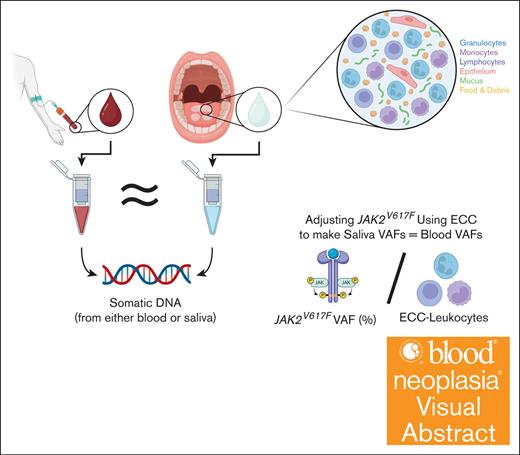
Visual Abstract
TO THE EDITOR:
Genetic analyses are expanding rapidly in availability and cost, allowing for increased clinical and research use.1 The COVID-19 pandemic has necessitated a rapid expansion of telemedicine, and patients will likely continue to demand the convenience of receiving clinical care without leaving the comfort of their homes.2-5 The demand for remote, user-friendly services applies to research studies as well. Although a blood draw is minimally invasive, a self-collected noninvasive technique would avail opportunities for frequent sampling with reduced cost. Saliva contains abundant leukocytes, making it an ideal source to noninvasively harvest hematopoietic cells. However, contaminating buccal epithelium can dilute out hematopoietic cells, thus potentially skewing interpretation of the variant allele frequency (VAF). Here, we demonstrate that saliva can be used to accurately quantify somatic variants in hematopoietic cells. Epigenetic cell counting (ECC) can be used to quantify contamination from epithelial cells. Unsupervised home saliva collection yields good DNA quality, even with intense daily collection. The study was conducted in accordance with the Declaration of Helsinki and approved by the institutional review board of University of California, Irvine, School of Medicine (UCI# 2014-9995, approved in 2014; UCI# 2018-4798, approved in 2018; and UCI# 23-32, approved in 2023). Informed consent was obtained from all individuals involved in the study.
Remote collection of samples relies on participant follow-through, so adequate return rates are required for a successful study. We are performing a fully online study to collect biological samples from affected and unaffected members of families with myeloproliferative neoplasm (MPN). After consenting and completing a questionnaire, participants are sent a sample collection kit, which contains a saliva sample kit, nail clippers, and an empty tube for blood collection. Participants are told that blood is ideal but if a blood draw is not feasible, then saliva (alternative somatic DNA source) and nail (germ line DNA source) samples will suffice. The return rate on sample kits was 67%, with the median time to return being 14 days. Of sample kits, 48.5% were returned with blood, saliva, and nails; 48.5% were returned with saliva and nails; and 3% were returned with nails only (supplemental Figure 1). This demonstrates that online remote home collection studies are a feasible and economical way to obtain biological samples. Moreover, alleviating the requirement for blood draw doubled the number of samples returned.
We confirmed that saliva collected by passive drool contains abundant leukocytes in both healthy controls and patients with hematologic malignancies. On direct histologic visualization in a patient with MPN (essential thrombocythemia) and healthy control, we observed leukocytes as well as epithelial cells (Figure 1A-B, bottom). On flow cytometry, both lymphoid and myeloid cells were observed in the saliva (Figure 1A, top). Similar results were seen with patients with polycythemia vera and primary myelofibrosis (supplemental Figure 2A). Saliva yields adequate DNA for assays such as targeted sequencing or whole exome sequencing (Figure 1B).
Saliva’s abundant leukocytes allow it to be an adequate noninvasive alternative to blood for genetic material. (A) Flow cytometry (top) and cytospin histology (bottom) of saliva from a healthy control and patient with essential thrombocythemia. Red arrows indicate leukocytes, and blue arrows indicate buccal epithelium. (B) Distribution of the genomic DNA concentrations isolated from blood (n = 570), saliva (n = 1670), and nails (n = 72); blood and saliva were eluted in 100 μL, and nails were eluted in 50 μL. Input amount of blood, saliva, and nails was 400 μL, 400 μL, and 20 mg respectively. (C) Concurrent blood and saliva samples were obtained from 49 patients with MPN, and JAK2V617F allele burden was measured by dPCR (r2 = 0.8931; P < .0001). Line of best fit, black; line of actual VAF, red.
Saliva’s abundant leukocytes allow it to be an adequate noninvasive alternative to blood for genetic material. (A) Flow cytometry (top) and cytospin histology (bottom) of saliva from a healthy control and patient with essential thrombocythemia. Red arrows indicate leukocytes, and blue arrows indicate buccal epithelium. (B) Distribution of the genomic DNA concentrations isolated from blood (n = 570), saliva (n = 1670), and nails (n = 72); blood and saliva were eluted in 100 μL, and nails were eluted in 50 μL. Input amount of blood, saliva, and nails was 400 μL, 400 μL, and 20 mg respectively. (C) Concurrent blood and saliva samples were obtained from 49 patients with MPN, and JAK2V617F allele burden was measured by dPCR (r2 = 0.8931; P < .0001). Line of best fit, black; line of actual VAF, red.
Next, to determine whether saliva is an equivalent to peripheral blood for detection of somatic hematopoietic mutations, we performed targeted next-generation sequencing with a myeloid panel (Archer VariantPlex Myeloid, 75 gene panel) on concurrently collected blood and saliva samples from a group of 8 patients with MPN and 1 patient with clonal cytopenia of undetermined significance. In all samples tested, we detected the known somatic mutations yielding nearly equivalent VAFs in blood and saliva, even with VAFs down to 3% (Table 1).
We also used microfluidic array plate digital polymerase chain reaction (dPCR; supplemental Figure 3A), which can accurately detect JAK2V617F VAFs of ∼ 0.1% (supplemental Figure 3B) to further compare the interchangeability of blood and saliva for somatic mutation detection. We also tested the JAK2V617F-dPCR system against JAK2V617F VAFs detected via the Illumina TruSight Myeloid Panel, showing the interchangeability of dPCR with an established next-generation sequencing panel (supplemental Figure 3C). We used dPCR to compare VAF in concurrently collected blood and saliva samples. In patients with low VAFs, the blood and saliva were essentially interchangeable; however, we noted that saliva slightly underestimated JAK2V617F VAF in patients with high VAFs (Figure 1C). This is likely because of contamination from buccal epithelial cells, which would be expected to skew VAF more if the sample has a high VAF (supplemental Figure 3D).
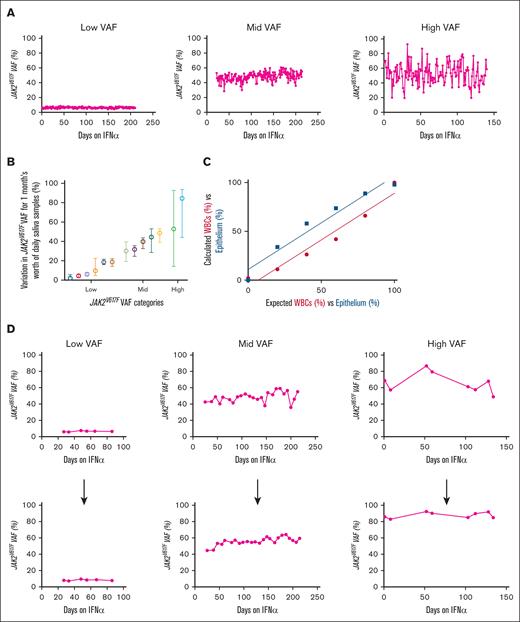
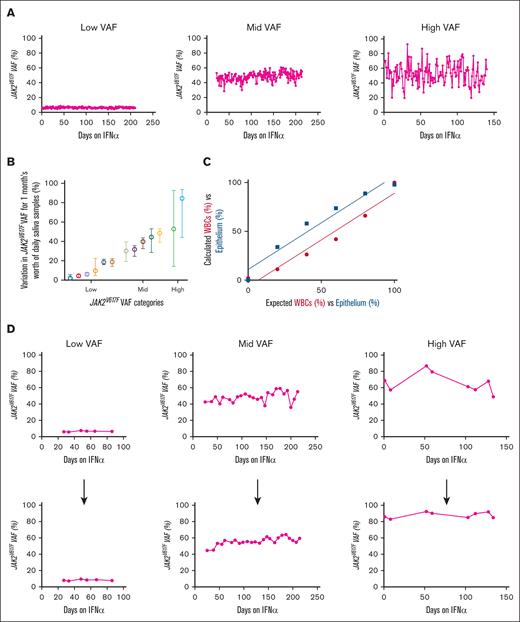
We are performing an observational study to intensely monitor patients initiating interferon alfa (IFN-α) for up to 1 year, with the goal of identifying early biomarkers that predict IFN-α success. This study includes frequent salivary sampling (daily to weekly) for JAK2V617F VAFs (via dPCR). Patients were able to provide and retain consistency in sample return rates (supplemental Figure 4A), DNA yields (supplemental Figure 4B), times (supplemental Figure 4C), and conditions (supplemental Figure 4D). When monitoring daily JAK2V617F VAFs in 3 representative patients with longitudinal samples, we noticed fluctuations, which are more pronounced in patients with higher baseline VAFs (Figure 2A). We also plotted daily sample–derived mean JAK2V617F VAFs with range for 1 month, from 13 patients with MPN, organized into 3 categories by baseline VAF: low = baseline VAF of <33%; mid = baseline VAF between 34% to 66%; and high = baseline VAF of >67% (Figure 2B). The highest variation in daily VAF was seen in the high VAF baseline VAF group.
Fluctuations in daily JAK2V617F allele burden with and without ECC to calculate epithelial cell contamination. (A) dPCR–derived JAK2V617F VAF was measured in daily passive drool saliva samples from patients since starting IFN-α treatment. Patients had either the following initial VAFs: low (8%), mid (45%), or high (78%). (B) Plot of daily sample–derived JAK2V617F VAFs (mean with range) for 1 month, from 13 patients with MPN. Low = baseline VAF of <33%; mid = baseline VAF between 34% to 66%; high = baseline VAF of >67%. (C) Detection sensitivity curves for leukocytes and epithelium when bisulfite converting different amounts of mixed blood and epithelium control DNA. (r2 = 0.9525; P = .0009). (D) Corrected JAK2V617F VAF in patients treated with IFN-α using the ECC system, weekly VAF points before (top) and after (bottom) ECC correction of patients treated with IFN-α.
Fluctuations in daily JAK2V617F allele burden with and without ECC to calculate epithelial cell contamination. (A) dPCR–derived JAK2V617F VAF was measured in daily passive drool saliva samples from patients since starting IFN-α treatment. Patients had either the following initial VAFs: low (8%), mid (45%), or high (78%). (B) Plot of daily sample–derived JAK2V617F VAFs (mean with range) for 1 month, from 13 patients with MPN. Low = baseline VAF of <33%; mid = baseline VAF between 34% to 66%; high = baseline VAF of >67%. (C) Detection sensitivity curves for leukocytes and epithelium when bisulfite converting different amounts of mixed blood and epithelium control DNA. (r2 = 0.9525; P = .0009). (D) Corrected JAK2V617F VAF in patients treated with IFN-α using the ECC system, weekly VAF points before (top) and after (bottom) ECC correction of patients treated with IFN-α.
To correct for buccal epithelium DNA in salivary samples, we explored methods of quantifying the contribution of leukocytes vs epithelium to the DNA. Because samples are collected directly in DNA/RNA preservative, direct counting of cells was not possible. We investigated genetic-based methods of cell counting, potentially via epigenetics5-9 or RNA.10,11 We found (ECC; using dPCR primers specific to differential promoter methylation status in hematopoietic vs epithelial cells) to be the best solution, combining the accuracy of epigenetic-based cellular quantification with the ease and cost of dPCR. Our system used dPCR probes specific for the methylated or unmethylated states of promoter 2 in Src homology-2-containing protein-tyrosine phosphatase 1 (SHP1P2). After bisulfited conversion of DNA, hematopoietic cells should only bind the unmethylated probe, whereas epithelial cells should only bind the methylated probe. We first confirmed the ability of the ECC probes for their respective cells by mixing different amounts of hematopoietic control DNA (from patient leukocytes) and epithelial control DNA (from 293T cell line) for bisulfite conversion and ECC dPCR (Figure 2C). By using ECC on the JAK2V617F VAFs of our patients who were treated with IFN-α (supplemental Figure 5), we were able to reduce the previously seen fluctuations seen in 3 patients shown in Figure 2A (Figure 2D), allowing for more precise interpretation of VAF trends. Therefore, ECC can serve as a tool to accurately quantify VAFs in hematopoietic cells from saliva samples.
To summarize, fully remote noninvasive collection of samples to assess somatic mutations in hematopoietic cells is feasible using saliva. These studies can be performed economically and without constraints of organizing remote blood draws. Furthermore, with saliva, intense daily monitoring of variants is feasible. Finally, we have addressed an important concern for accurate analysis of noninvasive salivary genomic material, epithelial contamination, which can be addressed efficiently with ECC. Although our focus is on rigorous monitoring of IFN-α–treatment initiation in patients with MPN with JAK2V617F, the same principles of accounting for buccal epithelium contamination via ECC should hold true for other hematologic malignancy mutation VAFs captured via saliva. We are currently optimizing dPCR protocols to monitor other MPN driver mutations (CALR and MPL) to use in other studies.
Acknowledgments: This study used core facilities from the University of California, Irvine’s Chao Family Comprehensive Cancer Center (5P30CA062203), including the Genomics Research and Technology Hub, Biobehavioral Core, and Flow Cytometry Core.
This research was funded by the National Institutes of Health (National Center for Research Resources and the National Center for Advancing Translational Sciences [grant T35DK128877-01]; the National Institute of General Medical Sciences [grant T32GM008620-23]; and the National Cancer Institute [grant T32CA009054-43]); and the Chao Family Comprehensive Cancer Center support grant (award number P30CA062203).
Contribution: E.M.S. developed the protocol for blood and saliva DNA purification, digital polymerase chain reaction (dPCR), salivary histological slide preparation/flow cytometry staining, interferon alfa (IFN-α) study, and epigenetic cell counting (ECC); G.R. developed the protocol for nail DNA purification; N.H. developed the Archer myeloid panel; A.G.F. developed the online study for families with myeloproliferative neoplasm; E.M.S., J.H.C., and J.C.H. processed blood and saliva samples; E.M.S., J.C.H., Y.J., E.A., H.H., and A.A. processed saliva samples from the patients who were treated with IFN-α; E.M.S., J.C.H., H.N., Y.J., E.A., A.A., and H.Y.L. purified DNA from blood and saliva; E.M.S. conducted bisulfite conversion on DNA for ECC analysis; G.R., H.N., H.Y.L., and S.B. processed and purified DNA from nail samples; E.M.S. collected and purified salivary leukocytes for histological slides and flow cytometry; E.M.S. analyzed samples on the dPCR platform and analyzed the data; A.G.F. supervised the study; E.M.S. and A.G.F. designed the figures; and E.M.S., G.R., and A.G.F. wrote the manuscript. L.C., E.M.M., and W.L.W. performed experiments and generated data figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Angela G. Fleischman, University of California, Irvine, 839 Medical Sciences Court, Sprague Hall 126, Irvine, CA 92617; email: agf@uci.edu.
References
Author notes
Data are available on request from the corresponding author, Angela G. Fleischman (agf@uci.edu).
The full-text version of this article contains a data supplement.