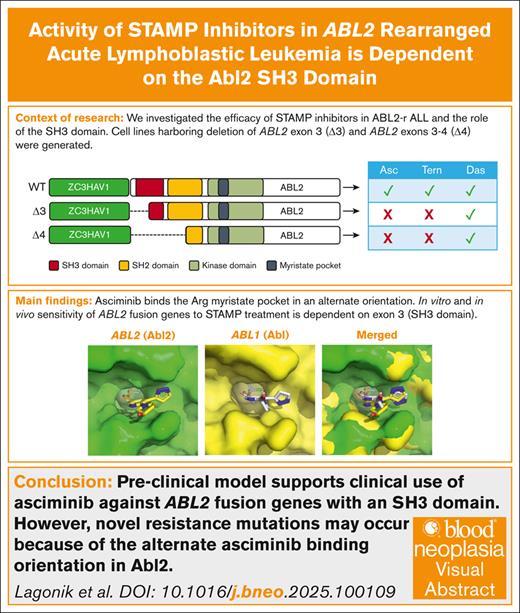
Key Points
Asciminib inhibits Abl2 (Abelson-related gene) by interacting with different myristate pocket residues when compared with Abl.
ABL2 exon 3–encoded region of the SH3 domain is critical for STAMP inhibitor activity against ABL2 rearrangements in vitro and in vivo.
Visual Abstract
ABL2 rearranged (ABL2r) acute lymphoblastic leukemia (ALL) is a subtype of high-risk Philadelphia chromosome–like ALL. Patients with ABL2r ALL are treated with high-dose multiagent chemotherapy, and the addition of tyrosine kinase inhibitors to their treatment regimen is currently being explored. We have previously demonstrated the in vitro sensitivity of cells harboring the ZC3HAV1::ABL2 fusion to asciminib, the first inhibitor that specifically targets the Abl myristate pocket (STAMP). In this study, we extended these in vitro findings to demonstrate similar sensitivity to the second-generation STAMP inhibitor, TERN-701, using ZC3HAV1::ABL2 ALL cells. In addition, using truncated ZC3HAV1::ABL2 isoforms, we identified that exon 3 of Abl2 (encoded by ABL2) is essential for the efficacy of both STAMP inhibitors. In an in silico model, we further demonstrated that different myristate pocket residues impact the effective binding of asciminib to Abl2 compared to Abl. Importantly, this suggests that, in the clinical setting, different asciminib binding site mutations may be anticipated with STAMP treatment for ABL2r ALL. Finally, we demonstrated the efficacy of both STAMP inhibitors against cells from patients with ZC3HAV1::ABL2 and asciminib as a novel treatment for ZC3HAV1::ABL2 disease in a preclinical in vivo study.
Introduction
Treatment advances have dramatically improved the survival of patients with acute lymphoblastic leukemia (ALL), most notably pediatric patients, with the 5-year survival rate currently exceeding 90%.1 In contrast, ALL remains a devastating disease in adolescents and young adults, as well as older adults. High-risk genomic lesions, high relapse rates, and treatment resistance are more common in older age groups, leading to lower overall survival rates.2-5 For example, Philadelphia chromosome–positive ALL (Ph+ ALL), a high-risk subtype, most frequently occurs in patients older than 40 years and is associated with high relapse rates despite the addition of tyrosine kinase inhibitors (TKIs) to conventional chemotherapy.3,6 Similarly, Philadelphia chromosome–like ALL (Ph-like ALL) is associated with treatment failure and overall poor survival outcomes in children and adults.4,5,7 Ph-like ALL represents ∼15% to 20% of ALL cases and prevalence increases with age, from 10% in pediatric patients to >25% in adolescents and adults.4,5 This subtype is driven by a heterogeneous group of lesions that cause dysregulation of kinase and cytokine signalling.4,5,7,8 This diversity of driver lesions leads to challenges in precision treatment options, with the ABL class being of particular interest given that TKIs are currently in clinical use with promising therapeutic effects.4
The ABL-class of Ph-like ALL is characterized by fusion genes in which the carboxyl-terminal of the ABL-class genes (ABL1, ABL2, CSF1R, PDGFRA/B) fuses with the amino-terminal of a partner gene, leading to the formation of a fusion protein with altered functions. The consequent Abl (encoded by ABL1) and Abl2 (encoded by ABL2) chimeric proteins, collectively referred to as ABL rearrangements (ABLr), account for ∼25% of the ABL class cases and share structurally homologous SH3, SH2, and kinase domains (KDs).8-10 The structural conservation of these domains exceeds 95% and includes the critical binding sites for TKIs, which bind the adenosine triphosphate (ATP) pocket, and the newer class of inhibitors, which specifically target the Abl myristate pocket (STAMP) inhibitors. In most cases, both Abl and Abl2 chimeric proteins can be successfully targeted with the same TKIs.4,10 In this study, we focus on ABL2 rearrangements (ABL2r), which lead to ABL2 exon 5 and exon 3 breakpoints. Patients with these lesions have the highest risk for treatment failure and the lowest overall survival (<40% after 2 years) of the ABL class group (pre-TKI era).9 Clinically, patients have a median age of 15 years, high white cell counts at diagnosis, coupled with high levels of measurable residual disease (MRD) at the end of the induction.9 Although the addition of TKIs to chemotherapy in these patients (including those with Ph+ and ABL-class Ph-like ALL in general) has improved the long-term outcomes,4,6,11,12 relapse caused by treatment resistance continues to be a clinically relevant concern. The most common mechanism of drug resistance observed in patients with chronic myeloid leukemia (CML) is the acquisition of KD point mutations, which lead to partial or complete abrogation of TKI sensitivity.13,14 KD mutations are the only reported mechanism of TKI resistance in patients with ALL.15 The most studied example is the Abl gatekeeper mutation, ABL1 p.T315I, which disrupts the binding of all ATP-competitive TKIs in clinical use, with the exception of ponatinib.16 The ABL1 p.T315I is one of the most common point mutations in patients with TKI-resistant CML and has also been observed in patients with Ph-like ALL with ABL1 rearrangements.14,15
One possible advantage of STAMP inhibitors is their potential to overcome TKI resistance caused by KD point mutations within the ATP pocket. Asciminib is currently the only US Food and Drug Administration–approved allosteric inhibitor in the class designated as STAMP inhibitors.17,18 The myristate pocket is located distally to the ATP pocket toward the C-lobe of the Abl/Abl2 KD.10,19 In the wild-type proteins, it functions as a negative regulator of kinase activity once bound by the myristoyl moiety, a polylipid modification present at the N-terminus of both Abl and Abl2 proteins (isoforms 1b).10,19,20 However, with the formation of ABLr, the myristoyl moiety is replaced with the 5′ partner, leading to loss of this autoinhibitory signal. Asciminib was designed to mimic the myristoyl moiety and thus binds the myristate pocket with high affinity. Upon binding, asciminib forces conformational changes in the Abl KD αI helix, thereby facilitating docking of the SH2 and SH3 domains to the KD and stabilizing the kinase in an inactive conformation.17,19,20 We previously have demonstrated the importance of the SH3 domain in asciminib-mediated inhibition of ABLr in Ph-like ALL in vitro.21,22 These results were also confirmed in asciminib-resistant rare isoforms of BCR::ABL1 with exon 2 missing in the Abl SH3 domain.23,24 Given the high structural homology between Abl and Abl2, resistance mechanisms applicable to ABL1r may also be relevant in the treatment of ABL2r disease; however, limited data are available regarding the efficacy of asciminib and STAMP inhibitors against ABL2r Ph-like ALL.22 In addition, the roles of the SH2 and SH3 domains in STAMP-mediated inhibition are poorly understood.
In this study, we developed an in silico homology model of Abl2 and the ZC3HAV1::ABL2 fusion gene was used as a representative lesion to further expand on and study the biology of ABL2r in in vitro and in vivo models (Figure 1). We report on the in vitro sensitivity of murine and patient leukemic cells that carry the ZC3HAV1::ABL2 fusion to a second-generation STAMP inhibitor, TERN-701,25 and provide in vitro data that confirm the critical role of the Abl2 SH3 domain for STAMP-mediated activity. In addition, the efficacy of asciminib was demonstrated against human leukemic cells that carry the ZC3HAV1::ABL2 fusion in a preclinical in vivo model. To our knowledge, this is the first time that STAMP inhibitor efficacy has been explored in detail in ABL2r ALL, and this will assist in the development of additional treatment strategies for ABLr ALL.
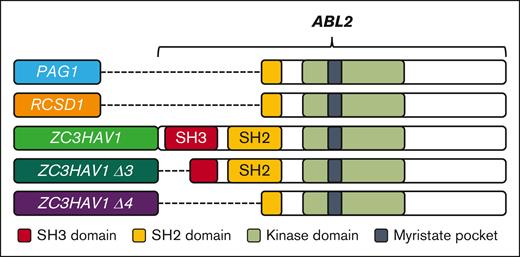
Schematic representation of the investigated ABL2 fusion genes, the breakpoint regions, and their relevant domains. Domain organization of ABL2 fusion genes. The 5′ partner genes are highlighted. The KD (light green) and myristate pocket (black) were retained in all fusions. The PAG1::ABL2 and RCSD1::ABL2 fusions retained a partial SH2 domain (truncation at exon 5), whereas the ZC3HAV1::ABL2 fusion retained the complete SH3 and SH2 domains (truncation at exon 3). The generated ZC3HAV1::ABL2 Δ3 (ZΔ3; exon 3 deletion; truncation of SH3 domain) and Δ4 isoforms (ΖΔ4; exon 3 & 4 deletion; deletion of SH3 and truncation of SH2 domain) are also shown. The SH3 and SH2 domains are shown in red and yellow, respectively. The PAG1::ABL2 and RCSD1::ABL2 fusion genes are provided for comparison.
Schematic representation of the investigated ABL2 fusion genes, the breakpoint regions, and their relevant domains. Domain organization of ABL2 fusion genes. The 5′ partner genes are highlighted. The KD (light green) and myristate pocket (black) were retained in all fusions. The PAG1::ABL2 and RCSD1::ABL2 fusions retained a partial SH2 domain (truncation at exon 5), whereas the ZC3HAV1::ABL2 fusion retained the complete SH3 and SH2 domains (truncation at exon 3). The generated ZC3HAV1::ABL2 Δ3 (ZΔ3; exon 3 deletion; truncation of SH3 domain) and Δ4 isoforms (ΖΔ4; exon 3 & 4 deletion; deletion of SH3 and truncation of SH2 domain) are also shown. The SH3 and SH2 domains are shown in red and yellow, respectively. The PAG1::ABL2 and RCSD1::ABL2 fusion genes are provided for comparison.
Methods
Patients with ZC3HAV1::ABL2 fusion genes
Leukemic blast cells isolated from the bone marrow (BM) and/or peripheral blood of 3 patients at diagnosis were referred to our laboratory for genomic analyses and ALL subtyping. The first patient was a male, aged 25 years, with a normal karyotype who was treated with the Australasian Leukaemia & Lymphoma Group (ALLG) ALL09 protocol, which intercalated chemotherapy with blinatumomab for induction.26 MRD, determined using allele-specific oligonucleotide polymerase chain reaction,27 was 2% at day 33 after a 4-drug induction regimen and was undetectable at day 79 after a 28-day infusion cycle of blinatumomab, which was used as consolidation before additional chemotherapy and maintenance. He was alive and in remission 36 months after diagnosis. The second patient was a woman, aged 61 years, with B-lineage ALL (B-ALL) who had the translocation t(1;7) and who underwent combination chemotherapy (hyper-fractionated cyclophosphamide, vincristine, anthracycline and dexamethasone) with imatinib for induction. MRD was measured using next-generation sequencing (LymphoTrack; Invivoscribe, San Diego, CA) and was determined to be 0.023% after cycle 2A and <0.01% after blinatumomab and before an allogeneic stem cell transplant with a sibling donor. She was alive at the time of this writing and in remission 3 years after the transplant. The third patient was a 13-year-old boy with the translocation t(1;7). The patient was treated in accordance with the very high-risk arm of the Children's Oncology Group AALL1131 protocol28 with incorporation of dasatinib. He achieved MRD-negative remission, as measured by flow cytometry, at the end of consolidation and was alive and in remission 6.5 years after diagnosis. The leukemia samples were subjected to our standard whole transcriptomic and bioinformatics analyses, as described elsewhere,22 the results of which were provided to the treating clinicians. Samples were used with informed consent under a human research ethics committee–approved research protocol.
In silico docking models
An Abl2 homology model was created using the X-ray crystal structure of Abl as a template (PDB:1OPK)20 and the DNA sequence of ABL2 (NM_001168237). The homology model was created using the Homology Model plugin of ICM Pro (Molsoft, San Diego, CA) and expanded from the Abl2 residue p.N110 to p.R558. No applicable crystal structure was available for Zc3hav1, thereby preventing the inclusion of the 5′ partner gene in the in silico modeling. After development, the homology model was subjected to 20 rounds of minimization, including side-chain refinement. The deleted regions of the ZC3HAV1::ABL2 Δ3 (ZΔ3) and ZC3HAV1::ABL2 Δ4 (ZΔ4) isoforms were modeled by deletion of the respective residues from the abovementioned homology model, followed by 20 rounds of minimization, including side-chain refinement.29-31 Docking of the STAMP inhibitor asciminib was specifically targeted to the Abl2 myristate pocket of the homology model using ICM Pro. Coordinates for the Abl inhibitor were created and minimized using phenix.elbow.32,33 A docking effort of 10 was used. The docking protocol allowed for flexible side chains.
Cell death assays
Transduced murine pro-B Ba/F3 (ATCC, Manassas, VA) cells were seeded at 8 × 104 cells per mL in a 24-well plate and were treated with vehicle control (dimethyl sulfoxide; DMSO) or increasing inhibitor concentrations that ranged from 0 to 50 000 nM for 72 hours. The cells were washed with a binding buffer composed of Hank’s balanced salt solution (Sigma-Aldrich, St Louis, MO), 5 mΜ calcium chloride (Sigma-Aldrich), and 1% HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; Sigma-Aldrich) and were stained for 20 minutes in the dark with 0.4 μL Annexin V-Phycoerythrin (PE) (BD Biosciences, Franklin Lakes, NJ) and 0.04 μL 7-aminoactinomycin D (7-AAD) (ThermoFisher, Waltham, MA) in 20 μL of binding buffer. The median lethal dose required to kill 50% of cells (LD50) was calculated as a measure of treatment-induced cytotoxicity using GraphPad Prism software (La Jolla, CA). The gating strategy is outlined in supplemental Figure 1A.
In vivo drug study
Female, 6-week-old NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory, Bar Harbor, ME) were subcutaneously injected with 0.1 mg/mL enrofloxacin (Baytril; Bayer, Leverkusen, Germany) in 0.9% sodium chloride per 10 g of body weight 3 days before sublethal gamma irradiation at 200 cGy. Approximately 1 × 106 patient blast cells (patient 1; supplemental Table 1) that expressed the ZC3HAV1::ABL2 fusion gene were IV injected into the tail vein. Mice were randomized into 4 cohorts, namely controlasciminib (n = 7), asciminib (n = 8), controldasatinib (n = 5), and dasatinib (n = 5). Leukemic engraftment was monitored by weekly tracking of human (h)CD45+ cells in the peripheral blood, and the percentage of hCD45+ was assessed by flow cytometry on a FACSCanto system (BD Biosciences). The gating strategy is provided in supplemental Figure 1C. Treatment commenced once the leukemic cell threshold of 5% was reached in the peripheral blood. Asciminib (30 mg/kg per day), dasatinib (20 mg/kg per day), and equivalent volumes of vehicle controls were administered via oral gavage (5 days on, 2 days off). Mice were maintained on water supplemented with 0.3 mg/mL enrofloxacin for the duration of the study and monitored daily for body weight loss and clinical signs of leukemia. Once the leukemic burden (hCD45+ cells) exceeded 50% in the peripheral blood, the mice were humanely killed, and the organs were harvested. Sections of the organs were fixed in formalin and stained with hematoxylin and eosin (Adelaide Health and Medical Sciences, The University of Adelaide). The experiments were conducted according to the South Australian Health and Medical Research Institute animal ethics committee guidelines.
Results
Computational modeling supports asciminib binding to the Abl2 myristate pocket
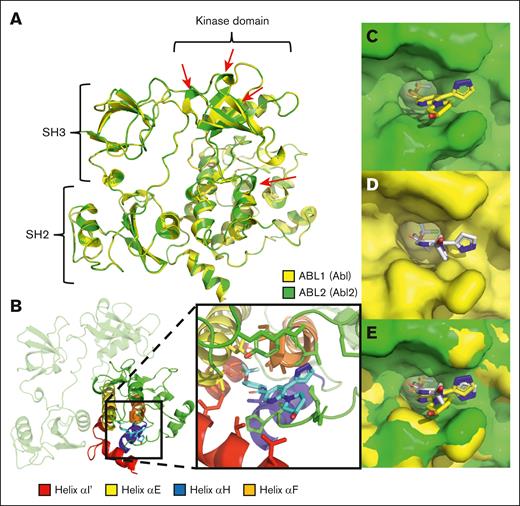
A crystal structure for the inactive conformation of Abl2 protein is currently unavailable, and thus a computational homology model of Abl2 (residue p.N110- p.R558) was developed to investigate in silico binding interactions between asciminib and the myristate pocket of Abl2. The complete SH3, SH2, and KD were present, and the results highlighted significant structural differences when compared with Abl that were observed at the top of the kinase N-lobe and the bottom of the ATP pocket (Figure 2A). Importantly, the in silico data confirmed the ability of asciminib to bind within the Abl2 myristate pocket (Figure 2B). However, despite the high degrees of structural similarity and conservation among the Abl asciminib binding residues, the homology model demonstrated a clear shift in the binding orientation of asciminib within the myristate pocket of Abl2 when compared with that of Abl (Figure 2C-E). This was primarily the consequence of interactions with a different set of Abl2 amino acids than seen with Abl. These residues were found predominantly at the extension of the helix αH, αF, and αΕ of the Abl2 myristate pocket (supplemental Figure 2).
Computational modeling of Abl2 (ABL2) supports the binding of asciminib to the Abl2 myristate pocket in an alternate orientation. (A) An overview of the complete homology model of Abl2 (green) superimposed on Abl (yellow; PDB:1OPK). Red arrows indicate regions of significant structural difference between Abl and Abl2. (B) Abl2 in complex with asciminib. The C-lobe of the Abl2 kinase is highlighted in green with the myristate pocket helixes colored. Helix αI′ (red), αE (yellow), αH (blue), and αF (orange) are shown. Surface representation of Abl2 (C), Abl (D), and the superimposition of Abl and Abl2 (E) myristate pocket in complex with asciminib.
Computational modeling of Abl2 (ABL2) supports the binding of asciminib to the Abl2 myristate pocket in an alternate orientation. (A) An overview of the complete homology model of Abl2 (green) superimposed on Abl (yellow; PDB:1OPK). Red arrows indicate regions of significant structural difference between Abl and Abl2. (B) Abl2 in complex with asciminib. The C-lobe of the Abl2 kinase is highlighted in green with the myristate pocket helixes colored. Helix αI′ (red), αE (yellow), αH (blue), and αF (orange) are shown. Surface representation of Abl2 (C), Abl (D), and the superimposition of Abl and Abl2 (E) myristate pocket in complex with asciminib.
STAMP-mediated inhibition of ABL2 fusion genes requires the presence of the Abl2 exon 3–encoded SH3 domain
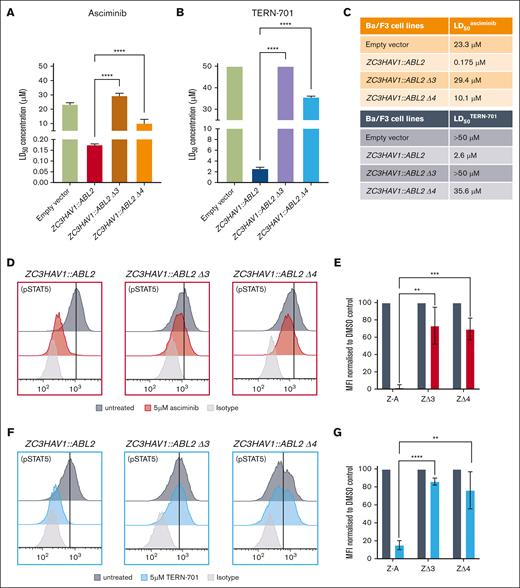
We have recently demonstrated the efficacy of asciminib against the ZC3HAV1::ABL2 fusion (LD50asciminib = 175 nM), the only ABL2 fusion in our patient cohort that harbors complete SH3 and SH2 domains22 (Figure 1; supplemental Figure 3). To investigate the role of the SH3 and SH2 domains in the STAMP-mediated inhibition, Ba/F3 cells expressing the ZC3HAV1::ABL2 Δ3 (ZΔ3; deletion of exon 3) and ZC3HAV1::ABL2 Δ4 (ZΔ4; deletion of exon 3 and 4) isoforms were generated to resemble clinically relevant ABLr breakpoints.22 The ZΔ3 isoform led to the truncation of the SH3 domain (loss of SH3 domain β1 strand and Receptor/Target (RT) loop; 34/54 amino acids present), whereas isoform ZΔ4 led to the deletion of the SH3 domain and truncation of the SH2 domain (33/94 amino acids present; supplemental Figure 4). The expression of the ΖΔ3 and ΖΔ4 isoforms in Ba/F3 cells with no additional mutations was confirmed using Sanger sequencing (data not shown), reverse transcriptase–quantitative polymerase chain reaction, and western blotting (supplemental Figure 5). Despite the deletion of the SH3 and SH2 domains, all transduced Ba/F3 cells became interleukin-3 independent (kinase active and capable of leukemogenesis). The Ba/F3 cells that expressed the ZΔ3 and ZΔ4 isoforms were resistant to asciminib, displaying an LD50asciminib of 29.4 and 10.1 μΜ, respectively, significantly higher than that of the full-length ZC3HAV1::ABL2 (LD50asciminib = 175 nM; both P < .0001; Figure 3A,C). These results highlight the importance of the SH3 domain for asciminib-mediated effect in cells carrying the ZC3HAV1::ABL2 fusion genes. Analogous results were obtained for the second-generation STAMP inhibitor, TERN-701 (Figure 3B-C). Ba/F3 cells that expressed the full-length ZC3HAV1::ABL2 fusion gene were sensitive to TERN-701 (LD50TERN-701 = 2.6 μΜ). As observed with asciminib, the ΖΔ3 and ZΔ4 isoforms were resistant to the inhibitor (LD50TERN-701 > 50 and 35.6 μΜ, respectively). The ability of asciminib and TERN-701 to inhibit kinase activity was investigated using flow cytometric analyses of the downstream effector protein, STAT5. Asciminib treatment (5 μM) had minimal effect on the levels of pSTAT5 in cells that expressed either of the isoform (ΖΔ3 and ΖΔ4) as opposed to the complete inhibition of STAT5 activity observed in the cells that expressed full-length ZC3HAV1::ABL2 (98% inhibition of STAT5 activity in full-length ZC3HAV1::ABL2 vs 27% [P < .01] in ΖΔ3 and 31% [P < .001] inhibition in ΖΔ4 cell lines) (Figure 3D-E). Similarly, the pSTAT5 levels were minimally affected in the ZΔ3 and ΖΔ4 cell lines in the presence of TERN-701 (5 μΜ) when compared with those with full-length ZC3HAV1::ABL2 (ΖΔ3: 14% decrease; P < .0001; and ΖΔ4: 24% decrease; P < .05; vs 85% decrease for the ZC3HAV1::ABL2 cell line; Figure 3F-G). All cell lines remained sensitive to dasatinib (supplemental Figure 6). These results demonstrate the critical role of the SH3 domain in STAMP-mediated inhibition of the ZC3HAV1::ABL2 fusion gene despite the unaffected binding of STAMP inhibitors to the Abl2 myristate pocket. However, the presence or absence of the SH3 domain is irrelevant to the inhibition of the ABL2 fusion genes by dasatinib and ATP-competitive TKIs.
Ba/F3 cells with the ZC3HAV1::ABL2 fusion gene are sensitive to STAMP inhibitors, and the SH3 domain is required for the STAMP-mediated in vitro effect. Sensitivity to asciminib (A) and TERN-701 (B) was assessed using the Annexin V-PE and 7-AAD cell death assays in Ba/F3 cells that expressed the empty vector (control), full-length ZC3HAV1::ABL2, and the ZC3HAV1::ABL2 Δ3 (ΖΔ3), ZC3HAV1::ABL2 Δ4 (ΖΔ4) isoforms. (C) The corresponding mean LD50 values are highlighted for each fusion gene. Representative histograms of the intracellular flow cytometry of Ba/F3 cell lines that were treated with 5 μM asciminib (D) or 5 μM TERN-701 (F) and stained with anti-pSTAT5 antibody. The corresponding column graphs of mean fluorescence intensity (MFI) values of pSTAT5 for asciminib (E) and TERN-701 treated cells (G), normalized to untreated control. All experiments were performed in biological replicates (n = 3) and statistical significance was calculated using unpaired Student t tests. Z-A, ZC3HAV1::ABL2; ΖΔ3, ZC3HAV1::ABL2 Δ3; ΖΔ4, ZC3HAV1::ABL2 Δ4. Significance is denoted with asterisks as follows: ∗∗∗∗P < .0001; ∗∗∗P < .001; ∗∗P < .01.
Ba/F3 cells with the ZC3HAV1::ABL2 fusion gene are sensitive to STAMP inhibitors, and the SH3 domain is required for the STAMP-mediated in vitro effect. Sensitivity to asciminib (A) and TERN-701 (B) was assessed using the Annexin V-PE and 7-AAD cell death assays in Ba/F3 cells that expressed the empty vector (control), full-length ZC3HAV1::ABL2, and the ZC3HAV1::ABL2 Δ3 (ΖΔ3), ZC3HAV1::ABL2 Δ4 (ΖΔ4) isoforms. (C) The corresponding mean LD50 values are highlighted for each fusion gene. Representative histograms of the intracellular flow cytometry of Ba/F3 cell lines that were treated with 5 μM asciminib (D) or 5 μM TERN-701 (F) and stained with anti-pSTAT5 antibody. The corresponding column graphs of mean fluorescence intensity (MFI) values of pSTAT5 for asciminib (E) and TERN-701 treated cells (G), normalized to untreated control. All experiments were performed in biological replicates (n = 3) and statistical significance was calculated using unpaired Student t tests. Z-A, ZC3HAV1::ABL2; ΖΔ3, ZC3HAV1::ABL2 Δ3; ΖΔ4, ZC3HAV1::ABL2 Δ4. Significance is denoted with asterisks as follows: ∗∗∗∗P < .0001; ∗∗∗P < .001; ∗∗P < .01.
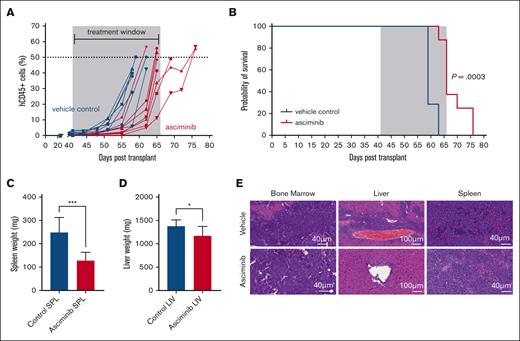
Asciminib treatment increases survival outcomes in ZC3HAV1::ABL2 PDX mice
Following the validation of asciminib efficacy in vitro, a patient-derived xenograft (PDX) model was developed, because there is currently no evidence of asciminib activity against human ALL cells carrying the ZC3HAV1::ABL2 gene fusion in vivo. Vehicle control mice (n = 7) developed an aggressive leukemic phenotype by median day 59 (median range, day 59-day 63), characterized by >50% blasts in the peripheral blood and clinical signs of disease. Asciminib-treated mice (n = 8) had significantly increased survival outcomes when compared with vehicle control mice (median day 66 vs day 59; P = .0003) (Figure 4A-B). The survival outcomes were comparable to those of dasatinib-treated ZC3HAV1::ABL2 mice (median: controldasatinib = day 57 vs dasatinib = day 67; P = .004; supplemental Figure 7A-B). Reverse transcriptase–quantitative polymerase chain reaction assays confirmed the expression of the ZC3HAV1::ABL2 fusion gene in control and asciminib-treated PDX mice (supplemental Figure 8), and no point mutations in SH3, SH2, and the KD were detected (data not shown). Asciminib-treated mice had significantly reduced spleen weights (130.5 ± 32.7 vs 247.8 ± 64.7 mg; P = .0002) and liver weights (1179 ± 193 vs 1373 ± 139.6 mg; P = .0382) when compared with vehicle-treated mice (Figure 4C-D). Surprisingly, dasatinib treatment did not reduce the spleen (dasatinib, 265 ± 24.8 vs controldasatinib, 291 ± 23.1) or liver weights (dasatinib, 1366 ± 69.4 vs controldasatinib, 1421 ± 101.7) when compared with the vehicle-treated mice (supplemental Figure 7C-D). Leukemic cells remained detectable in the BM, liver, and spleen of controlasciminib and asciminib-treated mice. All mice demonstrated leukemic blast infiltration in the BM with a minor decrease in the density of blasts in the asciminib-treated mice. Liver sections demonstrated the most notable reduction in leukemic blasts in asciminib-treated mice with a perivascular blastic infiltrate. Spleen sections from the asciminib cohort also exhibited a decrease in splenic ALL blast density (Figure 4E). These results indicate asciminib efficacy in PDX models of ZC3HAV1::ABL2 disease.
Asciminib reduces the leukemic burden and improves survival outcomes in vivo. NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice injected with patient cells that harbored the ZC3HAV1::ABL2 fusion gene were treated with vehicle control (blue curves) or asciminib (red curves). (A) The leukemic burden was evaluated by tracking hCD45+ cells in the peripheral blood. Each line represents an individual mouse. The treatment window is depicted in grey. (B) Kaplan-Meier curves of the control mice (n = 7) and asciminib-treated mice (n = 8) (30 mg/kg per day). ∗∗∗P = .0003. Statistical significance was measured using the log-rank test. (C) Spleen and (D) liver weights from the control and asciminib-treated mice at the experimental end point. Student t tests were used to determine significance. ∗P < .05; ∗∗∗P < .001. (E) Representative hematoxylin and eosin stains of BM, spleen, and liver sections. Images were analyzed using CaseViewer Software (version 2.2 RTM).
Asciminib reduces the leukemic burden and improves survival outcomes in vivo. NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice injected with patient cells that harbored the ZC3HAV1::ABL2 fusion gene were treated with vehicle control (blue curves) or asciminib (red curves). (A) The leukemic burden was evaluated by tracking hCD45+ cells in the peripheral blood. Each line represents an individual mouse. The treatment window is depicted in grey. (B) Kaplan-Meier curves of the control mice (n = 7) and asciminib-treated mice (n = 8) (30 mg/kg per day). ∗∗∗P = .0003. Statistical significance was measured using the log-rank test. (C) Spleen and (D) liver weights from the control and asciminib-treated mice at the experimental end point. Student t tests were used to determine significance. ∗P < .05; ∗∗∗P < .001. (E) Representative hematoxylin and eosin stains of BM, spleen, and liver sections. Images were analyzed using CaseViewer Software (version 2.2 RTM).
STAMP inhibitors halt ZC3HAV1::ABL2 signaling pathways ex vivo
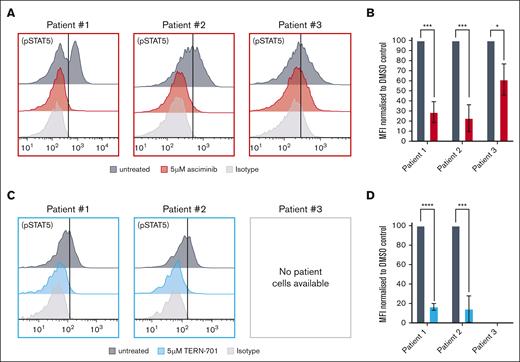
The efficacy of STAMP inhibitors was further assessed ex vivo in additional patient (n = 3) cells that harbored the full-length ZC3HAV1::ABL2 fusion gene. Asciminib treatment reduced pSTAT5 levels by 71% (patient 1; P = .003), 77% (patient 2; P = .006), and 40% (patient 3; P = .0125) when compared with untreated control (Figure 5A). Similarly, TERN-701 treatment reduced pSTAT5 levels by 84% (patient 1; P < .0001) and 86% (patient 2; P = .0004) (Figure 5B). The STAMP-induced reduction in pSTAT5 was comparable with that observed for dasatinib treatment (supplemental Figure 9).
STAMP-treated patient cells exhibited a reduction in pSTAT5 levels. Representative histograms of the intracellular flow cytometry of ZC3HAV1::ABL2 patient cells treated with 5 μM asciminib (A) or 5 μM TERN-701 (C) and stained with anti-pSTAT5 antibody. Corresponding column graphs of the MFI values for pSTAT5 for the asciminib (B) and TERN-701 treated cells (D) normalized to the untreated control. All experiments were performed in biological replicates (n = 3), and statistical significance was calculated using the unpaired Student t test. ∗P < .05; ∗∗∗P < .001; ∗∗∗∗P < .0001.
STAMP-treated patient cells exhibited a reduction in pSTAT5 levels. Representative histograms of the intracellular flow cytometry of ZC3HAV1::ABL2 patient cells treated with 5 μM asciminib (A) or 5 μM TERN-701 (C) and stained with anti-pSTAT5 antibody. Corresponding column graphs of the MFI values for pSTAT5 for the asciminib (B) and TERN-701 treated cells (D) normalized to the untreated control. All experiments were performed in biological replicates (n = 3), and statistical significance was calculated using the unpaired Student t test. ∗P < .05; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Discussion
ATP-competitive TKIs are currently used in treatment regimens for both patients with Ph+ or Ph-like ALL with ABLr.4-6,34 However, similar to BCR::ABL1 positive leukemias, the emergence of TKI resistance often occurs,13-15,35 highlighting the need for novel treatment options. The STAMP inhibitors asciminib and TERN-701, were designed to bind the myristate pocket, a previously unexploited feature of Abl and Abl2, forcing conformational changes that involve the αΙ′ helix (part of the myristate pocket; Figure 2B), SH2, SH3, and the KD, thereby stabilizing Abl and Abl2 in inactive conformations.18-20 The distant location of the myristate pocket from the ATP pocket led to a distinct asciminib resistance mutation profile, which may enable synergistic targeting of ABLr with agents from the 2 drug classes.17,20 Clinical trials (ClinicialTrials.gov identifiers: NCT03906292, NCT03578367, NCT02081378) that explored the combination of asciminib and ATP-competitive TKIs (imatinib, nilotinib, and dasatinib) in patients with BCR::ABL1 positive CML and those with Ph+ ALL have shown promising rates of deep molecular responses, with speculation of synergy or an additive effect between the 2 classes of inhibitors in targeting the Bcr::Abl1 chimeric protein.17,36,37 The role of asciminib as monotherapy or in combination with an ATP-competitive TKI in other ABLr settings remains to be determined, though any synergy between the 2 classes of drugs observed against Bcr::Abl1 is expected for other ABLr, though confirmation through future studies is needed.21,22
Given the extensive structural conservation of Abl and Abl2 (SH3, SH2, and KD, >95% similarity), ABL2r were highlighted as potential targetable lesions with asciminib. Although some domains of Abl2 have been independently crystalized, a complete crystal structure is not available.38,39 Consistent with the theoretical activity of asciminib against Abl2, our homology model supported the binding of asciminib to the myristate pocket of Abl2. Interestingly, despite the conservation of the Abl asciminib binding residues in Abl2, our model showed that asciminib bound in an alternate orientation, leading to a distinct asciminib contact residue network. These changes may have been a consequence of the minor structural differences observed at the Abl2 myristate pocket. The altered asciminib orientation and contact residues led to a lower affinity of the inhibitor for the Abl2 myristate pocket as observed by the reduction in sensitivity of the ZC3HAV1::ABL2 fusion when compared with native BCR::ABL1.17,22,40 The network of different contact residues used by asciminib in Abl2 is of potential clinical significance, because this could lead to novel asciminib-induced resistant point mutations within the myristate pocket of Abl2. In addition, the minor structural differences observed may be an important consideration for future specific drug design to target ABL1 and ABL2 gene fusions. However, crystallization of the complete Abl2 protein is required for the verification of the homology model.
After demonstrating asciminib efficacy against the ZC3HAV1::ABL2 fusion gene in vitro,22 we investigated efficacy in a preclinical PDX model. Because of the clinical success of dasatinib in targeting ABLr in Ph-like ALL, this TKI was used as a comparison measure for asciminib efficacy.4 To our knowledge, for the first time, we demonstrated the efficacy of asciminib against disease driven by the ZC3HAV1::ABL2 fusion gene in vivo in a preclinical drug study. Asciminib, administered as a single agent, slowed disease development and prolonged survival at a rate comparable with that of dasatinib treatment. The preclinical model established in this study provides strong evidence for the addition of asciminib to the treatment regimen of patients with ZC3HAV1::ABL2 while also addressing the lack of ABL2 preclinical models for use in clinically translatable studies.8
Of the 3 ABL2 fusion genes we previously investigated, ZC3HAV1::ABL2 was the only fusion with intact SH3 and SH2 domains.22 The roles of the SH2 and SH3 domains in Abl autoregulation and autoinhibition are well known.19,20,41-44 To investigate the contribution of these domains to STAMP inhibitor treatment, the ZC3HAV1::ABL2 isoforms ΖΔ3 and ΖΔ4 were developed to resemble clinical isoforms. The myristate pocket remained intact in all constructs. Ba/F3 cells that expressed the ΖΔ3 and ZΔ4 isoforms were resistant to asciminib and TERN-701. TERN-701 is a second-generation STAMP inhibitor currently in phase 1 clinical trials for patients with CML with resistance and/or intolerance to previous TKIs (ClinicalTrial.gov identifier: NCT06163430).25 These results highlight the importance of a functional SH3 domain in STAMP-induced allosteric inhibition of Abl2. This may be because of the involvement of the SH3 domain in the interaction between the myristate pocket and the N-lobe of Abl45 when in the inactive conformation. Importantly, high structural conservation between Abl and Abl2 would suggest that this is also the role of the SH3 domain in the inactive conformation of Abl2. During stabilization of the inactive conformation of both proteins, the SH3 domains recognize a proline-rich sequence, part of the SH2-kinase domain linker (SH2-KD linker), and adopt a polyproline type II helix configuration. The SH3-SH2-KD linker interface is essential for Abl autoinhibition.19,20,39,44,46 The RT-loop is of importance, because it is involved in the recognition of the SH2-KD linker for the formation of the SH3-SH2-KD linker interface and comprises important regulatory residues, such as p.Y116 (p.Y89 in Abl SH3). Other important regulatory residues include p.Y161 (p.Y134 in Abl SH3), which is part of the short helix, and p.Y139 (p.Y112 in Abl SH3 domain) of the N-Src loop.39 Phosphorylation of these residues disrupts the SH3-SH2-KD interface, preventing the recognition of the SH2-KD linker by the Abl/Abl2 SH3 domain, with minimal structural alterations,39 and thus the stabilization of the inactive conformation.
For these reasons, we hypothesize that disruption of the SH3-SH2-KD linker interface, either by loss of the SH3 domain from ABLr formation, partial deletions/insertions, or point mutations, would prevent the stabilization of the Abl or Abl2 inactive conformation, thereby leading to constitutive kinase activation. In addition, although deletion of the SH2-KD linker is unlikely to occur in Abl and Abl2 because of impaired kinase activity,41 SH2-KD linker point mutations that prevent the SH3 domain from binding could also disrupt the SH3-SH2-KD linker interface.17,47 This hypothesis is supported by recent studies that showed rare BCR::ABL1 isoforms (e14a3 and e13a3) that lack the SH3 RT-loop (exon 2) were resistant to asciminib despite the presence of asciminib binding by destabilization of the inactive conformation of Abl.23,24 Another ongoing study from our group has also demonstrated the importance of the SH3 domain in asciminib efficacy in ABL1r ALL.48
In conclusion, although previous studies have demonstrated the sensitivity of ABL2 fusion genes to ATP-competitive inhibitors,4,5,8,33 this study demonstrates, to our knowledge, for the first time, the in vitro and in vivo efficacy of STAMP inhibitors against the ZC3HAV1::ABL2 fusion gene. These data support the addition of asciminib to the treatment repertoire of patients who harbor ABL2 fusions in which the complete SH3 domain is present. The use of asciminib in patients with Ph-like ALL with an ABL2 fusion gene, as determined by fusion gene breakpoint screening, is warranted for patients who have acquired resistance mutations or who are refractory to other TKI therapies. The absence of the SH3 domain highlights a novel allosteric mechanism of primary resistance to STAMP inhibitors. Furthermore, the SH3 domain status in ABL2r disease could act as a surrogate in preclinical analysis to predict sensitivity to asciminib and future STAMP inhibitors, particularly in the current era in which next-generation sequencing is becoming the standard of care at diagnosis for patients with ALL.
Acknowledgments
Flow cytometry analysis and cell sorting were performed at the South Australian Health and Medical Research Institute (SAHMRI) in the Australian Cancer Research Foundation Cellular Imaging and Cytometry Core Facility. The facility is generously supported by the Detmold Hoopman Group, the Australian Cancer Research Foundation, and the Australian Government through the Zero Childhood Cancer Program. Animal model experiments were performed in the Bioresources Core Facility at SAHMRI, and the authors acknowledge the technical support provided.
This study was undertaken with the financial support of the SAHMRI BRIGHT Sparks Award and the National Health and Medical Research Council APP2007908 and an MRFF2007441 grants. E.L. was supported by a research scholarship from The University of Adelaide. D.T.Y. holds an early career clinical fellowship from the Beat Cancer project of the South Australian Cancer Council.
This study has been performed as partial fulfillment of the requirements for a PhD degree from The University of Adelaide Faculty of Sciences, Engineering and Technology by E.L.
Authorship
Contribution: E.L. designed the study, performed the experiments, analyzed the data, wrote the manuscript, and created the figures; E.C.P. designed the study and performed experiments; J.B.B. performed experiments; L.N.E. and S.L.H. designed the study; M.G., C.Y.F., and A.S.M. provided patient material for patient-derived xenograft and clinical data; D.T.Y. and T.P.H. designed the study and provided scientific and clinical insights; D.L.W. designed the study and provided scientific insight; and all authors critically reviewed the manuscript.
Conflict-of-interest disclosure: M.G. reports receiving research support from Servier and Amgen; and honoraria from Pfizer and Amgen. D.T.Y. reports receiving research support from Bristol Myers Squibb (BMS) and Novartis; and honoraria from BMS, Novartis, Pfizer, Ascentage, and Amgen. T.P.H. reports receiving research support from BMS and Novartis; and honoraria from BMS, Novartis, and Fusion Pharma. D.L.W. reports receiving research support from BMS; and honoraria from BMS and Amgen. The remaining authors declare no competing financial interests.
Correspondence: Deborah L. White, Precision Cancer Medicine, South Australian Health and Medical Research Institute, North Terrace, Adelaide, SA 5000, Australia; email: deborah.white@sahmri.com.
References
Author notes
The data are available on request from the corresponding author, Deborah L. White (deborah.white@sahmri.com).
The full-text version of this article contains a data supplement.