TO THE EDITOR:
Pediatric non-Down syndrome (non-DS) acute megakaryoblastic leukemia (AMKL) is characterized by chimeric fusion genes with poor outcomes.1 Colony-stimulating factor 1 receptor (CSF1R) is a type 3 receptor tyrosine kinase (RTK) playing pivotal roles in the survival, differentiation, and functions of myeloid lineage cells.2 An oncogenic RBM6::CSF1R fusion has previously been identified in the AMKL cell line MKPL1.3 Here, we characterize a new CSF1R fusion in a pediatric patient with non-DS AMKL and provide biological insights into CSF1R-driven AMKL pathogenesis.
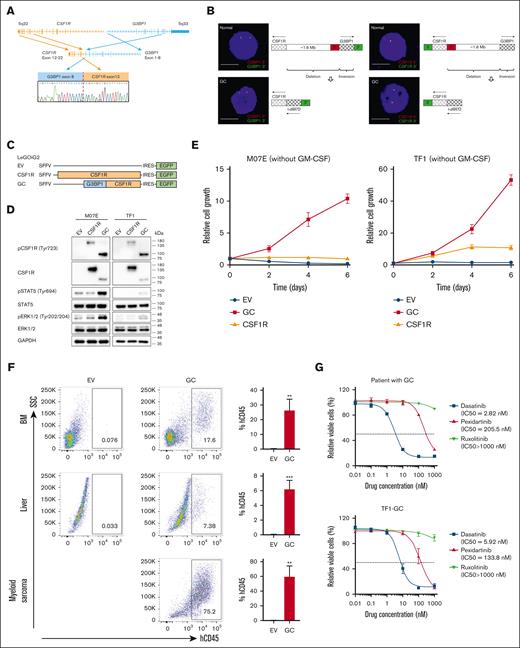
The patient was a 13-month-old girl who presented with easy bruising and pallor. Results of bone marrow (BM) examinations revealed 40% blasts with megakaryocytic morphology and immunophenotype (CD41a/CD42b/CD61+; supplemental Figure 1; supplemental Table 1). BM fibrosis, which is more frequent in AMKL,4 was evident in this patient. Results of cytogenetic analysis revealed monosomy 7 with a complex karyotype (45,XX,-1,-7,-9,+der(?)t(?;1)(?;p13),+der(?)(t?;1)(?;q25)[12]/44∼45,idem,+mar[cp2]/46,XX[2]). RNA sequencing identified a novel G3BP stress granule assembly factor 1 (G3BP1)::CSF1R (GC) fusion in her diagnostic BM,5 involving exon 8 of G3BP1 and exon 12 of CSF1R (Figure 1A). Fluorescence in situ hybridization revealed loss of 5' signal in both the G3BP1 and CSF1R loci (Figure 1B), suggesting that GC arises from a combination of interstitial deletion and inversion.
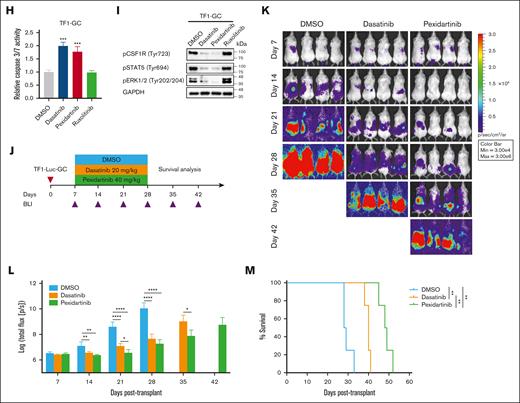
Oncogenic role of a novel GC fusion and its targetability. (A) Schematic diagram of G3BP1 and CSF1R rearrangement and Sanger sequencing validation of the GC fusion. Exons 1 to 8 of G3BP1 were fused to exons 12 to 22 of CSF1R. Red line, breakpoint. (B) Fluorescence in situ hybridization study and schematic diagram showing rearrangement of G3BP1 (left) and CSF1R (right) in the diagnostic BM of GC patient. Normal BM sample was used as the control. Scale bar, 10 μm. For both the G3BP1 and CSF1R loci, the normal cell has 2 red (5') and 2 green (3') signals, whereas the GC cell has 1 red (5') and 2 green (3') signals, indicating the loss of the red (5') signal in GC cell. The transcription of G3BP1 and CSF1R occurs in different directions, necessitating an inversion for the formation of a functionally effective gene fusion. (C) Schematic representation of the lentiviral vectors. (D) M07E and TF1 cells transduced with EV, CSF1R, or GC lentivirus (72 hours) were cultured without cytokine (GM-CSF) for 4 hours, and cell lysates were prepared for western blotting using indicated antibodies. Representative results from 3 independent experiments. (E) M07E and TF1 cells transduced with EV, CSF1R, or GC lentivirus were grown in medium without cytokine (GM-CSF), and cell viability was determined by CellTiter-Glo Luminescent Cell Viability Assay. Results are presented as mean ± standard deviation (SD) from 3 independent experiments. (F) Left, representative flow cytometric analysis of human CD45+ (hCD45+) cells in BM, liver, and sarcoma of non-obese diabetic-severe combined immunodeficiency IL2rγ null (NSG) mice intravenously injected with TF1-EV or TF1-GC cells (3e6 cells) for 28 days. Right, quantification of hCD45+ cells. ∗∗ and ∗∗∗ indicate P < .01 and P < .001 vs EV, respectively, by Student t test. EV, n = 3; GC, n = 3. (G) GC patient cells (from diagnostic BM) and TF1-GC cells were treated with dasatinib, pexidartinib, and ruxolitinib. DMSO was used as the control. After 72 hours, cell viability was determined using the CellTiter-Glo Luminescent Cell Viability Assay. IC50 was calculated by GraphPad Prism. (H) TF1-GC cells were treated with 100 nM dasatinib, 1000 nM pexidartinib, and 1000 nM ruxolitinib for 24 hours, and the induction of cell apoptosis was measured by Caspase-Glo 3/7 assay. ∗∗∗ indicate P < .001 vs DMSO by 1-way analysis of variance (ANOVA) followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (I) TF1-GC cells were treated with 100 nM dasatinib, 1000 nM pexidartinib, and 1000 nM ruxolitinib for 4 hours, and phosphorylation of CSF1R, STAT5, and ERK1/2 was detected by western blotting. Representative results of 2 independent experiments. (J) Experimental design of in vivo drug treatment. NSG mice were intravenously injected with 3e6 TF1-Luc-GC cells and treated with DMSO, 20 mg/kg dasatinib, and 40 mg/kg pexidartinib daily for 21 days by oral gavage starting from day 7 after cell injection. Leukemia burden was monitored by bioluminescence imaging (BLI) every 7 days from day 7 to day 42 after transplant. Survival of mice was measured after discontinuing treatment. (K) Follow-up of BLI of mice detected by in vivo imaging system spectrum. (L) Quantification of bioluminescence signal (total flux) of mice over time. ∗, ∗∗, and ∗∗∗∗ indicate P < .05, P < .01, and P < .0001 vs DMSO, respectively, by 1-way ANOVA followed by Dunnett test (days 7, 14, 21, and 28 after transplant) or Student t test (day 35 after transplant). Results are presented as mean ± SD after log transformation. n = 4 for each group. (M) Kaplan-Meier survival curves of mice. ∗∗ indicates P < .01 vs DMSO by log-rank test. n = 4 for each group. DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescent protein; GM-CSF, granulocyte-macrophage colony-stimulating factor; IRES, internal ribosome entry site; SFFV, spleen focus-forming virus.
Oncogenic role of a novel GC fusion and its targetability. (A) Schematic diagram of G3BP1 and CSF1R rearrangement and Sanger sequencing validation of the GC fusion. Exons 1 to 8 of G3BP1 were fused to exons 12 to 22 of CSF1R. Red line, breakpoint. (B) Fluorescence in situ hybridization study and schematic diagram showing rearrangement of G3BP1 (left) and CSF1R (right) in the diagnostic BM of GC patient. Normal BM sample was used as the control. Scale bar, 10 μm. For both the G3BP1 and CSF1R loci, the normal cell has 2 red (5') and 2 green (3') signals, whereas the GC cell has 1 red (5') and 2 green (3') signals, indicating the loss of the red (5') signal in GC cell. The transcription of G3BP1 and CSF1R occurs in different directions, necessitating an inversion for the formation of a functionally effective gene fusion. (C) Schematic representation of the lentiviral vectors. (D) M07E and TF1 cells transduced with EV, CSF1R, or GC lentivirus (72 hours) were cultured without cytokine (GM-CSF) for 4 hours, and cell lysates were prepared for western blotting using indicated antibodies. Representative results from 3 independent experiments. (E) M07E and TF1 cells transduced with EV, CSF1R, or GC lentivirus were grown in medium without cytokine (GM-CSF), and cell viability was determined by CellTiter-Glo Luminescent Cell Viability Assay. Results are presented as mean ± standard deviation (SD) from 3 independent experiments. (F) Left, representative flow cytometric analysis of human CD45+ (hCD45+) cells in BM, liver, and sarcoma of non-obese diabetic-severe combined immunodeficiency IL2rγ null (NSG) mice intravenously injected with TF1-EV or TF1-GC cells (3e6 cells) for 28 days. Right, quantification of hCD45+ cells. ∗∗ and ∗∗∗ indicate P < .01 and P < .001 vs EV, respectively, by Student t test. EV, n = 3; GC, n = 3. (G) GC patient cells (from diagnostic BM) and TF1-GC cells were treated with dasatinib, pexidartinib, and ruxolitinib. DMSO was used as the control. After 72 hours, cell viability was determined using the CellTiter-Glo Luminescent Cell Viability Assay. IC50 was calculated by GraphPad Prism. (H) TF1-GC cells were treated with 100 nM dasatinib, 1000 nM pexidartinib, and 1000 nM ruxolitinib for 24 hours, and the induction of cell apoptosis was measured by Caspase-Glo 3/7 assay. ∗∗∗ indicate P < .001 vs DMSO by 1-way analysis of variance (ANOVA) followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (I) TF1-GC cells were treated with 100 nM dasatinib, 1000 nM pexidartinib, and 1000 nM ruxolitinib for 4 hours, and phosphorylation of CSF1R, STAT5, and ERK1/2 was detected by western blotting. Representative results of 2 independent experiments. (J) Experimental design of in vivo drug treatment. NSG mice were intravenously injected with 3e6 TF1-Luc-GC cells and treated with DMSO, 20 mg/kg dasatinib, and 40 mg/kg pexidartinib daily for 21 days by oral gavage starting from day 7 after cell injection. Leukemia burden was monitored by bioluminescence imaging (BLI) every 7 days from day 7 to day 42 after transplant. Survival of mice was measured after discontinuing treatment. (K) Follow-up of BLI of mice detected by in vivo imaging system spectrum. (L) Quantification of bioluminescence signal (total flux) of mice over time. ∗, ∗∗, and ∗∗∗∗ indicate P < .05, P < .01, and P < .0001 vs DMSO, respectively, by 1-way ANOVA followed by Dunnett test (days 7, 14, 21, and 28 after transplant) or Student t test (day 35 after transplant). Results are presented as mean ± SD after log transformation. n = 4 for each group. (M) Kaplan-Meier survival curves of mice. ∗∗ indicates P < .01 vs DMSO by log-rank test. n = 4 for each group. DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescent protein; GM-CSF, granulocyte-macrophage colony-stimulating factor; IRES, internal ribosome entry site; SFFV, spleen focus-forming virus.
GC was predicted to encode a chimeric protein containing the NTF2 dimerization domain of G3BP1 (an RNA-binding protein involved in various cellular processes6) and the entire tyrosine kinase domain of CSF1R.5 Unlike the wide-type CSF1R which exhibited predominantly vesicular localization in transfected HeLa cells, GC and G3BP1 resided in the cytoplasm (supplemental Figure 2), suggesting that the G3BP1 moiety affects GC localization. Coimmunoprecipitation analysis in transfected HeLa and 293T cells showed that GC can self-associate as dimers (supplemental Figure 3), which may mediate constitutive CSF1R activation. Consistently, lentiviral transduction of GC induced phosphorylation of CSF1R and downstream effectors (STAT5 and ERK1/2) and conferred cytokine-independent growth in the following 2 cytokine-dependent human acute myeloid leukemia (AML) cell lines: M07E (megakaryocytic) and TF1 (bipotent with megakaryocytic/erythroid features) (Figure 1C-E). Non-obese diabetic-severe combined immunodeficiency IL2rγ null mice transplanted with GC-expressing TF1 (TF1-GC), but not empty vector (EV)-expressing TF1 (TF1-EV), exhibited leukemia symptoms such as limb paralysis with multiorgan myeloblast infiltration (Figure 1F; supplemental Figure 4). Together, these findings suggest that GC possesses oncogenic activity and can promote leukemia engraftment in vivo. Dasatinib is a dual SRC/ABL inhibitor.7 Pexidartinib is a US Food and Drug Administration–approved CSF1R inhibitor for treating tenosynovial giant cell tumor.8 GC patient cells, TF1-GC and MKPL1 cells harboring RBM6::CSF1R, were sensitive to dasatinib and pexidartinib, as demonstrated by reduced proliferation (Figure 1G; supplemental Figure 5A), increased apoptosis (Figure 1H; supplemental Figure 5B), and inhibition of kinase activation (Figure 1I; supplemental Figure 5C) in vitro. Furthermore, these kinase inhibitors significantly decreased leukemia burden and prolonged survival in non-obese diabetic-severe combined immunodeficiency IL2rγ null mice grafted with luciferase-GC-expressing TF1 cells (Figure 1J-M), pinpointing GC as a targetable lesion.
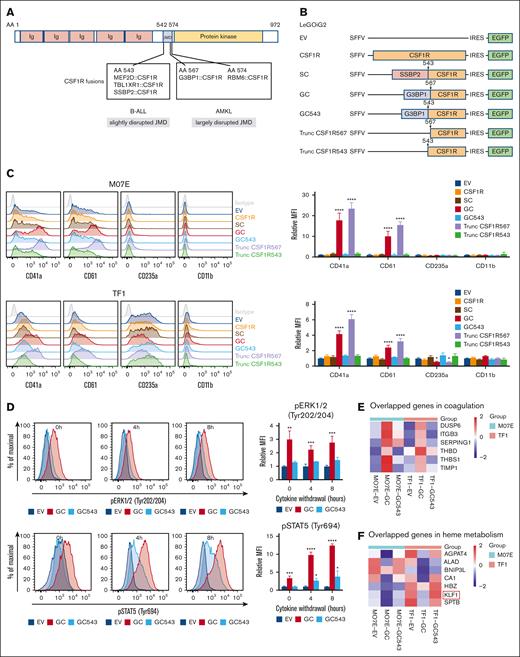
Notably, apart from AMKL, CSF1R fusions have been reported in B-cell acute lymphoblastic leukemia (B-ALL; supplemental Table 2).9-13 Although all these CSF1R fusions break at exon 12 of CSF1R, which encodes the juxta-membrane domain (JMD) (amino acids 542-574) that functions as a kinase autoinhibitory region,2 the extent of JMD disruption was found to be considerably different between AMKL (lacking a large part of JMD) and B-ALL (missing only amino acid 542; Figure 2A). This raises the possibility that the breakpoint location in CSF1R may influence leukemia phenotype. Using various CSF1R constructs with different extents of JMD disruption, we found that GC and truncated CSF1R567, but not wide-type CSF1R, SSBP2::CSF1R (a recurrent CSF1R fusion found in B-ALL with CSF1R breakpoint at amino acid 543), GC543 (an artificial fusion mimicking CSF1R breakpoint at amino acid 543 in B-ALL), and truncated CSF1R543, increased the expression of megakaryocytic markers (CD41a and CD61) in M07E and TF1 cells (Figure 2B-C), suggesting that GC could promote megakaryocytic commitment, and such effect required a largely disrupted JMD and was independent of the partner protein. Consistently, differential effects of GC and GC543 in inducing megakaryocytic skewing were observed in human hematopoietic stem/progenitor cells (supplemental Figure 6). Interestingly, GC and truncated CSF1R567 concomitantly downregulated CD235a (an erythroid marker) in the bipotent TF1 cells (Figure 2C), implicating that GC could drive abnormal megakaryopoiesis at the expense of erythroid development.
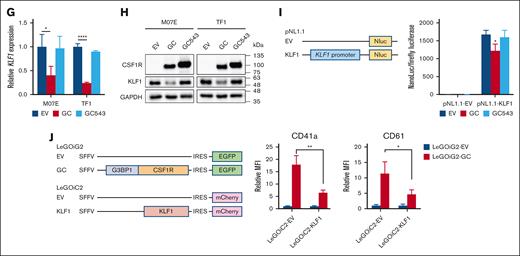
A largely disrupted CSF1R JMD in GC dictated the megakaryocytic phenotype. (A) CSF1R fusions identified in B-ALL and AMKL. The full length of CSF1R contains 972 amino acids (AAs). JMD is located between AAs 542 and 574. CSF1R fusions identified in B-ALL and AMKL are shown in the box. Ig, immunoglobulin domains. (B) Schematic representation of the lentiviral vectors. The arrows indicate different CSF1R breakpoints. Trunc, truncated. (C) Flow cytometric analysis of CD markers in M07E and TF1 cells transduced with the indicated lentivirus for 72 hours. Transduced (EGFP+) cells were gated for analysis. Left, representative results of 3 independent experiments. Right, MFI quantification. ∗ and ∗∗∗∗ indicate P < .05 and P < .0001 vs EV, respectively, by 1-way ANOVA followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (D) Flow cytometric analysis of ERK1/2 and STAT5 phosphorylation in M07E cells after GC and GC543 overexpression. M07E cells were transduced with EV, GC, and GC543 lentiviruses for 72 hours (0h). Subsequently, the cells were washed in phosphate-buffered saline 3 times and cultured in medium without cytokine (GM-CSF) for 4 hours (4h) and 8 hours (8h). Transduced (EGFP+) cells were gated for analysis. Left, representative results of 3 independent experiments. Right, MFI quantification. ∗, ∗∗, ∗∗∗, and ∗∗∗∗ indicate P < .05, P < .01, P < .001, and P < .0001 vs EV, respectively, by 1-way ANOVA followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (E) Heat map of GC-upregulated genes overlapped in coagulation hallmark gene set (systematic name M5946, which consists of genes encoding components of blood coagulation system and is also upregulated in platelets). Average expression values from the biological duplicates are shown. (F) Heat map of GC-downregulated genes overlapped in heme metabolism hallmark gene set (systematic name M5945, which includes genes related to the metabolism of heme, a cofactor consisting of iron and porphyrin, as well as erythroblast differentiation). Average expression values from the biological duplicates are shown. (G) RT-qPCR analysis of KLF1 mRNA levels in M07E and TF1 cells transduced with EV, GC, and GC543 lentiviruses. ∗ and ∗∗∗∗ indicate P < .05 and P < .0001 vs EV, respectively, by 1-way ANOVA followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (H) Western blotting analysis of KLF1 protein levels in M07E and TF1 cells transduced with EV, GC, and GC543 lentiviruses. Representative images of 3 independent experiments. (I) Left, schematic representation of the luciferase reporter vectors. Right, K562 cells were cotransfected with the reporter and overexpression vectors (LeGOiG2-EV, LeGOiG2-GC, or LeGOiG2-GC543). The reporter activity was measured by Nano-Glo Dual-Luciferase Reporter Assay 48 hours after transfection. ∗ indicates P < .05 vs EV by 1-way ANOVA followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (J) Left, schematic representation of the lentiviral vectors. Right, flow cytometric analysis of CD41a and CD61 in M07E cells cotransduced with the indicated lentivirus. Transduced (EGFP+ and mCherry+) cells were gated for analysis. ∗ and ∗∗ indicate P < .05 and P < .01 vs LeGOiC2-EV, respectively, by Student t test. Results are expressed as mean ± SD from 3 independent experiments.
A largely disrupted CSF1R JMD in GC dictated the megakaryocytic phenotype. (A) CSF1R fusions identified in B-ALL and AMKL. The full length of CSF1R contains 972 amino acids (AAs). JMD is located between AAs 542 and 574. CSF1R fusions identified in B-ALL and AMKL are shown in the box. Ig, immunoglobulin domains. (B) Schematic representation of the lentiviral vectors. The arrows indicate different CSF1R breakpoints. Trunc, truncated. (C) Flow cytometric analysis of CD markers in M07E and TF1 cells transduced with the indicated lentivirus for 72 hours. Transduced (EGFP+) cells were gated for analysis. Left, representative results of 3 independent experiments. Right, MFI quantification. ∗ and ∗∗∗∗ indicate P < .05 and P < .0001 vs EV, respectively, by 1-way ANOVA followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (D) Flow cytometric analysis of ERK1/2 and STAT5 phosphorylation in M07E cells after GC and GC543 overexpression. M07E cells were transduced with EV, GC, and GC543 lentiviruses for 72 hours (0h). Subsequently, the cells were washed in phosphate-buffered saline 3 times and cultured in medium without cytokine (GM-CSF) for 4 hours (4h) and 8 hours (8h). Transduced (EGFP+) cells were gated for analysis. Left, representative results of 3 independent experiments. Right, MFI quantification. ∗, ∗∗, ∗∗∗, and ∗∗∗∗ indicate P < .05, P < .01, P < .001, and P < .0001 vs EV, respectively, by 1-way ANOVA followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (E) Heat map of GC-upregulated genes overlapped in coagulation hallmark gene set (systematic name M5946, which consists of genes encoding components of blood coagulation system and is also upregulated in platelets). Average expression values from the biological duplicates are shown. (F) Heat map of GC-downregulated genes overlapped in heme metabolism hallmark gene set (systematic name M5945, which includes genes related to the metabolism of heme, a cofactor consisting of iron and porphyrin, as well as erythroblast differentiation). Average expression values from the biological duplicates are shown. (G) RT-qPCR analysis of KLF1 mRNA levels in M07E and TF1 cells transduced with EV, GC, and GC543 lentiviruses. ∗ and ∗∗∗∗ indicate P < .05 and P < .0001 vs EV, respectively, by 1-way ANOVA followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (H) Western blotting analysis of KLF1 protein levels in M07E and TF1 cells transduced with EV, GC, and GC543 lentiviruses. Representative images of 3 independent experiments. (I) Left, schematic representation of the luciferase reporter vectors. Right, K562 cells were cotransfected with the reporter and overexpression vectors (LeGOiG2-EV, LeGOiG2-GC, or LeGOiG2-GC543). The reporter activity was measured by Nano-Glo Dual-Luciferase Reporter Assay 48 hours after transfection. ∗ indicates P < .05 vs EV by 1-way ANOVA followed by Dunnett test. Results are expressed as mean ± SD from 3 independent experiments. (J) Left, schematic representation of the lentiviral vectors. Right, flow cytometric analysis of CD41a and CD61 in M07E cells cotransduced with the indicated lentivirus. Transduced (EGFP+ and mCherry+) cells were gated for analysis. ∗ and ∗∗ indicate P < .05 and P < .01 vs LeGOiC2-EV, respectively, by Student t test. Results are expressed as mean ± SD from 3 independent experiments.
It is conceivable that a largely disrupted CSF1R JMD may cause a more profound disruption of its autoinhibitory function, resulting in increased kinase activation and signal transduction. Concordantly, we observed that GC was more potent than GC543 to induce ERK1/2 and STAT5 phosphorylation (Figure 2D). To further explore the mechanisms underlying GC-driven megakaryocytic commitment, transcriptome analysis was performed in M07E and TF1 cells transduced with EV, GC, or GC543 lentivirus. Gene set enrichment analysis of hallmark gene sets revealed that GC-upregulated and GC-downregulated genes (top 500 genes with fold change >2 and adjusted P value <.05) were enriched in coagulation and heme metabolism, respectively (supplemental Figures 7 and 8). However, no such association was observed for GC543. Among the differentially expressed genes (Figure 2E-F), the downregulated Kruppel-like factor 1 (KLF1) is a known master erythroid transcription factor whose loss favors the megakaryocyte program.14,15 Using reverse transcription polymerase chain reaction, immunoblotting, and luciferase reporter assay, we found downregulation of KLF1 by GC (Figure 2G,I). Rescue experiments revealed that enforced KLF1 expression significantly impaired GC-induced megakaryocytic marker expression (Figure 2J), supporting the fact that KLF1 repression contributes to GC-mediated megakaryocytic commitment.
Pediatric non-DS AMKL is an aggressive disease requiring new treatment strategies to improve outcomes.1 Here, we characterized a novel and targetable CSF1R fusion in this AML subtype and provided mechanistic insights into malignant megakaryopoiesis. Similar to the previously identified RBM6::CSF1R,3 the JMD of CSF1R is also disrupted in GC. By comparing with an artificial GC (GC543) in which the JMD of CSF1R is largely intact, we showed that the authentic GC exerts stronger effects in STAT5 and ERK1/2 activation, and more importantly, promotes megakaryocytic commitment while suppressing an erythroid phenotype. Indeed, a negative regulatory role of CSF1R signaling in erythropoiesis has been reported.16 Together, our findings suggest that JMD disruption potentiates CSF1R kinase activity and that full CSF1R activation may be required to drive megakaryocytic skewing and mediate AMKL pathogenesis. More specifically, Trp550 and Tyr561 have been suggested to mediate the autoinhibitory function of the JMD of CSF1R.17 These residues were lost in both GC and RBM6::CSF1R, implying that their disruptions may be necessary for the maximization of kinase activity.
Transcription factors are key regulators of hematopoietic cell fate.18 The crucial role of KLF1 in determining erythroid vs megakaryocyte lineages has been well established.14,15 Our data suggested that GC represses KLF1 and perturbs erythroid/megakaryocytic balance. Interestingly, recent studies also revealed KLF1 downregulation by CBFAT3::GLIS2, the most common fusion in AMKL,19,20 implying that KLF1 repression may be a shared feature in non-DS AMKL pathogenesis. GATA1 transcriptionally activates KLF1 to regulate erythroid development.21 Notably, Gu et al reported that GATA1 is phosphorylated on several tyrosine residues, including some within the DNA binding domain in RBM6::CSF1R-expressing MKPL1 cells.3 However, its functional relevance in AMKL pathogenesis remains unexplored. It will be interesting to investigate whether GATA1 is also tyrosine phosphorylated in GC-expressing cells, leading to altered GATA1 activity, KLF1 downregulation, and megakaryocytic skewing.
Signaling through platelet-derived growth factor receptor (PDGFR), another type 3 RTK, has been found to enhance megakaryocyte proliferation and implicated in essential thrombocythemia,22 another myeloid neoplasm characterized by megakaryocytic hyperplasia. Interestingly, previous studies have suggested the presence of CSF1R and PDGFR in megakaryocytes22,23 and that PDGFR signaling regulates CSF1 expression.24 The potential interactions of these pathways in controlling megakaryocyte functions warrant further investigation.
CSF1R-targeted agents combined with standard chemotherapy may deepen induction response by targeting multiple AML survival pathways and minimizing drug resistance and serve as a bridge to allogeneic stem cell transplantation for curative treatment. Furthermore, the CSF1R inhibitor pexidartinib may simultaneously target the tumor microenvironment by eliminating the cancer-promoting tumor-associated macrophages,25 thereby eliciting a more potent anti-leukemia response. However, as exemplified by GC and RBM6::CSF1R, CSF1R driver fusions in AMKL may be cryptic and cannot be readily detected by conventional cytogenetics. Advanced molecular techniques are required to identify these druggable fusions for personalized therapy.
In summary, we characterized a novel targetable GC fusion in AMKL. These findings shed light on the role of CSF1R fusions in AMKL pathogenesis and provide a rationale for CSF1R-directed therapy for patients with AML with activating CSF1R alterations. Importantly, our novel observations that a largely disrupted JMD in CSF1R fusion dictates specific lineage commitment have provided founding insights into the molecular pathogenesis of various RTK fusions in myeloid neoplasms.
Acknowledgments: The authors thank the patient and her family, Charles G. Mullighan (St. Jude Children's Research Hospital) for providing the MSCV-SSBP2-CSF1R-IRES-GFP plasmid, and the Core Utilities of Cancer Genomics and Pathobiology (The Chinese University of Hong Kong) for providing the facilities and assistance in support of this study.
This study was partially supported by a grant from the General Research Fund program sponsored by the Research Grants Council in Hong Kong (CUHK M14108719).
Contribution: X.L. designed and performed the research and wrote the manuscript; C.-K.C. designed the research and advised on revision of the manuscript; H.-Y.C., N.Y.-F.C., H.A.P., and Y.-F.K. performed the research; K.-T.L., C.-K.L., and K.T. collected clinical samples; and M.H.L.N. designed and coordinated the research and advised on revision of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Margaret H. L. Ng, Blood Cancer Cytogenetics and Genomics Laboratory, Department of Anatomical and Cellular Pathology, 1/F Clinical Sciences Building, Prince of Wales Hospital, The Chinese University of Hong Kong, 30-32 Ngan Shing St, Shatin, New Territories, Hong Kong SAR, China; email: margaretng@cuhk.edu.hk.
References
Author notes
Data are available on reasonable request from the corresponding author, Margaret H. L. Ng (margaretng@cuhk.edu.hk).
The full-text version of this article contains a data supplement.