TO THE EDITOR:
A clonal relationship between nodal T-follicular helper cell lymphomas (nTFHLs) and myeloid neoplasms (MNs) is well-known. Identical mutations commonly seen in clonal hematopoiesis (CH) such as TET2 and DNMT3A mutations are frequently shared by both nTFHLs and MNs from the same patient, indicating a divergent clonal evolution of a founding CH clone to both neoplasms.1-3 In many cases, the MN occurs months or years after cytotoxic treatment of the preceding nTFHL, suggesting a potential role of selective treatment-related pressure and clonal expansion.4 However, there remains limited literature on the converse phenomenon of MN occurring before a nTFHL diagnosis, including the frequency, latency patterns, and patient- and disease-specific characteristics. This study aimed to address these knowledge gaps.
This study was approved by an institutional review board at the Memorial Sloan Kettering Cancer Center (MSKCC). The MSKCC pathology database was queried for all patients with concurrently or sequentially diagnosed MN and nTFHL from January 2010 to December 2024. In addition, we performed a systematic literature review to identify additional patients using PubMed and Google Scholar. We identified 41 patients with a history of MN and nTFHL, including 12 patients from MSKCC and 29 patients from a review of 19 published studies (Tables 1 and 2; supplemental Table 1).2,3,5-21 Of note, all nTFHL had features consistent with angioimmunoblastic T-cell lymphoma (AITL). Of these 41 total AITL cases identified, 29 patients (71%) had a MN diagnosed before or concurrently with AITL (MN-AITL cohort). None of these patients had received chemotherapy for AITL at the time of MN diagnosis. In the remaining 12 patients (29%), the MN was diagnosed after their AITL diagnosis (AITL-MN cohort).
Clinicopathologic characteristics of patients with concurrent or sequential MN and AITL diagnosis
| Patients . | Sex . | MN . | Clonal relationship . | Age (MN), y . | Age (AITL), y . | Interval between MN and AITL . | Biopsy site and DNA source (MN) . | Percent AITL involvement . | Biopsy site and DNA source (AITL) . | MN treatments . | AITL treatments . | Survival from AITL diagnosis (mo) . | Reference . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients with MN-AITL with paired NGS sequencing (n = 15) | |||||||||||||
| 1 | Male | CMML | Related | 76 | 78 | 16 months | PB | Not involved# | LN | allo-HSCT | BV-CHP, allo-HSCT | Alive (21) | MSKCC |
| 2 | Male | CMML | Related | 50 | 55 | 60 months | BM | Not involved# | LN | allo-HSCT | BV-CHP, allo-HSCT | Alive (29) | MSKCC |
| 3 | Male | MDS | Related | 75 | 75 | Synchronous | BM (FC sorted) | 2.1% by FC | BM (FC sorted) | EPOCH | Supportive care | Deceased (0.2) | 5 |
| 4 | Female | MDS | Related | 80 | 86 | 72 months | BM | NA | LN | Not treated | CHOP, azacitidine | Deceased (20) | 6 |
| 5 | NA | PV, post-PV MF | Related | NA | NA | NA | BM | NA | LN | NA | NA | NA | 7 |
| 6 | Female | PV | Related | 84 | 86 | 24 months | PB | NA | LN | Phlebotomy, hydroxyurea | BV | Deceased (12) | 8 |
| 7 | Male | MDS | Related | 78 | 81 | 42 months | BM | Not involved∗ | LN | NA | NA | NA | 9 |
| 8 | Male | CMML | Related | 83 | 83 | Synchronous | BM | ∼5%∗ | LN | NA | NA | NA | 9 |
| 9 | Female | MDS | Related | 73 | 73 | Synchronous | BM | Not involved∗ | LN | NA | NA | NA | 9 |
| 10 | Male | PMF | Related | 64 | 64 | Synchronous | BM | NA | LN | Not treated | CHOP | Deceased (64) | 10 |
| 11 | Female | ET | Related | 73 | 76 | 31 months | BM | NA | LN | Supportive care | CHOP | Deceased (98) | 10 |
| 12 | Male | ET | Related | 60 | 65 | 58 months | BM | NA | LN | Supportive care | CHOEP, ASCT | Deceased (72) | 10 |
| 13 | Female | PV | Unrelated | 72 | 76 | 42 months | BM | NA | LN | Phlebotomy, hydroxyurea | Supportive care | Deceased (1) | 10 |
| 14 | Female | MPN-U | NAΔ | 62 | 62 | Synchronous | BM | Involved∗ | LN | Not treated | CHOEP, ASCT | Alive (NA) | 10 |
| 15 | Male | CMML | Related | 70 | 70 | Synchronous | BM | Not involved## | LN | Azacitidine | Azacitidine, romidepsin | Alive (7) | 11 |
| Patients with AITL-MN with paired NGS sequencing (n = 10) | |||||||||||||
| 16 | Male | MDS | Related | 76 | 75 | 10 months | BM | 0.014% by FC | LN | Azacitidine | R-CHOP, ruxolitinib, duvelisib | Deceased (17) | MSKCC |
| 17 | Male | CMML | Related | 76 | 73 | 32 months | BM | 0.036% by FC | LN | NA | Lenalidomide-CHOEP, ASCT, and cerdulatinib | Deceased (61) | MSKCC |
| 18 | Male | MDS | Related | 79 | 74 | 66 months | BM | 1% by FC | LN | Luspatercept, romiplostim | CHOP, ASCT, romidepsin, bortezomib, duvelisib, ruxolitinib, and valemetostat | Deceased (109) | MSKCC |
| 19 | Female | AML, therapy related | Unrelated | 83 | 80 | 34 months | BM | 0.1% by FC | Tonsil | Azacitidine, venetoclax, | Mini-CHOP, duvelisib, romidepsin, and TTI-621 | Deceased (78) | MSKCC |
| 20 | Female | MPN (favored CNL) | Related | 75 | 72 | 36 months | BM | 0.004% by FC | LN | Not treated | CHOP, duvelisib, romidepsin, bortezomib, BV, and valemetostat | Deceased (43) | MSKCC |
| 21 | Male | MDS | Unrelated | 56 | 55 | 14 months | PB | 0.03% by FC | LN | Supportive care | CHOEP, ASCT | Deceased (16) | MSKCC |
| 22 | Male | AML MECOM rearranged | Related | 75 | 72 | 46 months | BM | Not involved by FC | LN | Azacitidine | CHOEP, romidepsin lenalidomide, and carfilzomib | Deceased (58) | MSKCC |
| 23 | NA | CMML | Related | NA | NA | NA | BM | NA | LN | NA | NA | NA | 7 |
| 24 | Male | MDS | Related | 67 | 64 | 36 months | BM | NA | LN | Azacitidine | CHOEP, ASCT | Deceased (48) | 12 |
| 25 | Male | AML NPM1 mutated | Related | 46 | 45 | 12 months | BM | NA | LN | FLAIE | CHOEP, ASCT | Deceased (18) | 3 |
| Patients . | Sex . | MN . | Clonal relationship . | Age (MN), y . | Age (AITL), y . | Interval between MN and AITL . | Biopsy site and DNA source (MN) . | Percent AITL involvement . | Biopsy site and DNA source (AITL) . | MN treatments . | AITL treatments . | Survival from AITL diagnosis (mo) . | Reference . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients with MN-AITL with paired NGS sequencing (n = 15) | |||||||||||||
| 1 | Male | CMML | Related | 76 | 78 | 16 months | PB | Not involved# | LN | allo-HSCT | BV-CHP, allo-HSCT | Alive (21) | MSKCC |
| 2 | Male | CMML | Related | 50 | 55 | 60 months | BM | Not involved# | LN | allo-HSCT | BV-CHP, allo-HSCT | Alive (29) | MSKCC |
| 3 | Male | MDS | Related | 75 | 75 | Synchronous | BM (FC sorted) | 2.1% by FC | BM (FC sorted) | EPOCH | Supportive care | Deceased (0.2) | 5 |
| 4 | Female | MDS | Related | 80 | 86 | 72 months | BM | NA | LN | Not treated | CHOP, azacitidine | Deceased (20) | 6 |
| 5 | NA | PV, post-PV MF | Related | NA | NA | NA | BM | NA | LN | NA | NA | NA | 7 |
| 6 | Female | PV | Related | 84 | 86 | 24 months | PB | NA | LN | Phlebotomy, hydroxyurea | BV | Deceased (12) | 8 |
| 7 | Male | MDS | Related | 78 | 81 | 42 months | BM | Not involved∗ | LN | NA | NA | NA | 9 |
| 8 | Male | CMML | Related | 83 | 83 | Synchronous | BM | ∼5%∗ | LN | NA | NA | NA | 9 |
| 9 | Female | MDS | Related | 73 | 73 | Synchronous | BM | Not involved∗ | LN | NA | NA | NA | 9 |
| 10 | Male | PMF | Related | 64 | 64 | Synchronous | BM | NA | LN | Not treated | CHOP | Deceased (64) | 10 |
| 11 | Female | ET | Related | 73 | 76 | 31 months | BM | NA | LN | Supportive care | CHOP | Deceased (98) | 10 |
| 12 | Male | ET | Related | 60 | 65 | 58 months | BM | NA | LN | Supportive care | CHOEP, ASCT | Deceased (72) | 10 |
| 13 | Female | PV | Unrelated | 72 | 76 | 42 months | BM | NA | LN | Phlebotomy, hydroxyurea | Supportive care | Deceased (1) | 10 |
| 14 | Female | MPN-U | NAΔ | 62 | 62 | Synchronous | BM | Involved∗ | LN | Not treated | CHOEP, ASCT | Alive (NA) | 10 |
| 15 | Male | CMML | Related | 70 | 70 | Synchronous | BM | Not involved## | LN | Azacitidine | Azacitidine, romidepsin | Alive (7) | 11 |
| Patients with AITL-MN with paired NGS sequencing (n = 10) | |||||||||||||
| 16 | Male | MDS | Related | 76 | 75 | 10 months | BM | 0.014% by FC | LN | Azacitidine | R-CHOP, ruxolitinib, duvelisib | Deceased (17) | MSKCC |
| 17 | Male | CMML | Related | 76 | 73 | 32 months | BM | 0.036% by FC | LN | NA | Lenalidomide-CHOEP, ASCT, and cerdulatinib | Deceased (61) | MSKCC |
| 18 | Male | MDS | Related | 79 | 74 | 66 months | BM | 1% by FC | LN | Luspatercept, romiplostim | CHOP, ASCT, romidepsin, bortezomib, duvelisib, ruxolitinib, and valemetostat | Deceased (109) | MSKCC |
| 19 | Female | AML, therapy related | Unrelated | 83 | 80 | 34 months | BM | 0.1% by FC | Tonsil | Azacitidine, venetoclax, | Mini-CHOP, duvelisib, romidepsin, and TTI-621 | Deceased (78) | MSKCC |
| 20 | Female | MPN (favored CNL) | Related | 75 | 72 | 36 months | BM | 0.004% by FC | LN | Not treated | CHOP, duvelisib, romidepsin, bortezomib, BV, and valemetostat | Deceased (43) | MSKCC |
| 21 | Male | MDS | Unrelated | 56 | 55 | 14 months | PB | 0.03% by FC | LN | Supportive care | CHOEP, ASCT | Deceased (16) | MSKCC |
| 22 | Male | AML MECOM rearranged | Related | 75 | 72 | 46 months | BM | Not involved by FC | LN | Azacitidine | CHOEP, romidepsin lenalidomide, and carfilzomib | Deceased (58) | MSKCC |
| 23 | NA | CMML | Related | NA | NA | NA | BM | NA | LN | NA | NA | NA | 7 |
| 24 | Male | MDS | Related | 67 | 64 | 36 months | BM | NA | LN | Azacitidine | CHOEP, ASCT | Deceased (48) | 12 |
| 25 | Male | AML NPM1 mutated | Related | 46 | 45 | 12 months | BM | NA | LN | FLAIE | CHOEP, ASCT | Deceased (18) | 3 |
allo-HSCT, allogeneic hematopoietic stem cell transplantation; AML, acute myeloid leukemia; ASCT, autologous stem cell transplantation; BM, bone marrow; BV, brentuximab vedotin; BV-CHP, brentuximab vedotin, cyclophosphamide, doxorubicin, and prednisolone; CHOEP, cyclophosphamide, doxorubicin hydrochloride, vincristine sulfate, etoposide, and prednisone; CMML, chronic myelomonocytic leukemia; CNL, chronic neutrophilic leukemia; EPOCH, etoposide, prednisolone, vincristine, cyclophosphamide, and hydroxydaunorubicin; ET, essential thrombocythemia; FC, flow cytometric study; FLAIE, fludarabine, cytarabine, etoposide, idarubicin; LN, lymph node; MDS, myelodysplastic syndrome; MF, myelofibros; MPN, myeloproliferative neoplasm; MPN-U, myeloproliferative neoplasm, unclassifiable; NA, not available; PB, peripheral blood; PMF, primary myelofibrosis; PV, polycythemia vera; R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone.
NGS studies for MNs in cases #1 and #2 were performed on samples obtained at the time of initial MN diagnosis, which occurred several years prior to the development of AITL. As a result, the MN samples were uninvolved by AITL based on flow cytometric and morphologic analysis.
The extent of bone marrow involvement by AITL was assessed by review of histomorphology and immunohistochemical staining.
The methodology to assess AITL involvement was not specified.
We excluded this patient from clonal relatedness assessment due to ambiguous genetic findings: the bone marrow sample was involved by AITL (extent of involvement was not specified in the original paper); the bone marrow and AITL samples shared only an IDH2 mutation with a 2% VAF in both samples and a RHOA mutation was only reported in the bone marrow but not in the AITL sample.
Clinicopathologic characteristics of patients with concurrent or sequential MN and AITL diagnosis and incomplete molecular information
| Patients . | Sex . | MN . | Clonal relationship . | Age (MN), y . | Age (AITL), y . | Interval between MN and AITL . | Molecular results . | MN treatments . | AITL treatments . | Survival from AITL diagnosis (mo) . | Reference . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients with MN-AITL with partial sequencing, paired Sanger sequencing, or no sequencing (n = 14) | |||||||||||
| 26 | Female | MPN | NA | 59 | 71 | 138 months | JAK2 p.V617F was detected in the MN by JAK2 single gene assay. 2 TET2 mutations detected in AITL by NGS. | Pegylated interferon, EPO mimetic | CEP | Deceased (23) | MSKCC |
| 27 | Male | AML | NA | 57 | 79 | 264 months | Not performed | HiDAC/IDR, cytarabine, and etoposide | Supportive care | Deceased (2) | MSKCC |
| 28 | Female | MDS | NA | 72 | 72 | 6 months | Not performed | NA | CHOP, pralatrexate, vorinostat, and BV | Deceased (64) | MSKCC |
| 29 | Female | CMML | Related | 70 | 80 | 120 months | Same TET2 mutation detected in both MN and AITL by Sanger sequencing DNMT3A, IDH2, and RHOA G17V were not detected in either neoplasm. | Not treated initially, later azacytidine | Azacytidine, rituximab | Alive (18) | 13 |
| 30 | Female | AML NPM1 mutation | NA | 65 | 75 | 120 months | TET2 mutation detected in MN No molecular data for AITL | Cytarabine, anthracycline, ASCT | BV-CHP | Alive (6) | 14 |
| 31 | Female | ET | NA | 78 | 78 | 6 months | Not performed | Hydroxyurea | CHOEP, R-ICE | Alive (5) | 15 |
| 32 | Male | ET | NA | 78 | 84 | 72 months | JAK2 p.V617F detected in MN No molecular data for AITL | Hydroxyurea | CEOP | Deceased (4) | 15 |
| 33 | Male | AML RUNX1::RUNX1T1 | NA | 47 | 47 | 4 months | Not performed | Idarubicin, cytarabine | Cyclophosphamide, vincristine, prednisolone | Deceased (1) | 16 |
| 34 | NA | CMML | NA | 55 | 56 | 7 months | Not performed | Hydroxyurea | MOPP, CHOP | Deceased (22) | 17 |
| 35 | Female | CMML | NA | 86 | 86 | Synchronous | Mutations in TET2, DNMT3A, ASXL1, SRSF2, NRAS, IDH1, CSF3R detected in MN by NGS No molecular data for AITL. | Azacytidine | Azacytidine, vinblastine | Alive (18) | 18 |
| 36 | NA | MDS | Related | NA | NA | NA | Same TET2 and DNMT3A mutations detected in both MN and AITL by Sanger sequencing. | NA | NA | NA | 19 |
| 37 | NA | MDS | Related | NA | NA | NA | Same DNMT3A mutation detected in both MN and AITL by Sanger sequencing. TET2 mutation detected only in AITL | NA | NA | NA | 19 |
| 38 | NA | MPN | NA | 58 | 62 | 48 months | Not performed | NA | NA | NA | 2 |
| 39 | NA | MDS | NA | 73 | 76 | 36 months | Not performed | NA | NA | NA | 2 |
| Patients with MN-AITL with partial sequencing, paired Sanger sequencing, or no sequencing (n = 2) | |||||||||||
| 40 | Female | AML | NA | 78 | 77 | 7 months | No molecular data for MN Negative for RHOA p.G17V and IDH2 p.R172 mutations in AITL by Sanger sequencing | NA | CHOP | Deceased (7) | 20 |
| 41 | Female | AML KMT2A rearranged | NA | 60 | 58 | 18 months | Not performed | Decitabine, cytarabine, and idarubicin | R-EPOCH, ASCT | Alive (21) | 21 |
| Patients . | Sex . | MN . | Clonal relationship . | Age (MN), y . | Age (AITL), y . | Interval between MN and AITL . | Molecular results . | MN treatments . | AITL treatments . | Survival from AITL diagnosis (mo) . | Reference . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients with MN-AITL with partial sequencing, paired Sanger sequencing, or no sequencing (n = 14) | |||||||||||
| 26 | Female | MPN | NA | 59 | 71 | 138 months | JAK2 p.V617F was detected in the MN by JAK2 single gene assay. 2 TET2 mutations detected in AITL by NGS. | Pegylated interferon, EPO mimetic | CEP | Deceased (23) | MSKCC |
| 27 | Male | AML | NA | 57 | 79 | 264 months | Not performed | HiDAC/IDR, cytarabine, and etoposide | Supportive care | Deceased (2) | MSKCC |
| 28 | Female | MDS | NA | 72 | 72 | 6 months | Not performed | NA | CHOP, pralatrexate, vorinostat, and BV | Deceased (64) | MSKCC |
| 29 | Female | CMML | Related | 70 | 80 | 120 months | Same TET2 mutation detected in both MN and AITL by Sanger sequencing DNMT3A, IDH2, and RHOA G17V were not detected in either neoplasm. | Not treated initially, later azacytidine | Azacytidine, rituximab | Alive (18) | 13 |
| 30 | Female | AML NPM1 mutation | NA | 65 | 75 | 120 months | TET2 mutation detected in MN No molecular data for AITL | Cytarabine, anthracycline, ASCT | BV-CHP | Alive (6) | 14 |
| 31 | Female | ET | NA | 78 | 78 | 6 months | Not performed | Hydroxyurea | CHOEP, R-ICE | Alive (5) | 15 |
| 32 | Male | ET | NA | 78 | 84 | 72 months | JAK2 p.V617F detected in MN No molecular data for AITL | Hydroxyurea | CEOP | Deceased (4) | 15 |
| 33 | Male | AML RUNX1::RUNX1T1 | NA | 47 | 47 | 4 months | Not performed | Idarubicin, cytarabine | Cyclophosphamide, vincristine, prednisolone | Deceased (1) | 16 |
| 34 | NA | CMML | NA | 55 | 56 | 7 months | Not performed | Hydroxyurea | MOPP, CHOP | Deceased (22) | 17 |
| 35 | Female | CMML | NA | 86 | 86 | Synchronous | Mutations in TET2, DNMT3A, ASXL1, SRSF2, NRAS, IDH1, CSF3R detected in MN by NGS No molecular data for AITL. | Azacytidine | Azacytidine, vinblastine | Alive (18) | 18 |
| 36 | NA | MDS | Related | NA | NA | NA | Same TET2 and DNMT3A mutations detected in both MN and AITL by Sanger sequencing. | NA | NA | NA | 19 |
| 37 | NA | MDS | Related | NA | NA | NA | Same DNMT3A mutation detected in both MN and AITL by Sanger sequencing. TET2 mutation detected only in AITL | NA | NA | NA | 19 |
| 38 | NA | MPN | NA | 58 | 62 | 48 months | Not performed | NA | NA | NA | 2 |
| 39 | NA | MDS | NA | 73 | 76 | 36 months | Not performed | NA | NA | NA | 2 |
| Patients with MN-AITL with partial sequencing, paired Sanger sequencing, or no sequencing (n = 2) | |||||||||||
| 40 | Female | AML | NA | 78 | 77 | 7 months | No molecular data for MN Negative for RHOA p.G17V and IDH2 p.R172 mutations in AITL by Sanger sequencing | NA | CHOP | Deceased (7) | 20 |
| 41 | Female | AML KMT2A rearranged | NA | 60 | 58 | 18 months | Not performed | Decitabine, cytarabine, and idarubicin | R-EPOCH, ASCT | Alive (21) | 21 |
AML, acute myeloid leukemia; ASCT, autologous stem cell transplantation; BV, brentuximab vedotin; BV-CHP, brentuximab vedotin, cyclophosphamide, doxorubicin, and prednisolone; CEOP, cyclophosphamide, etoposide, vincristine, and prednisone; CEP, cyclophosphamide, etoposide, and prednisone; CHOEP, cyclophosphamide, doxorubicin hydrochloride, vincristine sulfate, etoposide, and prednisone; CMML, chronic myelomonocytic leukemia; EPOCH, etoposide, prednisolone, vincristine, cyclophosphamide, and hydroxydaunorubicin; ET, essential thrombocythemia; HiDAC/IDR, high-dose cytarabine, idarubicin; MDS, myelodysplastic syndrome; MOPP, methylchloretamine, vincristine, procarbazine, and prednisone; MPN, myeloproliferative neoplasm; NA, not available; R-ICE, rituximab, ifosfamide, carboplatin and etoposide.
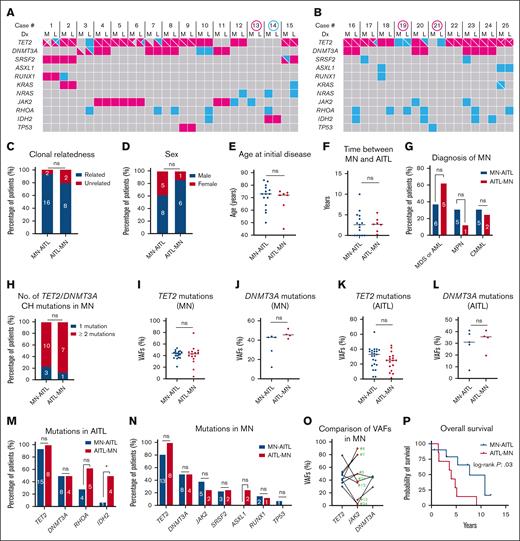
Of the 12 identified MSKCC patients, 9 (75%) had paired next-generation sequencing (NGS) performed on both neoplasms with our internal Memorial Sloan Kettering Integrated Mutation Profiling of Actionable Cancer Targets for Hematologic malignancies (MSK-IMPACT-HEME) panel.22 Of note, variant calls were restricted to a variant allele frequency (VAF) cutoff of 1% (with at least 5 supporting reads) for mutations previously reported in the patient, 2% for hot spot mutations, and 5% for non–hot spot mutations. Figure 1A-B illustrates the mutational profile of the 25 patients with paired targeted NGS, with or without a clonal relationship. There were also 16 patients from the literature review who had NGS data for both neoplasms and an additional 3 patients (patients 29, 36, and 37) with mutational data for both neoplasms by Sanger sequencing. In total, 28 patients had paired NGS (n = 25) or Sanger sequencing (n = 3) for subsequent comparisons.
Effects of temporal order on patients with clonally related MN and AITL. Oncoplots display common mutations in paired MNs (M) and AITLs (L) for patients with NGS data from the MN-AITL cohort (A) and the AITL-MN cohort (B). Mutations shared between the paired MNs and AITLs are highlighted in red color, whereas mutations not shared between the 2 neoplasms are highlighted in blue color. Multiple mutations within the same neoplasm are separated into different quadrants within a cell. Cases highlighted by a red circle indicate no clonal relatedness between MNs and AITLs. The case highlighted by a blue circle was excluded for clonal relatedness assessment due to ambiguous NGS result (explained in the note of Table 1). Although case 21 shared TET2 mutation (VAF, 1% in MN vs 41% in AITL) and case 23 shared IDH2 mutation (VAF, 0.3% in MN vs 7.5% in AITL) between the 2 neoplasms, the low VAFs in the MNs were most likely caused by contamination from infiltrating AITL in the bone marrow (confirmed by flow cytometry in case 21 which showed 0.03% abnormal T cells by flow cytometry). Therefore, MNs in these 2 cases were deemed to be negative/equivocal for these mutations. (C) Comparison of clonal relatedness between MN and AITL in both cohorts. All subsequent comparisons between the MN-AITL and AITL-MN cohorts were made among patients with clonally related neoplasms, including the comparisons for sex distribution (D), age at initial diagnosis for MN in the MN-AITL cohort or AITL in the AITL-MN cohort (E), the time (years) between MN and AITL diagnosis (F), MN types (G), number of patients with either 1 CH mutation or ≥ 2 CH mutations in TET2 and/or DNMT3A (H), VAFs of TET2 and DNMT3A mutations among MNs or AITLs (I-L), mutational rates in TET2, DNMT3A, RHOA, and IDH2 in AITLs (M), and mutational rates in TET2, DNMT3A, JAK2, SRSF2, ASXL1, RUNX1, and TP53 in MNs (N). (O) Comparison of paired VAFs for JAK2, TET2, and DNMT3A mutations in MNs from patients with clonally related neoplasms who had mutations in these genes; case numbers for each patient are marked in the graph. (P) Comparison of the overall survival between the patients with MN-AITL (from the time of MN diagnosis) and the patients with AITL-MN (from the time of AITL diagnosis) by Kaplan-Meier analysis. Numbers in the bars in panels C-D,G-H,M-N indicate the specific number of patients in each group. Black lines in the graphs in panels E-F,I-L indicate the median values. Dx, diagnosis; ns, not significant.
Effects of temporal order on patients with clonally related MN and AITL. Oncoplots display common mutations in paired MNs (M) and AITLs (L) for patients with NGS data from the MN-AITL cohort (A) and the AITL-MN cohort (B). Mutations shared between the paired MNs and AITLs are highlighted in red color, whereas mutations not shared between the 2 neoplasms are highlighted in blue color. Multiple mutations within the same neoplasm are separated into different quadrants within a cell. Cases highlighted by a red circle indicate no clonal relatedness between MNs and AITLs. The case highlighted by a blue circle was excluded for clonal relatedness assessment due to ambiguous NGS result (explained in the note of Table 1). Although case 21 shared TET2 mutation (VAF, 1% in MN vs 41% in AITL) and case 23 shared IDH2 mutation (VAF, 0.3% in MN vs 7.5% in AITL) between the 2 neoplasms, the low VAFs in the MNs were most likely caused by contamination from infiltrating AITL in the bone marrow (confirmed by flow cytometry in case 21 which showed 0.03% abnormal T cells by flow cytometry). Therefore, MNs in these 2 cases were deemed to be negative/equivocal for these mutations. (C) Comparison of clonal relatedness between MN and AITL in both cohorts. All subsequent comparisons between the MN-AITL and AITL-MN cohorts were made among patients with clonally related neoplasms, including the comparisons for sex distribution (D), age at initial diagnosis for MN in the MN-AITL cohort or AITL in the AITL-MN cohort (E), the time (years) between MN and AITL diagnosis (F), MN types (G), number of patients with either 1 CH mutation or ≥ 2 CH mutations in TET2 and/or DNMT3A (H), VAFs of TET2 and DNMT3A mutations among MNs or AITLs (I-L), mutational rates in TET2, DNMT3A, RHOA, and IDH2 in AITLs (M), and mutational rates in TET2, DNMT3A, JAK2, SRSF2, ASXL1, RUNX1, and TP53 in MNs (N). (O) Comparison of paired VAFs for JAK2, TET2, and DNMT3A mutations in MNs from patients with clonally related neoplasms who had mutations in these genes; case numbers for each patient are marked in the graph. (P) Comparison of the overall survival between the patients with MN-AITL (from the time of MN diagnosis) and the patients with AITL-MN (from the time of AITL diagnosis) by Kaplan-Meier analysis. Numbers in the bars in panels C-D,G-H,M-N indicate the specific number of patients in each group. Black lines in the graphs in panels E-F,I-L indicate the median values. Dx, diagnosis; ns, not significant.
Clonal relatedness was determined by mutations shared between MNs and AITL. The following methods were applied to identify the cellular compartment in which a mutation was present (neoplastic T cells or myeloid compartment): (1) immunohistochemical staining and/or flow cytometry was performed in a subset of the MN samples (mostly bone marrow) to rule out an involvement by AITL; (2) flow cytometric sorting was used to delineate myeloid cells from neoplastic T cells in 1 patient; (3) for cases with low levels of AITL involvement, mutations were considered present in the bone marrow myeloid compartment if their VAFs were ≥4 times the AITL percentage1; and (4) JAK2 mutations were excluded from the evaluation for clonal relatedness, even present in tissues with AITL.
After excluding 1 patient with ambiguous genetic results (patient 14), we identified a clonal relationship in 24 of 27 (89%) patients including 94% (16/17) of the MN-AITL cohort and 80% (8/10) of the AITL-MN cohort (Table 1 and Table 2; Figure 1A-C). All patients with clonally related neoplasms exhibited shared mutations in TET2 and/or DNMT3A, with or without additional shared aberrancies.
We only included patients with clonally related MNs and AITL for further analysis. There was no significant difference in patient demographics between the MN-AITL and AITL-MN cohorts, including sex (Figure 1D) and age at the time of initial diagnosis of either MN or AITL (Figure 1E). The median interval between AITL and MN diagnosis was 2 years (range, 0-10) in the MN-AITL cohort and 2.7 years (range, 0.2-5.5) in the AITL-MN cohort (Figure 1F). The types of MNs were not significantly different from the 2 cohorts (Figure 1G).
TET2 mutations, particularly nonsense or frameshift mutations, were the most frequent alterations in both the MN-AITL and AITL-MN cohorts (Figure 1A-B). Multiple TET2 mutations often coexisted within the same tumor. AITLs frequently acquired unique TET2 mutations not shared with their paired MNs, a phenomenon observed in both MN-AITL (patients 1, 6, and 7) and AITL-MN (patient 17 and 20) patients. Contrary to TET2, only a single DNMT3A mutation was detected in all but 1 neoplasm (patient 3). Seventeen (81%) of inferred CH clones had ≥2 CH mutations (either TET2 and/or DNMT3A; Figure 1H). The VAFs for TET2 and DNMT3A mutations in both the MNs and AITLs were comparably high, and there were no discernible differences in the median TET2 or DNMT3A VAFs when stratified by MN-AITL vs AITL-MN cohort (Figure 1I-L).
RHOA and IDH2 mutations are characteristic of AITL and are typically not shared with paired MNs.23 However, we observed a notable trend toward a higher frequency of these mutations in the AITL-MN cohort compared to the MN-AITL cohort. RHOA mutations occurred in 63% of the AITL-MN cohort vs 29% of the MN-AITL cohort (P = .2; Figure 1M). Similarly, IDH2 mutations were detected in 50% of the AITL-MN cohort vs only 7% of the MN-AITL cohort (P = .04; Figure 1M). RHOA and IDH2 mutations in both cohorts often involved hotspot loci, specifically p.G12V for RHOA and p.R172X for IDH2 (supplemental Table 1). On the contrary, the frequencies of recurrent mutations in MNs revealed no significant differences between the clinical cohorts including TET2, DNMT3A, JAK2, SRSF2, ASXL1, RUNX1, and TP53 (Figure 1N; supplemental Table 1). Notably, only 1 patient (5%) with clonally related neoplasms acquired a TP53 mutation as a subclonal event (patient 9). The VAFs of JAK2 mutations were slightly lower than those of TET2 or DNMT3A mutations in most cases (Figure 1O).
Survival data was available in 7 patients from the AITL-MN cohort and in 10 patients from the MN-AITL cohorts. All patients with AITL-MN died, whereas 40% of patients with MN-AITL were alive at time of last follow-up (P = .1). Interestingly, patients with AITL-MN appeared to have inferior outcomes with a median overall survival of 4 years from the time of their AITL diagnosis vs 7.7 years for patients with MN-AITL from the time of their MN diagnosis (P = .03; Figure 1P).
To our knowledge, this study uniquely details the largest cohort of patients with both a MN and nTFHL diagnosed concurrently or sequentially. A key finding is the demonstration of nearly 90% clonal relatedness between the 2 neoplasms when diagnosed in the same patients regardless of the temporal order. This further supports the concept of divergent clonal evolution from an ancestral CH clone, which may have potential implications for patient management. One immediate question is whether these MNs arising after an nTFHL diagnosis are truly therapy related. Several observations from this study argue against this pathogenesis. First, TP53 mutations, a critical genetic event enriched in therapy-related MNs, are nearly absent.24 Second, in patients with paired and clonally related neoplasms, their MNs were more commonly diagnosed before or concurrently with their AITL diagnosis. Lastly, the allelic burdens of CH mutations were similar in the AITL-MN cohort compared to the MN-AITL cohort. Collectively, the observations derived from this study support that the development of MN in the setting of AITL is driven by intrinsic genetic susceptibility, most likely secondary to CH mutations arising from ancestral clones, rather than by therapy-related selection. This is consistent with the observation that most neoplasms in our study have multiple CH mutations with high allelic burdens, carrying a higher risk for progression to MNs.25 In summary, this study offers novel and fundamental insights into the clinicopathological characteristics of concurrently or sequentially diagnosed MN and nTFHL, highlighting the need for further investigation.
Acknowledgments: The study was funded, in part, by the National Institutes of Health (NIH)/National Cancer Institute (NCI) Cancer Center Support Grant P30 CA008748. W.X. is supported by a Cycle for Survival’s Equinox Innovation Award in Rare Cancers, MSK Leukemia SPORE (Career Enhancement Program, NIH/NCI P50 CA254838), and an NCI grant K08CA267058.
Contribution: M.Y. identified and annotated the patients, analyzed the data, and drafted the manuscript; R.S., K.S.L., W.T.J., and S.M.H. provided and annotated patients; A.D. reviewed cases; and W.X. conceived and designed the study, identified, and annotated the patients, supervised the data analysis, and revised the manuscript.
Conflict-of-interest disclosure: W.X. has received research support from Stemline Therapeutics. R.S. has received research funding from Pfizer. W.T.J. has received consulting fees from BioNTech. S.M.H. has received grants from ADC Therapeutics, Affimed, Aileron, Celgene, CRISPR Therapeutics, Daiichi Sankyo, Forty Seven, Inc, Kyowa Hakko Kirin, Takeda, Secura Bio, Seattle Genetics, and Trillium Therapeutics; and personal fees from Yingli Pharma Limited, Takeda, Abcuro, Inc, Autolus, Auxilius Pharma, Corvus, Daiichi Sankyo, Dren Bio, Johnson & Johnson Medicine/Janssen Research & Development, Kyowa Hakko Kirin, March Bio, Ono Pharmaceuticals, Pfizer, Shoreline Biosciences, Inc, Secura Bio, and SymBio. A.D. has received research support from Roche/Genentech, Takeda, and AstraZeneca. The remaining authors declare no competing financial interests.
Correspondence: Wenbin Xiao, Department of Pathology and Laboratory Medicine, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10065; email: xiaow@mskcc.org.
Reference
Author notes
Original data are available on request from the corresponding author, Wenbin Xiao (xiaow@mskcc.org).
The full-text version of this article contains a data supplement.