Clinical resistance to imatinib mesylate is commonly observed in patients with advanced Philadelphia chromosome– positive (Ph+) leukemias. Acquired resistance is typically associated with reactivation of BCR-ABL due to kinase domain mutations or gene amplification, indicating that BCR-ABL remains a viable target for inhibition in these patients. Strategies for overcoming resistance can be envisioned through exploitation of other molecular features of the BCR-ABL protein, such as its dependence on the molecular chaperone heat shock protein 90 (Hsp90). To determine whether inhibition of Hsp90 could induce degradation of imatinib mesylate–resistant, mutant BCR-ABL proteins, hematopoietic cells expressing 2 mutant BCR-ABL proteins found in imatinib mesylate–resistant patients (T315I and E255K) were examined for sensitivity to geldanamycin and 17-allylaminogeldanamycin (17-AAG). Both compounds induced the degradation of wild-type and mutant BCR-ABL and inhibited cell growth, with a trend indicating more potent activity against mutant BCR-ABL proteins. These data support clinical investigations of 17-AAG in imatinib mesylate–resistant Ph+ leukemias.
Introduction
Chronic myeloid leukemia (CML) is a pluripotent hematopoietic stem cell disorder characterized by the Philadelphia (Ph) chromosome translocation.1,2 The resulting BCR-ABL fusion gene encodes a cytoplasmic protein with constitutive tyrosine kinase activity.3 Numerous experimental models have established that BCR-ABL is an oncogene and is sufficient to produce CML-like disease in mice.4 5 CML progresses through distinct clinical stages termed chronic phase, accelerated phase, and blast crisis. TheBCR-ABL oncogene is expressed at all stages, but blast crisis is characterized by multiple additional genetic and molecular changes.
Because the BCR-ABL kinase is deemed necessary for the deregulated growth of leukemic cells, it provides an ideal target for inhibition in the treatment of CML and other Ph+ leukemias. In clinical trials, imatinib mesylate (Gleevec), a 2-phenylamino pyrimidine that targets the adenosine triphosphate (ATP)–binding site of the kinase domain of ABL, induced remissions in patients with chronic phase CML as well as blast crisis.6,7 However, while responses in chronic phase were durable, remissions observed in blast crisis patients were typically short-lived with relapse occurring within 6 months despite continued therapy.7 In patients with transient responses to imatinib mesylate, acquired resistance has typically been associated with failure to maintain effective inhibition of BCR-ABL kinase activity—an indication that BCR-ABL remains a viable target for inhibition in these patients.8 Mechanisms of resistance in relapsed patients include BCR-ABL gene amplification and kinase domain mutations, such as T315I and E255K, which alter the targeted region of the protein and therefore abrogate imatinib mesylate binding.8-12
Strategies for overcoming resistance associated with kinase domain mutations will likely require targeting other molecular features of the BCR-ABL protein. Heat shock protein 90 (Hsp90) is a molecular chaperone which affects the stability and function of multiple oncogenic proteins including BCR-ABL.13,14 Geldanamycin (GA) is a benzoquinone ansamycin which specifically inhibits Hsp90 by competitively binding to an ATP-binding pocket in the amino-terminus of Hsp90.15-17 Disruption of Hsp90 function by GA or its less toxic analog, 17-allylaminogeldanamycin (17-AAG), in BCR-ABL–expressing leukemia cells has been shown to induce BCR-ABL protein degradation and suppress cell proliferation.13,18 19 17-AAG is currently in phase I clinical trials.
To determine whether inhibition of Hsp90 could induce degradation of imatinib mesylate–resistant, mutant BCR-ABL proteins, hematopoietic cells expressing 2 mutant BCR-ABL proteins found in imatinib mesylate–resistant patients (T315I and E255K) were derived and tested for sensitivity to GA and 17-AAG. We found that both compounds induced the degradation of wild-type and mutant BCR-ABL proteins as well as inhibited cell growth. The data also suggest a trend indicating a greater potency against mutant BCR-ABL proteins. These results provide a rationale for the use of 17-AAG in the clinical setting of imatinib mesylate–resistant Ph+ leukemia.
Study design
Chemicals
Stock solutions of GA (Sigma, St Louis, MO), 17-AAG (NSC 330507, National Cancer Institute, Bethesda, MD), and imatinib mesylate (Novartis, Basel, Switzerland) were prepared as 10 mM dimethylsulfoxide solutions and stored at −20°C.
Plasmids and cell lines
Full-length P210 T315I and P210 E255K BCR-ABL in pBluescript (Stratagene, La Jolla, CA) were generated using site-directed mutagenesis and confirmed by sequencing as described previously.8 Wild-type and mutant P210 BCR-ABL were subsequently subcloned into the EcoRI site of pMSCVpuro (Clontech, Palo Alto, CA) for retrovirus generation. Ecotropic retroviruses were generated by cotransfection of pMSCVpuro DNA and Ecopac retroviral packaging vector (kindly provided by R. Van Etten) into 293T cells using the CaCl2 method.20 The murine hematopoietic cell line Ba/F3 was maintained in RPMI1640 supplemented with 10% fetal bovine serum, l-glutamine, and 1 ng/mL recombinant murine IL-3 (R&D, Minneapolis, MN). Ba/F3 populations with stable BCR-ABL expression were derived by retroviral infection of Ba/F3 cells in the presence of IL-3, and subsequent selection by puromycin. IL-3–independent BCR-ABL–expressing cells were derived by culturing in IL-3–free media at low densities in 96-well tissue culture plates. Multiple IL-3–independent populations were assayed for comparable BCR-ABL protein expression by Western blot.
In vitro drug exposure assays
Results and discussion
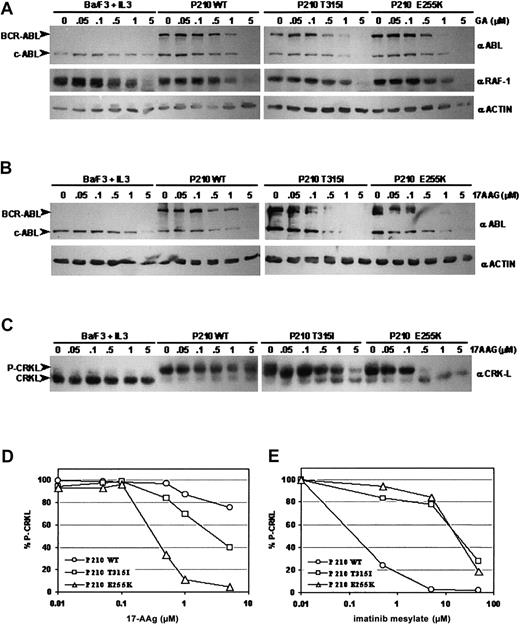
Previous studies have shown that the Hsp90 inhibitors GA and its derivative, 17-AAG, disrupt Hsp90 function and induce BCR-ABL protein degradation.13,18,19 To determine whether GA can similarly cause the degradation of BCR-ABL proteins carrying imatinib mesylate–resistant point mutations, populations of interleukin-3 (IL-3)–dependent Ba/F3 murine hematopoietic cells were engineered to express either wild-type, T315I, or E255K P210 BCR-ABL and exposed to varying concentrations of inhibitor. Consistent with previous reports, both mutant BCR-ABL alleles rendered the cells independent of IL-3, and cells expressing either mutant contained high levels of phosphotyrosine on BCR-ABL and other substrate proteins (data not shown).8,11 Western blot analyses using ABL-specific antibodies demonstrated that GA caused BCR-ABL protein levels to decrease significantly in cells expressing wild-type BCR-ABL after treatment for 24 hours at a dose of 1.0 μM, as expected.13,18 19 BCR-ABL protein was also degraded in cells expressing either T315I or E255K BCR-ABL, but this degradation occurred at a lower GA concentration (0.5 μM) (Figure1A). This apparently enhanced degradation of the 2 mutant BCR-ABL proteins was specific because degradation of another Hsp90 client protein, RAF-1, was comparable in all cells tested. These data suggest that GA may have greater potency against mutant BCR-ABL proteins compared with wild type.
Geldanamycin and 17-AAG induce degradation of wild-type and imatinib mesylate–resistant, mutant BCR-ABL proteins and inhibit BCR-ABL signaling.
(A) Ba/F3 cells expressing wild-type, T315I, or E255K BCR-ABL were incubated in the presence of increasing concentrations of geldanamycin (GA) for 24 hours. Immunoblotting of cell lysates was performed with anti-ABL (Ab3, Oncogene, San Diego, CA) (upper panels), anti–RAF-1 (Santa Cruz Biotechnology, Santa Cruz, CA) (middle panels), and antiactin (ac-15, Sigma) as a control for protein loading (lower panels). (B) Ba/F3 cells expressing wild-type, T315I, or E255K BCR-ABL were incubated in the presence of increasing concentrations of 17-AAG for 24 hours. Immunoblotting of these lysates was performed with anti-ABL (upper panels) and antiactin as a control for protein loading (lower panels). (C) Immunoblotting of the same lysates from (B) was performed with anti-CRKL (Santa Cruz). CRKL, when tyrosine-phosphorylated, migrates more slowly on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) resulting in an upper band representing phosphorylated CRKL (P-CRKL) and a lower band representing nonphosphorylated CRKL. (D) Densitometric analysis of CRKL immunoblot shown in (C) using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). Quantified CRKL phosphorylation is expressed as percentage of phosphorylated CRKL over total CRKL protein (% P-CRKL). (E) Densitometric analysis of CRKL immunoblot using lysates from the same Ba/F3 cell lines incubated in the presence of increasing concentrations of imatinib mesylate for 24 hours.
Geldanamycin and 17-AAG induce degradation of wild-type and imatinib mesylate–resistant, mutant BCR-ABL proteins and inhibit BCR-ABL signaling.
(A) Ba/F3 cells expressing wild-type, T315I, or E255K BCR-ABL were incubated in the presence of increasing concentrations of geldanamycin (GA) for 24 hours. Immunoblotting of cell lysates was performed with anti-ABL (Ab3, Oncogene, San Diego, CA) (upper panels), anti–RAF-1 (Santa Cruz Biotechnology, Santa Cruz, CA) (middle panels), and antiactin (ac-15, Sigma) as a control for protein loading (lower panels). (B) Ba/F3 cells expressing wild-type, T315I, or E255K BCR-ABL were incubated in the presence of increasing concentrations of 17-AAG for 24 hours. Immunoblotting of these lysates was performed with anti-ABL (upper panels) and antiactin as a control for protein loading (lower panels). (C) Immunoblotting of the same lysates from (B) was performed with anti-CRKL (Santa Cruz). CRKL, when tyrosine-phosphorylated, migrates more slowly on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) resulting in an upper band representing phosphorylated CRKL (P-CRKL) and a lower band representing nonphosphorylated CRKL. (D) Densitometric analysis of CRKL immunoblot shown in (C) using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). Quantified CRKL phosphorylation is expressed as percentage of phosphorylated CRKL over total CRKL protein (% P-CRKL). (E) Densitometric analysis of CRKL immunoblot using lysates from the same Ba/F3 cell lines incubated in the presence of increasing concentrations of imatinib mesylate for 24 hours.
We next tested 17-AAG—a GA derivative currently in phase I clinical trials—for its ability to induce BCR-ABL protein degradation in the same Ba/F3 cell lines. Western blot analyses of lysates from cells cultured in 17-AAG showed a similar trend to that seen with GA. Wild-type BCR-ABL protein levels fell gradually after 24 hours of exposure to 0.5 to 1.0 μM 17-AAG. Although BCR-ABL protein levels in both the T315I and E255K BCR-ABL–expressing cells began to decline at a similar concentration of 17-AAG as wild-type BCR-ABL (0.5 μM), the magnitude of decrease was more dramatic in cells expressing the BCR-ABL mutants. Virtually no BCR-ABL protein was detectable at 1.0 μM of 17-AAG for both mutants (Figure 1B). This trend was confirmed when we assessed the effect of 17-AAG on downstream BCR-ABL signaling by measuring the phosphorylation status of CRKL, a direct BCR-ABL substrate with functional relevance in CML.22-25 Western blot analysis using CRKL-specific antisera on lysates from cells incubated in the presence of increasing concentrations of imatinib mesylate confirmed that the BCR-ABL mutants conferred resistance to imatinib mesylate (Figure 1E). CRKL Western blot analysis on lysates from17-AAG–treated cells revealed that lower doses of 17-AAG were needed to inhibit BCR-ABL activity in cells expressing the BCR-ABL mutants when compared with wild-type BCR-ABL (Figure 1C-D). Significant changes in CRKL phosphorylation were not observed in wild-type BCR-ABL–expressing cells until a 17-AAG concentration of 5.0 μM was reached, whereas CRKL phosphorylation in T315I and E255K BCR-ABL–expressing cells was significantly inhibited at 0.5 μM of drug (Figure 1C-D). Although we cannot rule out the possibility that 17-AAG may affect another kinase which plays a role in CRKL phosphorylation in these cells, the fact that 17-AAG also reduced the level of BCR-ABL protein, together with previously published data showing that constitutively elevated CRKL phosphorylation is relatively specific for CML,22 strongly suggests that BCR-ABL is the target.
Previous studies have also shown that GA and 17-AAG inhibit growth and induce apoptosis of BCR-ABL–positive leukemic cell lines.18 19 To determine whether GA could inhibit growth in cells expressing imatinib mesylate–resistant BCR-ABL mutants, Ba/F3 cells transformed by wild-type, T315I, and E255K BCR-ABL were cultured in a range of GA concentrations. Trypan blue dye exclusion assessments of viability and corresponding IC50 calculations (the concentration of inhibitor required to reduce the number of viable cells by 50%) indicated that the growth of all 3 BCR-ABL–positive cell lines was inhibited by GA at lower doses when compared with BCR-ABL–negative parental cells (Table1). The enhanced sensitivity of the imatinib mesylate–resistant BCR-ABL mutants compared with wild-type BCR-ABL observed in the biochemical analyses was also recapitulated in the growth inhibition assays. Similar results were observed with 17-AAG–treated cells. All BCR-ABL–expressing cells were more sensitive to 17-AAG than Ba/F3 parental cells, with the imatinib mesylate–resistant BCR-ABL–expressing cells again displaying a heightened sensitivity to inhibition compared with wild-type BCR-ABL–expressing cells (Table 1).
In summary, targeted inhibition of Hsp90 with either GA or 17-AAG induced the degradation of wild-type BCR-ABL and 2 imatinib mesylate–resistant BCR-ABL mutants, T315I and E255K. Both compounds also inhibited the growth of hematopoietic cells transformed by wild-type and mutant BCR-ABL. The results also suggest that the imatinib mesylate–resistant mutants are more sensitive to Hsp90 inhibition than wild-type BCR-ABL. One potential explanation could be that these 2 mutant proteins are less stable than wild-type BCR-ABL, and therefore more dependent on molecular chaperones. A better understanding of the variables that determine the relative dependence of client proteins on Hsp90 function is required to fully evaluate this question. Nevertheless, these data provide support for clinical investigations of 17-AAG in imatinib mesylate–resistant Ph+ leukemia.
We wish to thank Randy Chen for expert technical assistance.
Prepublished online as Blood First Edition Paper, June 7, 2002; DOI 10.1182/blood-2002-05-1361.
Supported by grants from the Leukemia and Lymphoma Society, the National Cancer Institute (C.L.S.) and USPHS National Research Service Award GM07185 (M.E.G.). C.L.S. is a Doris Duke Distinguished Clinical Scientist.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Charles L. Sawyers, David Geffen School of Medicine at UCLA, Division of Hematology and Oncology, 11-934 Factor Bldg 10833 Le Conte Ave, Los Angeles, CA 90095-1678; e-mail:csawyers@mednet.ucla.edu.