Protoporphyria is generally inherited as an autosomal dominant disorder. The enzymatic defect of protoporphyria is a deficiency in ferrochelatase, which chelates iron and protoporphyrin IX to form heme. Patients with protoporphyria have decreased ferrochelatase activities that range from 5% to 30% of normal caused by heterogeneous mutations in the ferrochelatase gene. The molecular mechanism by which the ferrochelatase activity is decreased to less than an expected 50% is unresolved. In this study, we assessed the effect of a ferrochelatase exon 10 deletion, a common mutation in human protoporphyria, introduced into the mouse by gene targeting. F1 crosses produced (+/+), (+/−), and (−/−) mice at a ratio of 1:2:0; (−/−) embryos were detected at 3.5 days postcoitus, consistent with embryonic lethality for the homozygous mutant genotype. Heterozygotes demonstrated equivalent levels of wild-type and mutant ferrochelatase messenger RNAs and 2 immunoreactive proteins that corresponded to the full-length and an exon 10–deleted ferrochelatase protein. Ferrochelatase activities in the heterozygotes were an average of 37% of normal, and protoporphyrin levels were elevated in erythrocytes and bile. Heterozygous mice exhibited skin photosensitivity but no liver disease. These results lend support for a dominant-negative effect of a mutant allele on ferrochelatase activity in patients with protoporphyria.
Introduction
Ferrochelatase catalyzes the formation of heme by chelating ferrous iron to protoporphyrin IX. High ferrochelatase activity is required during erythropoiesis to provide heme for hemoglobin, but low constitutive activity is required in all cell types to provide heme for cytochromes and other essential heme-containing proteins. The ferrochelatase gene is located on chromosome 18 and consists of 11 exons spanning about 40 to 50 kilobases (kb) in both mice and humans.1-4 Mammalian ferrochelatase is a metalloenzyme that possess a 2-iron/2-sulfur cluster, a structural component of the protein that is located near the carboxyl terminus and is required for enzymatic activity.5 The ferrochelatase protein is translated with a mitochondrial targeting peptide, which is cleaved off after translocation of the enzyme to its active site in the inner mitochondrial membrane.6,7 The mammalian enzyme is active in situ as an 80- to 82-kd homodimer as suggested by radiation inactivation studies8 and confirmed by x-ray crystallography.9 10
Protoporphyria is a disease caused by a deficiency in ferrochelatase activity.11 The principal clinical manifestation of protoporphyria is severe photosensitivity and in rare cases hepatobiliary disease, which can advance to hepatic failure and require liver transplantation.12 The symptoms of protoporphyria are due to the partial enzymatic block, which results in excess protoporphyrin that accumulates in tissues, serum, and bile.13,14 Approximately 80% of the excess protoporphyrin originates from developing erythroid cells and 20% from the liver.15
Cutaneous photosensitivity occurs when the excess protoporphyrin in the skin is excited by sunlight (or wavelengths of 400-410 nm), which concomitantly results in tissue-damaging production of free radicals and reactive oxygen species.16,17 Hepatic protoporphyria results from the accumulation of protoporphyrin in the liver. Excess protoporphyrin generated in the developing erythrocytes leaks out of the cells into the serum and is then extracted by the liver.18,19 Microcrystals of protoporphyrin form in hepatocytes and protoporphyrin aggregates occlude biliary ducts.14,20-22 Hepatocyte damage from the protoporphyrin crystals and chronic cholestasis ultimately leads to fibrosis and cirrhosis.12 14
Therapies for protoporphyria are primarily prophylactic.20,23-25 Avoidance of the sun, UV-blocking creams, and a high-dose β-carotene regimen may aid in mitigating the effects of free radicals and reactive oxygen species. Liver transplantation is indicated for end-stage hepatic failure in patients with protoporphyria.12,26-28 Although liver transplantation provides a temporary therapy for severe cases of hepatic protoporphyria, the excess protoporphyrin derived from the bone marrow is ultimately able to surpass the clearance capability of the transplanted liver, producing recurrent protoporphyric liver disease.29 Bone marrow transplants and gene therapies using recombinant viruses to express therapeutic levels of ferrochelatase in erythroid-lineage cells have been successfully accomplished in a mouse model of recessive protoporphyria.30-32
Mutations affecting ferrochelatase activity are heterogeneous. Missense and nonsense mutations have been reported in several protoporphyric patients33,36,37,39-43; however, more common are splice-donor and acceptor-site mutations, which lead to exon skipping.36,44 In vitro studies have demonstrated that ferrochelatases with individual exon deletions (3-11) all lose their 2-iron/2-sulfur cluster and retain no enzymatic activity.45 46
Protoporphyria is generally transmitted as an autosomal dominant disorder with variable penetrance. Rare, recessive cases of protoporphyria have also been reported.33-36 A puzzling feature of dominantly inherited protoporphyria is that enzyme activities are consistently reported at 15% to 30% of normal and not 50% as might be expected for a dominantly inherited disorder. Two models have been proposed to explain this less-than-50% activity. The dominant-negative model maintains that a mutant ferrochelatase monomer interacts with a wild-type monomer in a dominant-negative manner to decrease enzymatic activity levels lower than 50% of normal.37 The “low-expressing allele” model suggests that a null allele in combination with a low-expressing allele (with a wild-type coding region) decreases ferrochelatase activities to lower than 50% of normal.38 39
In a previous study, the dominant-negative model was tested by introducing an exon 10 deletion into the mouse genome via homologous recombination in mouse embryonic stem (ES) cells.47 In ferrochelatase heterozygous ES cells, ferrochelatase protein levels and activities were consistently 50% of normal, suggesting that a second mutation is required to bring the enzymatic activity levels to those observed in human protoporphyria.
In this study, the molecular, biochemical, and pathophysiologic effects of the exon 10 deletion mutation were assessed in vivo by generating mice that were heterozygous for the mutation. Exon 10–deleted messenger RNA (mRNA) expression patterns in erythroid and nonerythroid tissues were determined; the wild-type and exon 10–deleted ferrochelatase protein was characterized by immunoblotting and mitochondrial activity assays. Erythrocyte and bile protoporphyrin levels were determined and the clinical phenotype was assessed in heterozygous mice. Unlike studies in ES cells, in vivo ferrochelatase activities in heterozygous mice were less than 50% of normal, resulting in protoporphyrin accumulation and photosensitivity.
Materials and methods
ES targeting and breeding
To introduce an exon 10 deletion into the mouse ferrochelatase gene, a targeting construct was designed that on homologous recombination with the ferrochelatase gene would replace exon 10 with the neomycin gene-selectable marker (Figure1A). The splice donor site for exon 9 and splice acceptor site for exon 11 remained intact so that the neomycin gene would be spliced out of the pre-mRNA resulting in the in-frame joining of exon 9 to exon 11. ES cells heterozygous for the exon 10 deletion were generated by electroporating the targeting construct into the BK4 cell line (a subclone of E14TG2a), and selecting with geneticin (200 μg/mL) and ganciclovir (0.5 μg/mL) for 10 days. Individual geneticin-resistant and ganciclovir-insensitive clones were expanded and tested for correct targeting by Southern blotting.47 48 Heterozygous ES cells were injected into day 3, C57B6 strain blastocysts. Chimeric mice were crossed to C57B6 mice, and agouti, F1 offspring were tested for the genomic deletion of exon 10 using the polymerase chain reaction (PCR). F1 heterozygous mice were then intercrossed to obtain F2 progeny for developmental analysis. For all other experiments, F1 heterozygotes were crossed to C57B6 strain mice. Wild-type littermates were used as controls for all experiments.
Structure of the mouse ferrochelatase locus and exon 10–deleted protein.
(A) Diagram indicates the structure of the native ferrochelatase locus (top) and the targeted ferrochelatase locus (bottom). EcoRI restriction endonuclease sites, which can be used for Southern blot genotyping, are indicated with an E. The position of the gene-specific primers used for PCR genotyping of the targeted locus are indicated by bent arrows. The Pr1 primer sequence is located in the 3′ end of the neomycin cDNA, and the Pr2 primer sequence is located in the 5′ end of exon 11. The Pr3 primer sequence is located in intron 10 in a region that has been deleted in the homologous recombinant. (B) Schematic of the ferrochelatase protein and known functional domains (top). Amino acids conferred by exon 10 and approximate location in the protein (bottom). The underlined glycine is a valine in the human sequence.
Structure of the mouse ferrochelatase locus and exon 10–deleted protein.
(A) Diagram indicates the structure of the native ferrochelatase locus (top) and the targeted ferrochelatase locus (bottom). EcoRI restriction endonuclease sites, which can be used for Southern blot genotyping, are indicated with an E. The position of the gene-specific primers used for PCR genotyping of the targeted locus are indicated by bent arrows. The Pr1 primer sequence is located in the 3′ end of the neomycin cDNA, and the Pr2 primer sequence is located in the 5′ end of exon 11. The Pr3 primer sequence is located in intron 10 in a region that has been deleted in the homologous recombinant. (B) Schematic of the ferrochelatase protein and known functional domains (top). Amino acids conferred by exon 10 and approximate location in the protein (bottom). The underlined glycine is a valine in the human sequence.
Genotyping
For PCR genotyping of F1 and F2 progeny, primers were designed that would either amplify the wild-type allele (PR3 and PR2) or the exon 10-deleted allele (PR1 and PR2; Figure 1A). Genomic DNA (gDNA) was isolated from mouse tails using standard methods,49 and 1 μg gDNA was added to a PCR reaction containing the gene-specific primers: PR1(NEO) 3′-CTG CTC GAC ATT GGG TGG AAA-5′, PR2(exon 11) 3′-GAA GGA TTT AGT CTT CCT GCA-5′, or PR3(intron 10) 5′-CAG TCT GCT TCT CAG TGA ACA-3′. Successful amplification of the exon 10–deleted allele required a buffer containing 10 mM Tris-Cl, pH 8.8, 1.5 mM MgCl2, and 75 mM KCl. Amplification of the wild-type allele produced a 332-bp band, and amplification of the exon 10 deletion produced a 459–base pair [bp] product after 35 thermocycles (97°C, 15 seconds; 54°C, 15 seconds; 72°C, 1.5 minutes).
For embryo studies, heterozygous mice were crossed to generate exon 10−/− progeny. Preimplantation embryos were collected at 3.5 days of gestation. The blastocysts were flushed from the uterus, collected in Dulbecco modified Eagle medium-high glucose (DMEM-H) containing 10% fetal calf serum (FCS), 2 mMl-glutamine, 100 U/mL penicillin, and 1 mg/mL streptomycin, and transferred twice to new medium to separate blastocysts from contaminating maternal cells. Individual blastocysts were placed in 20 μL lysis buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl [pH 8.3], 0.45% Tween-20, 0.45% NP-40, and 100 μg/mL proteinase K) and incubated at 50°C for 2 hours, and then incubated at 100°C for 10 minutes to destroy the proteinase K. PCR genotyping was carried out using the primers and buffer already described. To amplify the wild-type and mutant alleles, 2 μL blastocyst lysate was used as template in a 34-cycle reaction (94°C, 15 seconds; 54°C, 15 seconds; 72°C, 30 seconds) for the wild-type allele and a 45-cycle reaction (97°C, 15 seconds; 54°C, 15 seconds; 72°C, 1.5 minutes) for the exon 10–deleted allele.
Late-gestation embryos were collected in utero at 9 to 10 days postcoitus (dpc). The entire uterus was fixed in formalin for 24 hours, then transferred to 70% ethanol before sectioning. To collect tissue for genotyping day 9 to 10 embryos, the coverslips from the stained sections were removed by soaking in xylene for 24 hours. Then, an area of tissue about 1 to 2 mm in diameter from each embryo was scraped from the slide using a tuberculin needle. The tissue was then placed in 50 μL proteinase K buffer (see above) and incubated at 56°C for 30 minutes. The DNA mixture was heated to 100°C for 10 minutes to inactivate the proteinase K and then centrifuged at 12 000 rpm in a microfuge to pellet debris. A PCR amplification reaction, using 2.5 μL of the DNA mixture as template, was conducted as described above.
mRNA analysis
To assess the expression of the exon 10–deleted ferrochelatase allele in a nonerythroid and erythroid tissue, we amplified complementary DNA (cDNA) made from liver (nonerythroid) and spleens (erythroid) induced for extramedullary erythropoiesis by treating mice with 200 μL of a 0.1% (wt/vol) solution ofN-acetylphenylhydrazine (PHZ)50,51 for 7 consecutive days. PHZ treatment was withdrawn 2 days before the mice were killed. Total RNA was isolated from liver and spleens by centrifugation through a CsCl gradient.52 Total RNA (1 μg) was added to a standard reverse transcriptase (RT) reaction,52 and one fifth of the RT reaction (about 120 ng template) was used for PCR amplification using a primer set that amplified a 340-bp region from exon 8 (PR3: 5′-CCG ACT GGT TTG GCA GTC CA-3′) to exon 11 (PR4: 5′-GAA GGA TTT AGT CTT CCT GCA-3′). Temperatures were cycled 30 times at 94°C for 15 seconds, 58°C for 15 seconds, and 72°C for 45 seconds. Amplification products were analyzed on a 5% tris borate EDTA (TBE)–acrylamide gel stained with ethidium bromide.
Immunoblotting
Mitochondria were isolated from normal and ferrochelatase heterozygous mice by homogenizing two thirds of a mouse liver in 10 mL ice-cold (0°C) 0.25 M sucrose containing 10 mM Tris-acetate (pH 8.0) and 1 mM phenylmethylsulfonyl fluoride. The homogenate was centrifuged at 2500g for 10 minutes to pellet debris. The supernatant was transferred to a clean Corex tube, and centrifuged again at 9000g for 15 minutes at 4°C. The mitochondrial pellet was washed once in 10 mL sucrose buffer, and the mitochondria were pelleted at 9000g for 15 minutes at 4°C. The mitochondrial pellet was suspended in 0.75 mL sucrose buffer containing 20% glycerol, and frozen at −80°C until assayed.
To determine ferrochelatase protein levels in mitochondrial extracts, 50 μg mitochondrial protein was separated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel. The proteins were electrophoretically transferred from the gel to pure nitrocellulose membrane (S & S, Keene, NH). Equal loading of protein was assessed by Ponceau S staining. The membrane was then blocked with BLOTTO (5% wt/vol nonfat dried milk in water) for 15 minutes at room temperature. Then, an antirecombinant human ferrochelatase polyclonal antibody53 was applied at a 1:500 dilution in BLOTTO to the membrane and allowed to incubate at room temperature for 16 hours. The membrane was then washed once with BLOTTO and twice in TBS-Tween (Tris-buffered saline containing 0.1% Tween-20). A goat antirabbit, horseradish peroxidase–conjugated, secondary antibody (Santa Cruz Biotechnologies, Santa Cruz, CA) was then applied at a 1:1000 dilution in BLOTTO and allowed to incubate for 30 minutes at room temperature. The membrane was washed once in TBS-Tween, once in BLOTTO, and twice in TBS-Tween for 10 minutes each at room temperature. Enhanced chemiluminescence reagent (Amersham, Arlington Heights, IL) was applied. Kodak BIOMAX MR (Eastman Kodak, Rochester, NY) film was exposed to the membrane for 20 minutes.
Ferrochelatase activity
Ferrochelatase activity was determined essentially as described.47 Liver mitochondrial lysates were sonicated twice in a Branson sonicator (Danbury, CT) for 10 seconds and incubated on ice. The enzymatic chelation of zinc and mesoporphyrin was carried out in a reaction that contained 25 μg mitochondrial lysate, 50 μM zinc acetate, and 50 μM mesoporphyrin in palmitic acid buffer.47 The reaction was incubated at 37°C in the dark. After 30 minutes, 1.0 mL ice-cold 8:2 methanol/dimethyl sulfoxide was added to stop the reaction. The reaction mixture was centrifuged for 10 minutes at 12 000g. To detect the zinc-mesoporphyrin reaction product, 20 μL supernatant was loaded onto a Delta PAK, 5 μm, C18 300 Å, high-performance liquid chromatography (HPLC) column (Waters, Milford, MA) and separated in an 88% methanol, 12% 1 M ammonium acetate (pH 5.16) mobile phase with a 1.25 mL/min flow rate. The column flow-through was directed into the flow cell adapter of the LS50B spectrofluorometer (Perkin-Elmer, Shelton, CT), which was set to an excitation wavelength of 403 nm and an emission wavelength of 574 nm. Concentrations of zinc-mesoporphyrin were determined by comparing samples to a standard curve derived from zinc-mesoporphyrin standards (Porphyrin Products, Logan, UT).
Determination of free erythrocyte and bile protoporphyrin levels
Levels of protoporphyrin in erythrocytes and bile were determined using HPLC separation of pigments followed by spectrofluorometry.54 55 After a 12-hour fast to induce bile accumulation, mice were euthanized by cervical dislocation. Bile and whole blood were collected, and 10 μL whole blood (in sodium heparin) was set aside for the determination of free erythrocyte protoporphyrin levels. To extract protoporphyrin from the samples, 2.5 μL bile or whole blood was added to 250 μL 0.9 N perchloric acid/methanol (1:1 vol/vol), briefly vortexed, and centrifuged at 12 000g to pellet the precipitated protein. Then, 200 μL of the supernatant was applied to a reverse-phase HPLC column and separated in an elution gradient consisting of 0.1 M sodium phosphate (pH 3.5)/methanol mobile phase. The flow rate was 1.25 mL/min, and the injections were initiated when the gradient was 45% methanol/55% sodium phosphate. Immediately after sample injection, a linear gradient was initiated and held for 2 minutes. The gradient ended at 95% methanol/5% sodium phosphate; then, a reverse gradient was initiated, which brought the mobile phase back to original conditions (45% methanol/55% sodium phosphate) over a 1-minute period. Protoporphyrin was quantified fluorometrically by directing the flow-through into an LS50B spectrofluorometer (Perkin-Elmer) set to an excitation wavelength of 400 nm (slit width 15 nm) and emission wavelength of 624 nm (slit width 20 nm). Protoporphyrin concentrations in the samples were determined using a standard curve derived from protoporphyrin standards (Porphyrin Products).
Fluorescence microscopy was used to visually assess protoporphyrin accumulation in erythrocytes. A smear was made with 1 μL tail vein blood. The erythrocytes were observed using an Olympus IX70, ×200 magnification, 405-nm excitation filter, and a 620-nm emission filter (Olympus, Melville, NY).
Photosensitization
Normal and heterozygous mice (approximately 14 months of age) were depilated using Nair (New York, NY) and were exposed to mercury vapor lamp light at a distance of 20 cm.56 57 A piece of shielding glass was placed between the lamp and the mouse to block UV-B and UV-C light. Initial experiments, using a 20-minute exposure time, produced no photosensitivity. Thirty-minute exposures were determined to be the minimum time required to elicit photosensitivity, which was first observed 48 to 72 hours after exposure. Images of light exposed skin were captured using a Nikon Cool Pix-990 digital camera (Nikon, Melville, NY).
Results
Homozygosity of the exon 10 deletion is embryonic lethal
To introduce an exon 10 deletion into the mouse ferrochelatase gene, we designed a targeting construct that on homologous recombination with the ferrochelatase gene would replace exon 10 with the neomycin gene-selectable marker (Figure 1A). The splice donor site for exon 9 and splice acceptor site for exon 11 remained intact so that the neomycin gene would be spliced out of the pre-mRNA resulting in the in-frame adjoining of exon 9 to exon 11 (Figure 1B). The targeting ratio for this region of the mouse ferrochelatase locus was 1 homologous recombination to 4 random integrations.
Chimeric mice were generated by injecting heterozygous ES cells into day 3, C57B6 strain, blastocysts. Mice heterozygous for the ferrochelatase exon 10 deletion were crossed to obtain an F2 generation in which the observed genotype was a ratio of 1 wild-type to 2 heterozygotes. No homozygous mice were observed in 10 separate breedings (producing over 100 progeny).
To further investigate whether there were any embryos homozygous for the mutation, embryos that were at 9 to 10 days of gestation were collected and histologically analyzed. In 2 separate breedings of heterozygous mice, 1 of 4 embryos was being reabsorbed. There were no anatomic structures that could be used to determine the day of lethality. No embryonic abnormalities were observed in control breedings using wild-type mice. To confirm the presence of early-stage homozygous embryos, preimplantation embryos were collected at day 3.5 of gestation. PCR genotype analysis demonstrated that 4 of 26 blastocysts were homozygous for the exon 10 deletion.
The ferrochelatase exon 10–deleted allele and the wild-type allele are expressed equally in heterozygous mice
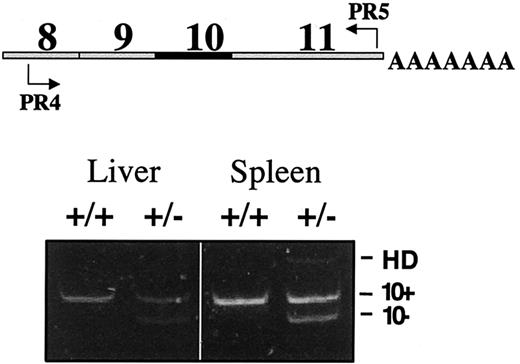
To assess the ratios of wild-type and mutant ferrochelatase mRNAs in heterozygous mice, we amplified cDNA using semiquantitative RT-PCR. The gene-specific primers amplified a region that spanned from exon 8 to exon 11. Separation of the PCR fragments on a 4% TBE-PAGE gel revealed 2 bands. A slower migrating band corresponded to the size of ferrochelatase cDNA containing exon 10, and a faster migrating band corresponded to the size of an exon 10–deleted cDNA as previously confirmed by DNA sequencing (Figure2).47 The slowest migrating band observed in the amplification products from heterozygous mice is a heteroduplex (HD) consisting of wild-type and exon 10–deleted DNA strands.47 The intensities of the 2 bands were quantified by scanning densitometry. The exon 10–deleted band is 96% and 90% of the wild-type band in liver and spleen mRNA, respectively.
RT-PCR analysis demonstrates equivalent levels of wild-type and exon 10 mRNA.
Levels of wild-type and mutant ferrochelatase mRNA in liver and spleen were determined using semiquantitative RT-PCR. Schematic indicates the location of the primers in the ferrochelatase cDNA used for RT-PCR analysis (top). Exon status of each PCR fragment is indicated (bottom). The slowest migrating band observed in the spleen +/− lane is a heteroduplex (HD) consisting of a wild-type DNA strand and an exon 10-deleted DNA strand. Genotypes of mice are indicated by +/+ (wild type) or +/− (heterozygous).
RT-PCR analysis demonstrates equivalent levels of wild-type and exon 10 mRNA.
Levels of wild-type and mutant ferrochelatase mRNA in liver and spleen were determined using semiquantitative RT-PCR. Schematic indicates the location of the primers in the ferrochelatase cDNA used for RT-PCR analysis (top). Exon status of each PCR fragment is indicated (bottom). The slowest migrating band observed in the spleen +/− lane is a heteroduplex (HD) consisting of a wild-type DNA strand and an exon 10-deleted DNA strand. Genotypes of mice are indicated by +/+ (wild type) or +/− (heterozygous).
Equal amounts of mutant and wild-type ferrochelatase protein but reduced enzyme activity in heterozygous mice
To examine ferrochelatase protein in the heterozygous mice, we conducted immunoblotting assays using an antihuman ferrochelatase antibody that cross-reacts with mouse ferrochelatase (Figure3A). An immunoreactive protein that corresponds to the size of wild-type ferrochelatase appeared in extracts from both normal and heterozygous mice. However, in heterozygous mice there was also a unique, fast migrating band that corresponded to the size of an exon 10–deleted ferrochelatase protein.
Detection of an exon 10–ferrochelatase protein and decreased ferrochelatase activity heterozygous mice.
(A) Immunoblot analysis was used to determine ferrochelatase levels in liver mitochondrial extracts using an antirecombinant human ferrochelatase antibody that cross-reacts with mouse ferrochelatase. A 10% SDS-PAGE gel was used to separate the wild-type (+/+) ferrochelatase from the exon 10–deleted (+/−) ferrochelatase. (B) Liver mitochondrial ferrochelatase activities were determined by measuring the chelation rate of zinc and mesoporphyrin. The genotypes and number of mice represented in the graph are indicated below the x-axis. Average activities (cross-bars) and SEs (vertical lines) are indicated (SEM for +/+ = 9.6 and +/−= 8.8; P < .001). Units of activity are expressed as picomoles zinc-mesoporphyrin per milligram protein per hour.
Detection of an exon 10–ferrochelatase protein and decreased ferrochelatase activity heterozygous mice.
(A) Immunoblot analysis was used to determine ferrochelatase levels in liver mitochondrial extracts using an antirecombinant human ferrochelatase antibody that cross-reacts with mouse ferrochelatase. A 10% SDS-PAGE gel was used to separate the wild-type (+/+) ferrochelatase from the exon 10–deleted (+/−) ferrochelatase. (B) Liver mitochondrial ferrochelatase activities were determined by measuring the chelation rate of zinc and mesoporphyrin. The genotypes and number of mice represented in the graph are indicated below the x-axis. Average activities (cross-bars) and SEs (vertical lines) are indicated (SEM for +/+ = 9.6 and +/−= 8.8; P < .001). Units of activity are expressed as picomoles zinc-mesoporphyrin per milligram protein per hour.
To assess ferrochelatase activity in heterozygous and wild-type mice, we measured the enzymatic chelation rate of zinc and mesoporphyrin (Figure 3B). The hepatic mitochondrial ferrochelatase activity in heterozygous mice was an average of 37% ± 8% of normal control mice activities. The ferrochelatase activities in heterozygous mice were significantly decreased compared to 50% activities of wild-type mice (P < .002), consistent with the mutant allele having a dominant-negative effect on the wild-type allele.
Increased protoporphyrin levels in ferrochelatase heterozygous mice
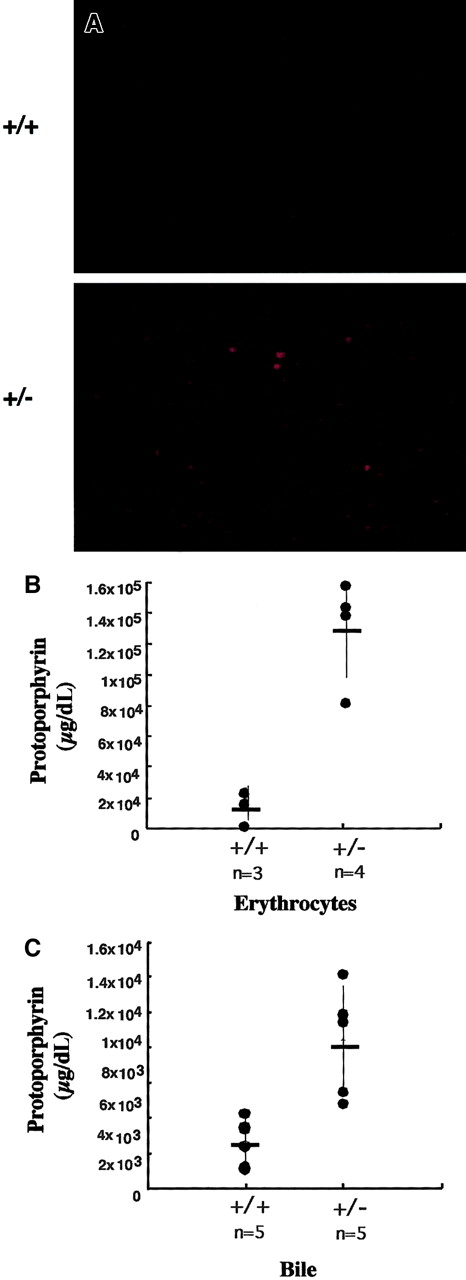
Fluorescence microscopy and spectrofluorometry were used to determine if there was an accumulation of protoporphyrin in heterozygous mice. Peripheral whole blood was observed microscopically using wavelengths that excite protoporphyrin (405 nm). Red fluorescence was observed in the erythrocytes of heterozygous mice, but not observed in the erythrocytes of normal control mice (Figure4).
Erythrocyte and bile protoporphyrin levels are elevated in heterozygous mice.
(A) Whole blood was isolated from tail vein of normal and heterozygous mice. Fluorescence of erythrocytes was detected by exciting an blood smear with 405-nm light. Images were captured with a Spot digital camera using an Olympus IX70, × 200 magnification. Genotypes are indicated to the left of the figures. (B) Erythrocyte and (C) bile protoporphyrin levels were determined by HPLC separation of pigments with subsequent spectrofluorometric analysis. The genotypes and number of mice represented in the data are indicated below the x-axis. Average activities (cross-bars) and SEs (vertical lines) are indicated. (SEM for free erythrocyte protoporphyrin +/+ = 1.8 × 104, +/−= 3.38 × 104; for bile, +/+ = 1.36 × 103, +/−= 3.14 × 103;P < .001).
Erythrocyte and bile protoporphyrin levels are elevated in heterozygous mice.
(A) Whole blood was isolated from tail vein of normal and heterozygous mice. Fluorescence of erythrocytes was detected by exciting an blood smear with 405-nm light. Images were captured with a Spot digital camera using an Olympus IX70, × 200 magnification. Genotypes are indicated to the left of the figures. (B) Erythrocyte and (C) bile protoporphyrin levels were determined by HPLC separation of pigments with subsequent spectrofluorometric analysis. The genotypes and number of mice represented in the data are indicated below the x-axis. Average activities (cross-bars) and SEs (vertical lines) are indicated. (SEM for free erythrocyte protoporphyrin +/+ = 1.8 × 104, +/−= 3.38 × 104; for bile, +/+ = 1.36 × 103, +/−= 3.14 × 103;P < .001).
To quantify the accumulated protoporphyrin in the erythrocytes, we extracted erythrocyte porphyrins from wild-type and heterozygous mice and determined protoporphyrin levels using HPLC combined with spectrofluorometry (Figure 4B). Protoporphyrin levels were an average of 9 ± 2-fold higher in erythrocytes of heterozygous mice compared to normal mouse erythrocytes (P < .001).
Secretion of protoporphyrin into bile is the primary elimination route of excess protoporphyrin.18 19 To determine if there were increased levels of protoporphyrin in the bile of heterozygous mice, we assessed protoporphyrin levels by separating the bile porphyrins using HPLC with subsequent spectrofluorometric quantification. Bile protoporphyrin levels in heterozygous mice were found to be an average of 2.2 ± 0.7-fold higher (P < .001) than that of normal mice (Figure 4C).
Heterozygous mice exhibit skin photosensitivity
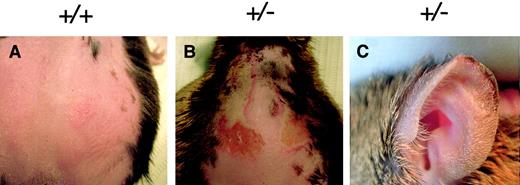
Patients with protoporphyria exhibit photosensitivity of varying levels and severity. To assess whether heterozygous mice exhibited photosensitivity, both heterozygous and normal control mice (14 months of age) were depilated and exposed to light from a UV-shielded mercury vapor lamp. There was no acute photosensitivity observed immediately following light exposure in either the heterozygous mice or controls. However, 48 to 72 hours after light treatment, heterozygous mice exhibited severe edema and necrosis of the skin on their depilated backs (Figure 5). Additionally, tissue necrosis was observed on the ear tips of heterozygous mice, but not in normal control mice (Figure 5C).
Heterozygous mice exhibit photosensitivity.
(A) Control or (B) heterozygous mice were exposed to mercury vapor lamp light (that was UV shielded) for 30 minutes. Lesions were observed on the depilated backs and (C) ear tips of heterozygous mice 72 hours after light exposure. Genotypes are indicated above each figure.
Heterozygous mice exhibit photosensitivity.
(A) Control or (B) heterozygous mice were exposed to mercury vapor lamp light (that was UV shielded) for 30 minutes. Lesions were observed on the depilated backs and (C) ear tips of heterozygous mice 72 hours after light exposure. Genotypes are indicated above each figure.
No hepatic damage in heterozygous mice
In approximately 5% to 10% of reported cases, protoporphyric patients have clinically significant liver disease.15,58To determine if there was hepatic damage in heterozygous mice (2 months and 8-12 months of age), livers were resected, fixed, sectioned, and stained with hematoxylin-eosin, a collagen-specific stain, and a bile-specific stain. In both heterozygous and control mice, the liver architecture was normal, and there were no signs of cholestasis or fibrosis. Results from the electron microscopy studies indicated no protoporphyrin microcrystals, a hallmark feature of hepatic protoporphyria,59-61 in hepatocytes of heterozygous mice. Additionally, there were no significant differences in the aspartate aminotransferase and alanine aminotransferase levels in heterozygous mice compared to controls.
Discussion
The aim of this study was to introduce a well-described mutation identified in human protoporphyria into the mouse ferrochelatase gene to determine the in vivo effect of the mutation at the molecular, biochemical, and pathophysiologic levels.
A breeding scheme designed to generate mice that were homozygous for the exon 10 deletion failed to produce progeny that possessed a homozygous mutant genotype. It was predicted that exon 10−/− mice would not be viable, because nonenzymatic heme production would be insufficient for the synthesis of hemoproteins. It was a question, however, whether nonenzymatic chelation of iron and protoporphyrin would provide sufficient levels of heme for viability prior to a developmental stage that required erythrocytes. In matings of heterozygotes, we made an observation that embryos were being reabsorbed at a ratio of 1:4 in the uteri of mice (9-10 dpc). The proportion of nonviable (−/−) to viable (+/+ or +/−) embryos based on mendelian genetics suggested all exon 10–deleted homozygotes were dead by day 9 postcoitus. Because tissue from embryos being reabsorbed was contaminated by maternal cells, we were unable to genotype these remnants. Studies on embryos from an earlier gestational stage indicated that exon 10−/− embryos were viable until at least day 3.5 of gestation, suggesting that lethality occurs between day 4 (implantation) and day 9. Thus, we speculate that a homozygous mutant embryo could implant and develop for a finite period.
Protoporphyria is inherited as an autosomal dominant trait, but the enzymatic activity is 5% to 30% of normal and not the expected 50%. Thus, we have hypothesized that the mutant protein has a dominant-negative effect on the wild-type protein. For a mutant ferrochelatase monomer to exert a dominant-negative effect on the ferrochelatase dimer, there would need to be sufficient levels of mutant monomers to dimerize with wild-type monomers. Nearly equivalent amounts of the mutant and wild-type mRNAs were observed in the livers and spleens of exon 10+/− mice, and immunoblot analysis of mitochondrial extracts demonstrated immunoreactive material that corresponded to the size of the wild-type and exon 10–deleted ferrochelatases at a 1:1 ratio (Figure 3A). These results indicate that the exon 10–deleted ferrochelatase is in a stoichiometric quantity that could produce a measurable dominant-negative effect.
We were able to rule out that the 37% ferrochelatase activities were due to differences in the wild-type allele expression from the hybrid mouse strain. The exon 10 deletion was on the ferrochelatase allele derived from the 129/Ola strain and the wild-type ferrochelatase allele was derived from C57B6 strain. Because the hybrid heterozygotes 129/C57B6 were always crossed to the C57B6 pure strain, wild-type ferrochelatase expression was derived from the wild-type C57B6 allele in both normal and heterozygous mice. Therefore, the 37% ferrochelatase activities observed in the heterozygous mice suggests that the exon 10–deleted ferrochelatase is decreasing ferrochelatase activity by a dominant-negative mechanism and not by lower allelic expression.
In a previous study we analyzed ferrochelatase mRNA and protein from exon 10+/− ES cells in culture.47 In contrast to the results in mice, studies in ES cells demonstrated that more than 90% of the ferrochelatase mRNA originated from the wild-type allele. Additionally, no exon 10–deleted protein was observed using immunoblot assays. These observations indicate there is a differential stability of the mutant ferrochelatase mRNA or protein (or both) in different cell lineages. The lack of an exon 10–deleted ferrochelatase provides a reasonable explanation as to why we did not observe less-than-50% ferrochelatase activity in exon 10+/− ES cells.
In patients with protoporphyria the erythrocyte protoporphyrin levels are increased 9.3- to 98-fold higher than normal controls,62 and bile protoporphyrin concentrations are increased 5- to 161-fold higher than normal controls.63 Likewise, in heterozygous mice, we observed increased levels of protoporphyrin in erythrocytes (9-fold) and bile (2.2-fold; Figure 4). Data in this study show that erythrocytic protoporphyrin levels from heterozygous mice are within the range of erythrocyte protoporphyrin levels observed in patients with protoporphyria. Although bile protoporphyrin levels in the 10+/− mice are increased above controls, the accumulation is less than that observed in patients with protoporphyria.
Biochemical and histologic analysis of exon 10+/−mouse livers demonstrated that protoporphyrin had not reached sufficient levels to produce hallmark features of hepatic protoporphyria. Because the 37% ferrochelatase activities in the exon 10–deleted mouse are generally higher than those observed in patients with protoporphyria, it is likely that protoporphyrin accumulation is less than that in patients with protoporphyria; thus, serum protoporphyrin can be efficiently extracted by the liver and secreted into bile, without accumulating and causing liver injury. Serum protoporphyrin levels in exon 10–deleted mice were below the detection limit of the assay, consistent with the proposed complete biliary excretion.
Hepatic protoporphyria has been demonstrated in a homozygous mouse model for protoporphyria (Fechm1Pas).64 The mutagen-induced defect in these mice was characterized as a point mutation that decreased ferrochelatase activity to approximately 50% in heterozygous mice and 2.5% to 6% in homozygous mice.65 Heterozygous mice showed no signs of liver disease, whereas homozygous mice presented with cholestasis and severe hepatic dysfunction. The levels of ferrochelatase activity in theFechm1Pas mouse are approximately one eighth of the levels in mice with the exon 10 deletion. These data suggest that ferrochelatase activities of 37% and the resulting protoporphyrin production are not sufficient to accumulate in the liver and induce liver injury during the short lifespan of the mouse.
Protoporphyrin levels were sufficiently high in the heterozygous mice to elicit photosensitivity. Mice that were 14 months of age were exposed to mercury vapor lamp light, which has a high output in the 400-nm range. Three of 3 heterozygous mice showed light-induced lesions that were absent in control mice (Figure 5) indicating that ferrochelatase activities of 37% of normal result in protoporphyrin accumulation that is adequate to produce light-induced lesions.
Theoretically, if a dominant-negative interaction existed that abolished all enzymatic activity from a heterodimer, there would still remain 25% of normal activity from the wild-type homodimers. Because some patients with protoporphyria exhibit activities less than 25% of normal, it is probable that a second mechanism exists that would decrease enzymatic activity to less than 25%. Studies of ferrochelatase mRNA conducted in some families with protoporphyria have demonstrated that inheritance of a low-expressed allele is generally involved in clinical expression of the disease.66
A recent study has identified a T to C transition located in the third intron (IVS3-48T/C) of the human ferrochelatase gene.67This single nucleotide polymorphism modulates the use of a constitutive aberrant splice site leading to a decrease in steady-state levels of wild-type ferrochelatase mRNA. Lymphoblastoid ferrochelatase activities from individuals with a C/T genotype were decreased by approximately 16% compared to ferrochelatase activities from individuals with a T/T genotype. Hypothetically, the low-expressed allele (IVS3-48T/C) in combination with a null allele (such as a exon deletion) would produce ferrochelatase activities of about 34%. A low-expressing allele combined with a dominant-negative mutant allele could explain the low ferrochelatase activities observed in patients with protoporphyria.
Because highly variable ferrochelatase activities are described in patients with protoporphyria, it is probable that multiple modulating factors are involved in the molecular pathogenesis of this disease. The combined effects of both a dominant-negative ferrochelatase and a low-expressed allele could explain the wide variations in ferrochelatase activities observed in patients with protoporphyria. It is possible that only certain mutant ferrochelatase monomers are able to exert a dominant-negative effect on the ferrochelatase dimer, or that the level of the dominant-negative effect varies between different exon-deleted ferrochelatases. Therefore, it will be necessary to functionally study the interaction of various mutant monomers with wild-type ferrochelatase monomers to verify the existence of, and contribution to, a dominant-negative interaction.
The exon 10–deleted mouse model demonstrates that a heterozygous exon 10 deletion is sufficient to cause the enzymatic, biochemical, and some clinical manifestations of protoporphyria. Because the ferrochelatase exon 10–deleted mouse model demonstrates ferrochelatase activities that are near the threshold of phenotypic expression of protoporphyria, the heterozygous animal model will be useful for studying the effect of contributory genetic or environmental factors that modulate the protoporphyric phenotype.
We would like to thank Kim Kluckman for blastocyst injection, Dwight Debree and Slade Trabucco for technical assistance, and Victoria Madden from the University of North Carolina (UNC) Microscopy Services Laboratory for assistance with electron microscopy.
Prepublished online as Blood First Edition Paper, May 31, 2002; DOI 10.1182/blood-2001-12-0283.
Supported by National Institutes of Health-National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants DK47361 and DK34987 (to D.A.B.), P30-DK34987 (to S.T.M.), and a grant from the American Porphyria Foundation (to S.T.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David A. Brenner, University of North Carolina at Chapel Hill, CB#7038, Department of Medicine, Chapel Hill, NC 27599; e-mail: dab@med.unc.edu.