthy donors with IFN-α (Figure 3). The activation of the MBN promoter by IFN-α in monocytic U937 cells and also, though less pronounced, in Jurkat T cells provided a molecular mechanism for this regulation (Figure 4). However, IFN-α had no transactivating effects when the same luciferase reporter constructs were tested in erythroleukemic K562 cells and in Raji B cells. The exact reason for a cell line–specific potential of IFN-α to activate MBN transcription cannot be deduced from this study. The MBN promoter does not contain a bona fide interferon-stimulated response element (ISRE) for type I alpha/beta interferons.29 However, theMBN promotor harbors a PU.1–, a CCAAT/ enhancer-binding protein (C/EBP)–, and a c-Myb–binding site, and IFN-α activates PU.1 transcription specifically in a myelo-monocytic cell context.30,31 A U937-specific, IFN-α–dependentMBN transcription may thus be mediated indirectly, for example via the PU.1-binding site of the MBNpromoter.31 Another reason could be that interferon regulatory factor (IRF) expression and interaction as well as induction of IRFs by interferons are cell line specific.29Therefore, it is conceivable that cells of erythroleukemic, monocytic, or T-cell background respond differently to IFN-α. Notably, imatinib did not stimulate MBN transcription either in vivo or in vitro.
As demonstrated by others, IFN-α appears to induce remissions from CML by eliciting an MBN-specific T-cell response.26 Our results suggest that the induction ofMBN expression may be a part of this response mechanism, because sole expression of tumor-specific genes in APCs or loading of tumor peptides onto APCs has been shown to induce a potent HLA class I–restricted, tumor-specific CTL response.21,22,32Indirect evidence for a central role of MBN expression in the generation of a T-cell response is provided by the fact that all tested HLA-A*0201+ IFN-α–treated patients, but only 2 of 11 imatinib-treated patients, developed PR1-specific CTLs (Table 2). Notably, the presence of PR1-CTLs in these 2 imatinib patients may be explained by a previous long-term exposure to IFN-α (3 and 3.5 years, respectively). Mainly owing to IFN-α intolerance, 7 of the remaining 9 imatinib patients were not significantly pre-exposed to IFN-α. Besides inducing MBN expression, IFN-α up-regulates MHC class I and II expression.33 MHC class I proteins are specifically needed to present peptides to T cells.34 Finally, IFN-α promotes the maturation of monocytes to APCs with T-cell costimulatory potential.35These 3 properties of IFN-α can explain why this drug, and not imatinib, supports a PR1-CTL response. An autologous presentation of MBN peptides on monocytes/APCs would be in line with the finding that PR1-CTLs are detectable even in the absence ofMBN-expressing CML cells, that is, after reaching a major or complete cytogenetic remission (Table 2: UPN 30).26 This also suggests that the presence of BCR/ABL is not a conditio sine qua non for the beneficial effects of IFN-α, whose action on the nonmalignant cell population may be as important. This hypothesis is strongly supported by our own experiments showing that patients treated with IFN-α for reasons other than CML (namely hepatitis C) develop frequencies of PR1-CTLs comparable to those seen in IFN-α–treated CML patients (not shown). This indicates a novel mode of action of IFN-α: induction of MBN may promote the generation of autoimmune PR1-CTLs that have the ability to kill CML cells.
In summary, different molecular regulations and effector mechanisms are associated with remissions under imatinib and IFN-α. The induction of an antileukemic MBN-specific T-cell response is restricted primarily to IFN-α treatment. From this perspective, the therapeutic long-term efficacy and prognostic predictive value of a response may not be equivalent under IFN-α and imatinib. This, however, will become obvious only in the future, when the long-term efficacy of imatinib therapy can be assessed. Until then, the extraordinary good initial treatment responses of imatinib have to be followed closely for their durability. For example, very recent data demonstrating a failure of imatinib to kill cell cycle–arrested CML precursors advise us to be attentive to arising escape mechanisms of even chronic phase CML.36 Therefore, a concurrent or sequential combination therapy of IFN-α and imatinib, using their different effector mechanisms, may be more effective in the treatment of CML than any current monotherapy. Large multicenter trials addressing this question are presently being planned or are underway. An additive cytotoxic effect of these 2 drugs has—at least in vitro—already been demonstrated.37
We wish to thank Drs Beyer, Kim, Ritter, and Reckzeh for their help collecting patients samples. We are grateful to M. Rehn for her excellent technical assistance and to Dr Jaques for performing the cell sorting. We thank T. Kroll for his help analyzing the array data.
Prepublished online as Blood First Edition Paper, June 28, 2002; DOI 10.1182/blood-2002-02-0659.
Supported by the Deutsche José Carreras Leukämie-Stiftung (A.N., M.S, and A.H.); by the P. E. Kempkes Stiftung (A.B.); by the H. W. & J. Hector Stiftung (A.N. and M.S.); by the Wilhelm-Sander-Stiftung (A.N. and M.S.); by the Deutsche Forschungsgemeinschaft (A.N.); and by a grant from the German Ministry of Education and Research (BMBF), Kompetenznetz: Akute und chronische Leukämien, 01 GI9980/6.
H.G. is employed by Novartis Pharma, whose product was used in the present study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Andreas Neubauer, Klinikum der Philipps Universität Marburg, Klinik für Hämatologie, Onkologie und Immunologie, Baldinger Strasse, 35033 Marburg, Germany; e-mail: neubauer@mailer.uni-marburg.de.