Key Points
Combining CRISPR drug screens and outcome in AML reveals genetic modulators predictive of treatment outcomes.
BCL2, CLIP2, and VAV3 were identified as drug-resistant and unfavorable biomarkers in pAML treated with ADE at high gene expression level.
Visual Abstract
Cytarabine, daunorubicin, and etoposide (ADE) have been the standard backbone of induction chemotherapy regimen for patients with pediatric acute myeloid leukemia (pAML) for >5 decades. However, chemoresistance is still a major concern, and a significant proportion of pAML becomes resistant to ADE treatment and relapse, leading to poor survival. Therefore, there is a considerable need to identify mechanisms mediating drug resistance for overcoming chemoresistance. Herein, we performed synthetic lethal CRISPR/Cas9 screens using the ADE components to identify response markers. We further integrated significant markers in 3 independent pAML clinical cohorts treated with only an ADE regimen to identify drug response biomarkers with prognostic significance. We were able to identify several mediators that represent clinically and biologically significant marker genes for ADE treatment, such as BCL2, CLIP2, and VAV3, which are resistant markers to ADE, with high expression associated with poor outcomes in pAML treated with ADE, and GRPEL1, HCFC1, and TAF10, which are sensitive markers to ADE, with high expression showing beneficial outcomes. Notably, BCL2, CLIP2, and VAV3 knockdowns in their expression in AML cell lines sensitized the cells more to the ADE components, suggesting that these modulators should be further studied as potential therapeutic targets to overcome chemoresistance.
Introduction
Cytarabine (also known as AraC), daunorubicin, and etoposide (ADE) have been the standard backbone of induction chemotherapy regimens for patients with pediatric acute myeloid leukemia (AML; pAML) for >5 decades, despite the introduction of new AML drugs. These new agents are typically administered with or subsequent to ADE regimens when patients relapse. Mechanistically, cytarabine (AraC) is an antimetabolic drug that binds to DNA polymerase and inhibits DNA synthesis (specific to the S phase of the cell cycle). Daunorubicin intercalates and forms a complex with DNA, inhibiting DNA and RNA synthesis. Daunorubicin’s production of free radicals after its metabolism also contributes to its anticancer activity (cell cycle phase nonspecific). Etoposide inhibits DNA synthesis before mitosis by creating a complex with topoisomerase II and DNA (cycle dependent and phase specific). However, ADE fails to induce remission in ∼15% of pAML. A significant proportion of patients who achieve remission after ADE will become resistant or relapsed to treatment, and >90% of resistant or relapsed patients die within 3 years.1,2 Consequently, AML remains the major contributor to deaths due to leukemia, with a 5-year survival rate of ∼68% in pediatric patients.3 Despite ongoing research into the molecular mechanisms by which treatments affect AML outcomes, chemoresistance remains a significant concern.
Previously, we performed a genome-wide CRISPR/Cas9 synthetic lethal screening to identify functional modulators of etoposide response in a leukemic cell line.4 The study sought to integrate CRISPR-screen results with gene expression and clinical outcomes in patients with pAML treated with an etoposide-containing regimen. Although we confirmed the involvement of well-studied markers in etoposide pharmacology (TOP2A and ABCC1) and identified novel markers as potential drug targets (RAD54L2, PRKDC, and ZNF451), one limitation of the study was the lack of investigation of cytarabine and daunorubicin because they are given in combination with etoposide in clinical cohorts.
In this study, we performed genome-wide CRISPR/Cas9-knockout (KO) screens in leukemic cells exposed to the individual components of ADE to identify functional modulators associated with sensitivity or resistance to ADE components. K-562 edited mutant pools were exposed to the 3 drugs, of ADE, for 4 days to identify markers contributing to early increased sensitivity or resistance. To identify drug response biomarkers with prognostic significance, the results of CRISPR screens were integrated with 3 independent pAML cohorts exclusively treated with the ADE-containing regimen. Top significant CRISPR drug-resistant markers with unfavorable prognostic significance at high expression in diagnostic pAML samples (BCL2, CLIP2, and VAV3) were further orthogonally validated with siRNA-mediated knockdown in AML cell lines with MOLM-13 and ML-2 to confirm these findings. BCL2 encodes for B-cell lymphoma 2 (BCL2) protein, a key apoptosis regulator. BCL2 has been shown to be highly expressed in many hematological malignancies, including adult AML, and protects against cell death induced by oncogenic and external stresses.5,6CLIP2 encodes the CAP-Gly domain–containing linker protein 2, which regulates the cytoskeleton to preserve cell shape, size, and motility, whereas VAV3 encodes Vav guanine nucleotide exchange factor 3, with little evidence of involvement in AML and/or treatment response.
Methods
Generation of CRISPR-sgRNA library pool and in vitro CRISPR screening
The generation of the CRISPR–single-guide RNA (sgRNA) library pool and in vitro CRISPR screening were previously described.4 Briefly, the genome-wide CRISPR/Cas9-KO Brunello library (catalog no. 73179; Addgene) targeting 19 114 genes with 76 441 unique sgRNAs was used in K-562 cells, in which K-562 myeloid cell line has been studied extensively as one of the models for AML.7 Details on transduction are included in supplemental Note N1.
After puromycin selection, the transduced cell population was split into vehicle and drug groups (dimethyl sulfoxide vs 37, 0.85, or 0.1 μM [IC30 at 48 hours after treatment] of cytarabine, daunorubicin, or etoposide, respectively; catalog nos. S1648, S1225, or S3035; Selleck Chemicals). IC30 (30% inhibitory concentrations) were used to ensure selective pressure occured while maintaining enough live cells for genomic coverage by the end of each drug treatment. Screens were performed in triplicates of 30 × 106 cells to provide genomic coverage up to 400×. Cells were counted and subcultured, and fresh media and drugs were replenished every 2 days. From each drug and control condition, 30 × 106 cells from day 0 (after puromycin selection) and day 4 (end of treatment) were collected for sequencing, similar to our previous study.4 More details on next generation sequencing are included in supplemental Note N3.
CRISPR screen hits identification and in silico functional pathway analysis
After demultiplexing, the abundance of each sgRNA was determined between samples using the MAGeCK (version 0.5.9.4) algorithm. All comparisons and visualizations were performed using the MAGeCK-MLE pipeline and the MAGeCK-Flute R package.8,9 MAGeCK-MLE generated β scores and associated statistics for multiple conditions, including drug conditions (cells cocultured with AraC, daunorubicin, or etoposide) and control conditions (cells cocultured without drug). In addition, differential β (diff-β) scores were calculated to determine treatment-related screen significant markers by subtracting β scores between treatment and control conditions. The diff-β scores described how a gene is selected; a positive score indicates positive selection or markers associated with drug sensitivity, whereas a negative score indicates negative selection or markers associated with drug resistance. The significant cutoff for top marker gene hits identification was defined as a diff-β standard deviation (SD) of 2, whereas markers with diff-β <2 SD were associated with drug resistance (negative selection; CRISPR-KO led to cell depletion), and markers with diff-β >2 SD were associated with increased drug sensitivity (positive selection; CRISPR-KO led to cell enrichment).
Functional pathway analysis via the Reactome database was performed using gene set enrichment analysis (GSEA) for genome-wide gene sets and overrepresentation analysis (ORA) for only significant genes from the CRISPR drug screens. GSEA and ORA were performed for each drug using the ClusterProfiler 4.0 R package to provide insights into the biological pathways of sensitive and resistant genes.10 For GSEA, a positive normalized enrichment score represented gene sets enriched in drug sensitivity, and a negative normalized enrichment score represented sets enriched in drug resistance. For ORA, marker genes significant in the CRISPR screen were compared and contrasted across all 3 drugs in either a sensitive or resistant directionality to assess any overlapping and unique biological pathways. Gene sets with false discovery rate <0.1 were considered significant.
Clinical integration with CRISPR screen significant genes in 3 independent pAML patient cohorts treated with ADE
Significant marker genes for each drug CRISPR screen were further investigated with diagnostic gene expression levels in 3 independent pAML cohorts treated with ADE during induction 1. Gene expression levels obtained from pAML diagnostic specimens were evaluated for association with various clinical outcome end points. To be considered as clinically and biologically significant biomarkers for ADE treatment, we closely evaluated genes identified as markers with drug resistance in CRISPR screens, with their high expression in pAML associated with detrimental outcomes, and genes identified as markers with drug sensitivity in CRISPR screens, with their high expression in pAML associated with beneficial clinical outcomes.
Cohort 1 included 163 patients treated with ADE on the St. Jude AML02 clinical trial (ClinicalTrials.gov identifier: NCT00136084) with available gene expression (Affymetrix U133A microarray).11,12 Cohorts 2 and 3 included RNA sequencing from patients with pAML treated with only ADE on Children’s Oncology Group trials AAML0531 and AAML1031 from the TARGET-AML database from Bolouri et al.13,14 Full details of TARGET-AML cohort generation are listed in supplemental Note N2.
Continuous diagnostic gene expression level association analyses were performed with multiple clinical end points, such as minimal residual diseases after induction 1 (MRD1), event-free survival (EFS), and overall survival (OS). Specifically, logistic regression was used to estimate the odds ratio (OR) of MRD1, whereas Cox proportional hazards (Cox-PH) were used to calculate the hazard ratio (HR) of EFS and OS, and forest plots were used to visualize the OR or HR results for each clinical end points with risk adjustments. Genes with OR/HR of >1 were defined as detrimental, and OR/HR of <1 were defined as beneficial outcomes. Association analysis with P value < .05, with or without risk adjustment, was considered significant. The study was conducted in accordance with the institutional review boards of both the University of Florida and St. Jude Children's Research Hospital.
In vitro siRNA-mediated knockdown functional validation of BCL2, CLIP2, and VAV3
Neon NxT Electroporation System (Thermo Fisher Scientific) system was used to transfect genes of interest and nontarget control (NTC) into MOLM-13 and ML-2 AML cell lines, respectively, based on the adapted manufacturer-recommended protocol. The protocol and quantitative polymerase chain reaction confirmation details are listed in supplemental Note N4.
After transfection of siRNA, ∼10 000 cells were treated with cytarabine, daunorubicin, etoposide, or dimethyl sulfoxide (control). The viability of the cells was assessed 48 hours after drug exposure using a CellTiter-Glo 2.0 assay viability kit (Promega). One-way analysis of variance tests were used to assess the significance of sensitivity in viability assays, with a significant threshold of P value < .05. All experiments were done in triplicates.
Results
CRISPR genome-wide screening identifies chemotherapy response determinants for ADE
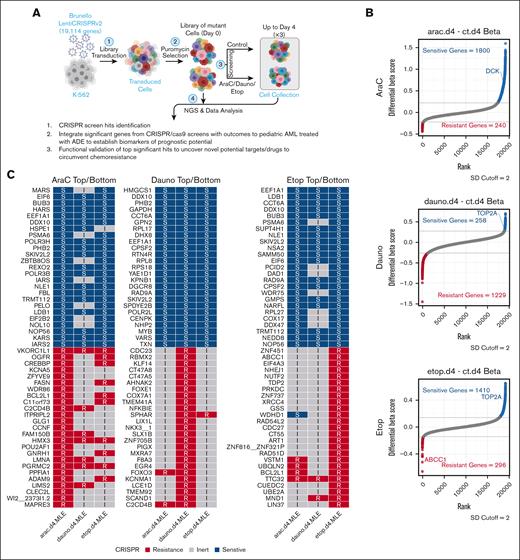
The overall study schema is summarized in Figure 1A, in which pooled CRISPR cells were exposed to the individual components of ADE for up to 4 days, and a raw summary statistic with diff-β for CRISPR is included in supplemental Table 1. At the 2 SD cutoff of the diff-β score from MAGeCK-MLE algorithm comparing treated conditions vs control for each drug, 240, 1229, and 296 genes were associated with AraC, daunorubicin, or etoposide resistance (sgRNA depletion in negative selection were defined as drug-resistance genes), and 1800, 258, and 1410 genes were associated with increased AraC, daunorubicin, or etoposide sensitivity (sgRNA enrichment in positive selection were defined as drug-sensitive genes), respectively (Figure 1B). The well-characterized pharmacology functional genes for AraC, daunorubicin, and etoposide are also highlighted in the plots. Furthermore, the top 25 drug-sensitive marker genes and the bottom 25 drug-resistant marker genes for each drug are shown in Figure 1C, and a complete list of significant sensitive and resistant marker genes that met the 2 SD cutoff for each CRISPR screens is included in supplemental Table 2. Among the top sensitive marker genes with knockout conferring cell survival or a positive drug score, they showed multiple positive diff-β scores with increased sensitivity for all 3 drugs, suggesting these sensitive markers among the 3 drugs might share common mechanistic pathways and functions for drug sensitivity. On the contrary, resistant marker genes with knockout conferring cell death or negative diff-β scores showed significant negative selection toward more unique pathways in its drug screen than other drugs, suggesting that each of our CRISPR screen drugs possessed a distinct mechanism for its drug resistance. Genes not meeting the 2 SD significance cutoff were deemed as inert marker genes to the drug.
Overall significant gene markers from ADE CRISPR screens. (A) CRISPR/Cas9 loss-of-function screening overall schema. (B) Significant marker genes were identified by CRISPR screens in response to AraC, Dauno, or Etop exposures, with sensitive marker genes for each drug highlighted in blue and resistant genes highlighted in red (SD cutoff, 2), whereas nonsignificant marker genes or inert marker genes highlighted in gray. (C) Top 25 sensitive and bottom 25 resistant markers for each drug screen CRISPR results. Dauno, daunorubicin; Etop, etoposide; I, inert markers for each drug; R represents resistant markers for each drug; S, sensitive markers for each drug. Diagram was created with BioRender.com.
Overall significant gene markers from ADE CRISPR screens. (A) CRISPR/Cas9 loss-of-function screening overall schema. (B) Significant marker genes were identified by CRISPR screens in response to AraC, Dauno, or Etop exposures, with sensitive marker genes for each drug highlighted in blue and resistant genes highlighted in red (SD cutoff, 2), whereas nonsignificant marker genes or inert marker genes highlighted in gray. (C) Top 25 sensitive and bottom 25 resistant markers for each drug screen CRISPR results. Dauno, daunorubicin; Etop, etoposide; I, inert markers for each drug; R represents resistant markers for each drug; S, sensitive markers for each drug. Diagram was created with BioRender.com.
ADE drug pathway analysis identifies shared functions among significant sensitive genes and distinct functions among resistant genes
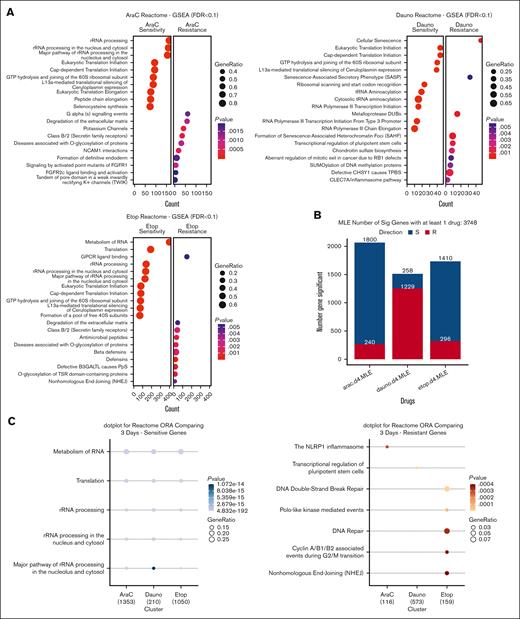
CRISPR library results containing 19 114 genes with descending rank by the diff-β score for each drug were subjected to GSEA with the Reactome database. GSEA dot plot results in Figure 2A showed the top 10 significantly enriched gene sets for drug sensitivity or resistance, and the full summary result is included in supplemental Table 3. Enriched sensitive gene sets from each ADE drug show similar enrichment in shared functions such as ribosomal RNA and transfer RNA processing, eukaryotic translation initiation, cap-dependent translation initiation, and other typical functions in translational and transcription processes in responding to ADE drugs. Interestingly, the top significant resistant gene sets in this analysis for each ADE component showed distinct enrichment pathways. We further narrowed down to the significant CRISPR genes that met the 2 SD diff-β cutoff criteria for each drug, as shown in the bar plot of Figure 2B, and compared their enriched functions via ORA with Reactome pathways across all 3 drugs for both resistant and sensitive markers separately, as shown in Figure 2C; and the full summary result is included in supplemental Table 4. Sensitive marker gene sets across the 3 drugs were significantly enriched for RNA metabolism, cell translation, and rRNA processes; and resistant marker genes for each drug showed distinct enriched pathways. AraC significant resistant markers showed enrichment in NLRP1 inflammasome; daunorubicin resistant markers were enriched in transcriptional regulation of pluripotent stem cells; and etoposide resistant markers revealed enrichment in various pathways involved in DNA damage repairs, such as DNA double-strand break repair and nonhomologous end joining.
Biological pathway enrichment analysis with GSEA for each drug and ORA comparison among all drugs. (A) GSEA dot plot with Reactome biological pathway database for the whole gene set of CRISPR screens with decreasing ranking in diff-β scores for each drug. (B) Bar plots showed several significant genes that met each drug's 2 SD cutoff for resistant or sensitive genes. (C) ORA dot plot showed CRISPR significant gene comparing across 3 drugs; significant pathway gene sets have FDR <0.1; P value represents adjusted P value with FDR. FDR, false discovery rate; GPCR, G protein-coupled receptors; GTP, guanosine triphosphate; PpS, Peters plus syndrome; TSR, thrombospondin type 1 repeat; TPBS, temtamy preaxial brachydactyly syndrome; tRNA, transfer RNAs.
Biological pathway enrichment analysis with GSEA for each drug and ORA comparison among all drugs. (A) GSEA dot plot with Reactome biological pathway database for the whole gene set of CRISPR screens with decreasing ranking in diff-β scores for each drug. (B) Bar plots showed several significant genes that met each drug's 2 SD cutoff for resistant or sensitive genes. (C) ORA dot plot showed CRISPR significant gene comparing across 3 drugs; significant pathway gene sets have FDR <0.1; P value represents adjusted P value with FDR. FDR, false discovery rate; GPCR, G protein-coupled receptors; GTP, guanosine triphosphate; PpS, Peters plus syndrome; TSR, thrombospondin type 1 repeat; TPBS, temtamy preaxial brachydactyly syndrome; tRNA, transfer RNAs.
Clinical integration with CRISPR screens’ significant genes in pAML revealed sensitive and resistant genes with prognostic potential
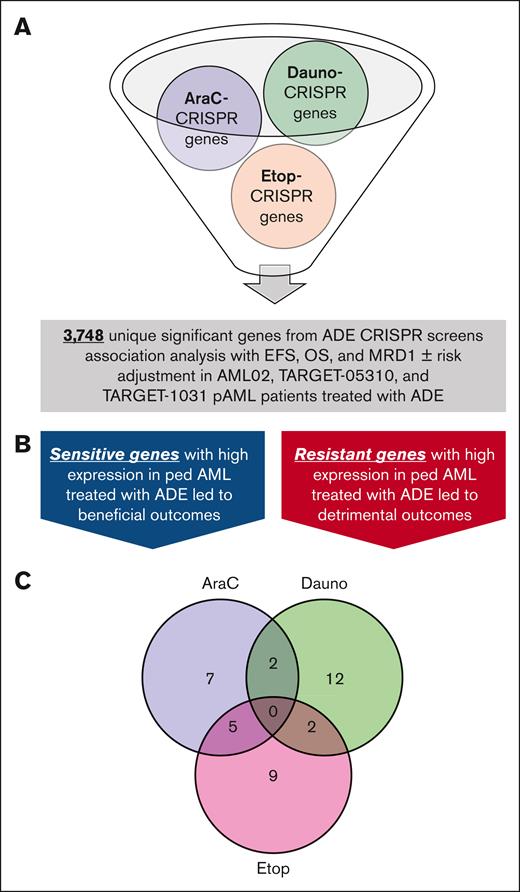
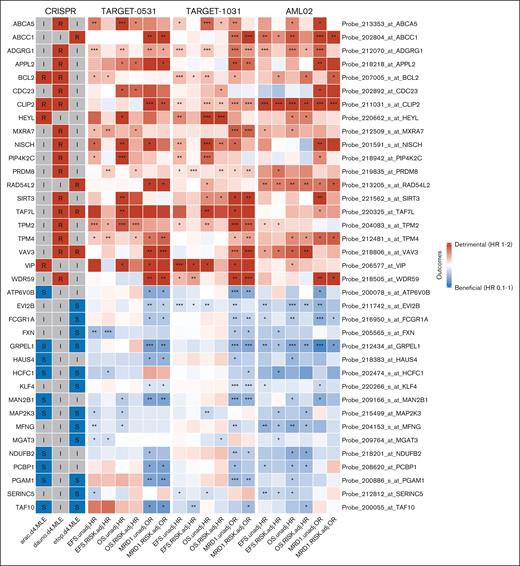
A total of 3748 genes passing the threshold of 2 SD cutoff described above for ADE CRISPR screens (supplemental Table 2) were further evaluated for their prognostic potential in 3 pAML cohorts: St. Jude AML02 (n = 163), TARGET-0531 (n = 201), and TARGET-1031 (n = 411; patient’s characteristics are shown in Table 1). Although there were no major differences between the 3 clinical cohorts of pAML treated with ADE-containing regimens, it should be noted that the TARGET-1031 cohort had less representation of patients in the high-risk group category, primarily due to FLT3-ITD–positive patients treated with sorafenib being in a separate arm that is not included in this study. Gene expression was treated as continuous in the association analyses with EFS, OS, and MRD1, with or without risk adjustment. Sensitive and resistant markers from the CRISPR screen with consistent association with clinical outcome (for sensitive genes, high expression associated with favorable outcomes; and for resistant genes, high expression associated with detrimental outcomes) across all 3 cohorts were deemed to be marker genes of biological and prognostic relevance (Figure 3B). As a result of this analysis, 14 genes met the criteria for sensitive or resistant markers with potential prognostic significance for AraC, 16 marker genes for daunorubicin, and 16 marker genes for etoposide, as shown in Figure 3C. There were 5 significant markers shared between AraC and etoposide, 2 between AraC and daunorubicin, and 2 between daunorubicin and etoposide. Figure 4 shows these results in the form of a heat map of the 37 sensitive or resistant marker genes; of these, 20 markers were resistant to at least 1 drug from CRISPR screens, with high expression indicating detrimental outcomes, and 17 markers were sensitive to at least 1 drug from CRISPR screens, with high expression indicating beneficial outcomes (also included in supplemental Table 5). Interestingly, BCL2, CLIP2, and VAV3 were significant markers of drug resistance for at least 2 drugs in our CRISPR screens, and their high expressions were associated with detrimental outcomes in all 3 pAML cohorts (Figure 4). GRPEL1, HCFC1, MAP2K3, PGAM1, and TAF10 were significant markers for drug sensitivity for both AraC and etoposide in our CRISPR screens, and their high expressions were associated with improved outcomes in all 3 clinical trials. Of these significant markers with prognostic potential, resistant markers with BCL2, CLIP2, and VAV3 demonstrated significantly poor outcomes at high expression levels in pAML diagnostic samples, whereas sensitive markers GRPEL1, HCFC1, and TAF10 showed improved outcomes at high expression levels in pAML treated with ADE in all 3 cohorts, regardless of the risk group of the patients, with at least 1 outcome (EFS, OS, or MRD1; Table 2). Note that BCL2, CLIP2, and VAV3 met significant thresholds as resistant markers for 2 drugs, but they all had negative diff-β scores across all ADE components; whereas GRPEL1, HCFC1, and TAF10 met significant thresholds as sensitivity for 2 drugs, but they all had positive diff-β scores across all ADE components (supplemental Table 1).
Patient characteristics of 3 independent pAML cohorts with St. Jude AML02, COG’s TARGET-0531, and COG’s TARGET-1031 clinical cohorts
| Ped-AML with only ADE . | AML02 (N = 163) . | TARGET-0531 (N = 201) . | TARGET-1031 (N = 411) . | P value∗ . |
|---|---|---|---|---|
| Age, median (range) | 9.3 (0.0-21.2) | 10.1 (0.1-20.4) | 10.0 (0.1-29.6) | .14 |
| Sex, n/N (%) | .37 | |||
| Female | 74/163 (45%) | 105/201 (52%) | 194/411 (47%) | |
| Male | 89/163 (55%) | 96/201 (48%) | 217/411 (53%) | |
| Race, n/N (%) | .023 | |||
| White | 115/162 (71%) | 150/201 (75%) | 296/409 (72%) | |
| Black | 31/162 (19%) | 17/201 (8.5%) | 52/409 (13%) | |
| Other/Unknown | 16/162 (9.9%) | 34/201 (17%) | 61/409 (15%) | |
| Risk group, n/N (%) | <.001 | |||
| Low | 55/163 (34%) | 90/196 (46%) | 165/405 (41%) | |
| Standard | 65/163 (40%) | 76/196 (39%) | 231/405 (57%) | |
| High | 43/163 (26%) | 30/196 (15%) | 9/405 (2.2%) |
| Ped-AML with only ADE . | AML02 (N = 163) . | TARGET-0531 (N = 201) . | TARGET-1031 (N = 411) . | P value∗ . |
|---|---|---|---|---|
| Age, median (range) | 9.3 (0.0-21.2) | 10.1 (0.1-20.4) | 10.0 (0.1-29.6) | .14 |
| Sex, n/N (%) | .37 | |||
| Female | 74/163 (45%) | 105/201 (52%) | 194/411 (47%) | |
| Male | 89/163 (55%) | 96/201 (48%) | 217/411 (53%) | |
| Race, n/N (%) | .023 | |||
| White | 115/162 (71%) | 150/201 (75%) | 296/409 (72%) | |
| Black | 31/162 (19%) | 17/201 (8.5%) | 52/409 (13%) | |
| Other/Unknown | 16/162 (9.9%) | 34/201 (17%) | 61/409 (15%) | |
| Risk group, n/N (%) | <.001 | |||
| Low | 55/163 (34%) | 90/196 (46%) | 165/405 (41%) | |
| Standard | 65/163 (40%) | 76/196 (39%) | 231/405 (57%) | |
| High | 43/163 (26%) | 30/196 (15%) | 9/405 (2.2%) |
Kruskal-Wallis rank-sum test; Pearson χ2 test.
Clinical integration of significant CRISPR marker genes across ADE drugs in pAML cohorts treated with only ADE regimen. (A) All significant CRISPR signature marker genes for each drug with 3748 unique genes were further investigated in 3 independent pAML cohorts treated with ADE to determine the association between diagnostic gene expression and clinical outcomes (EFS, OS, and MRD1). (B) Each gene was evaluated to determine whether sensitive markers from CRISPR results with high expression led to beneficial (HR/OR <1) or resistant markers from CRISPR results with high expression led to detrimental outcomes (HR/OR >1) in all 3 clinical cohorts. Collectively, these marker genes were deemed to be biologically important genes with prognostic potential. (C) Significant markers genes with CRISPR screens and at least 1 significant clinical outcome in pAML cohorts treated with ADE either with or without risk adjustment. Significant outcomes were defined as P value < .05 in HR or OR.
Clinical integration of significant CRISPR marker genes across ADE drugs in pAML cohorts treated with only ADE regimen. (A) All significant CRISPR signature marker genes for each drug with 3748 unique genes were further investigated in 3 independent pAML cohorts treated with ADE to determine the association between diagnostic gene expression and clinical outcomes (EFS, OS, and MRD1). (B) Each gene was evaluated to determine whether sensitive markers from CRISPR results with high expression led to beneficial (HR/OR <1) or resistant markers from CRISPR results with high expression led to detrimental outcomes (HR/OR >1) in all 3 clinical cohorts. Collectively, these marker genes were deemed to be biologically important genes with prognostic potential. (C) Significant markers genes with CRISPR screens and at least 1 significant clinical outcome in pAML cohorts treated with ADE either with or without risk adjustment. Significant outcomes were defined as P value < .05 in HR or OR.
Heat map results of significant CRISPR screen markers and their association analysis from 3 independent pAML cohorts. CRISPR column showed significant markers of the gene with either a R, I, or S feature for each drug’s CRISPR screen. TARGET-0531, TARGET1031, and AML02 column showed association analysis of EFS, OS, and MRD1 with (RISK.adj) and without risk adjustment (unadj). HR was estimated for EFS and OS using the Cox proportional model, and OR was calculated for MRD1 using logistic regression. Genes with OR/HR >1 were defined as detrimental, and OR/HR <1 were defined as beneficial outcomes. Resistant features and detrimental outcomes are in red; sensitive features and beneficial outcomes are in blue; and inert features are in gray. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001. I, inert; R, resistant; S, sensitive.
Heat map results of significant CRISPR screen markers and their association analysis from 3 independent pAML cohorts. CRISPR column showed significant markers of the gene with either a R, I, or S feature for each drug’s CRISPR screen. TARGET-0531, TARGET1031, and AML02 column showed association analysis of EFS, OS, and MRD1 with (RISK.adj) and without risk adjustment (unadj). HR was estimated for EFS and OS using the Cox proportional model, and OR was calculated for MRD1 using logistic regression. Genes with OR/HR >1 were defined as detrimental, and OR/HR <1 were defined as beneficial outcomes. Resistant features and detrimental outcomes are in red; sensitive features and beneficial outcomes are in blue; and inert features are in gray. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001. I, inert; R, resistant; S, sensitive.
Significant gene in at least 2 drug CRISPR screens with prognostic significance in all AML pediatric cohorts treated with ADE, irrespective of patients’ risk group
| RNA-seq symbol . | U133A probes . | CRISPR . | Clinically relevant . | AML02 (U133A) . | TARGET-0531 (RNA-seq) . | TARGET-1031 (RNA-seq) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AraC . | Dauno . | Etop . | EFS adj . | OS adj . | MRD1 adj . | EFS adj . | OS adj . | MRD1 adj . | EFS adj . | OS adj . | MRD1 adj . | |||
| BCL2 | 207005_s_at_BCL2 | R | R | I | Detrimental | 1.51 (1.06-2.16)∗ | 1.73 (1.11-2.67)∗ | 1.84 (1.07-3.16)∗ | 1.33 (1.02-1.73)∗ | 1.01 (0.75-1.36) | 1.07 (0.69-1.65) | 1.2 (1.01-1.42)∗ | 1.13 (0.92-1.4) | 1.28 (0.93-1.75) |
| CLIP2 | 211031_s_at_CLIP2 | R | R | I | Detrimental | 2.02 (1.36-2.99)∗∗∗ | 2.09 (1.29-3.37)∗∗ | 3.16 (1.68-5.94)∗∗∗ | 0.89 (0.71-1.11) | 0.88 (0.68-1.15) | 1.92 (1.27-2.91)∗∗ | 1.13 (0.99-1.29) | 1.28 (1.08-1.52)∗∗ | 1.53 (1.19-1.97)∗∗∗ |
| VAV3 | 218806_s_at_VAV3 | I | R | R | Detrimental | 1.72 (1.12-2.66)∗ | 2.22 (1.28-3.83)∗∗ | 1.38 (0.81-2.36) | 1.25 (0.95-1.65) | 1.18 (0.85-1.65) | 3.08 (1.69-5.64)∗∗∗ | 1.03 (0.87-1.22) | 1.17 (0.94-1.44) | 2.21 (1.55-3.15)∗∗∗ |
| GRPEL1 | 212434_at_GRPEL1 | S | I | S | Beneficial | 0.6 (0.36-0.98)∗ | 0.44 (0.25-0.76)∗∗ | 0.33 (0.12-0.9)∗ | 1.05 (0.73-1.52) | 1.29 (0.83-1.99) | 0.35 (0.18-0.69)∗∗ | 0.81 (0.64-1.02) | 0.79 (0.59-1.05) | 0.4 (0.26-0.61)∗∗∗ |
| HCFC1 | 202474_s_at_HCFC1 | S | I | S | Beneficial | 0.36 (0.13-0.97)∗ | 0.32 (0.1-1.08) | 0.44 (0.1-1.98) | 0.65 (0.44-0.96)∗ | 0.78 (0.52-1.17) | 1.39 (0.68-2.82) | 0.95 (0.75-1.19) | 0.95 (0.72-1.27) | 0.66 (0.43-1)∗ |
| TAF10 | 200055_at_TAF10 | S | I | S | Beneficial | 0.82 (0.47-1.43) | 0.52 (0.31-0.88)∗ | 0.97 (0.39-2.39) | 1.76 (0.9-3.44) | 1.29 (0.61-2.72) | 0.28 (0.09-0.84)∗ | 0.63 (0.46-0.86)∗∗ | 0.81 (0.55-1.2) | 0.52 (0.29-0.94)∗ |
| RNA-seq symbol . | U133A probes . | CRISPR . | Clinically relevant . | AML02 (U133A) . | TARGET-0531 (RNA-seq) . | TARGET-1031 (RNA-seq) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AraC . | Dauno . | Etop . | EFS adj . | OS adj . | MRD1 adj . | EFS adj . | OS adj . | MRD1 adj . | EFS adj . | OS adj . | MRD1 adj . | |||
| BCL2 | 207005_s_at_BCL2 | R | R | I | Detrimental | 1.51 (1.06-2.16)∗ | 1.73 (1.11-2.67)∗ | 1.84 (1.07-3.16)∗ | 1.33 (1.02-1.73)∗ | 1.01 (0.75-1.36) | 1.07 (0.69-1.65) | 1.2 (1.01-1.42)∗ | 1.13 (0.92-1.4) | 1.28 (0.93-1.75) |
| CLIP2 | 211031_s_at_CLIP2 | R | R | I | Detrimental | 2.02 (1.36-2.99)∗∗∗ | 2.09 (1.29-3.37)∗∗ | 3.16 (1.68-5.94)∗∗∗ | 0.89 (0.71-1.11) | 0.88 (0.68-1.15) | 1.92 (1.27-2.91)∗∗ | 1.13 (0.99-1.29) | 1.28 (1.08-1.52)∗∗ | 1.53 (1.19-1.97)∗∗∗ |
| VAV3 | 218806_s_at_VAV3 | I | R | R | Detrimental | 1.72 (1.12-2.66)∗ | 2.22 (1.28-3.83)∗∗ | 1.38 (0.81-2.36) | 1.25 (0.95-1.65) | 1.18 (0.85-1.65) | 3.08 (1.69-5.64)∗∗∗ | 1.03 (0.87-1.22) | 1.17 (0.94-1.44) | 2.21 (1.55-3.15)∗∗∗ |
| GRPEL1 | 212434_at_GRPEL1 | S | I | S | Beneficial | 0.6 (0.36-0.98)∗ | 0.44 (0.25-0.76)∗∗ | 0.33 (0.12-0.9)∗ | 1.05 (0.73-1.52) | 1.29 (0.83-1.99) | 0.35 (0.18-0.69)∗∗ | 0.81 (0.64-1.02) | 0.79 (0.59-1.05) | 0.4 (0.26-0.61)∗∗∗ |
| HCFC1 | 202474_s_at_HCFC1 | S | I | S | Beneficial | 0.36 (0.13-0.97)∗ | 0.32 (0.1-1.08) | 0.44 (0.1-1.98) | 0.65 (0.44-0.96)∗ | 0.78 (0.52-1.17) | 1.39 (0.68-2.82) | 0.95 (0.75-1.19) | 0.95 (0.72-1.27) | 0.66 (0.43-1)∗ |
| TAF10 | 200055_at_TAF10 | S | I | S | Beneficial | 0.82 (0.47-1.43) | 0.52 (0.31-0.88)∗ | 0.97 (0.39-2.39) | 1.76 (0.9-3.44) | 1.29 (0.61-2.72) | 0.28 (0.09-0.84)∗ | 0.63 (0.46-0.86)∗∗ | 0.81 (0.55-1.2) | 0.52 (0.29-0.94)∗ |
∗, ∗∗, and ∗∗∗ indicate significant terms at the P < .05; P < .01; and P < .001 statistical levels, respectively. Boldface values indicate significant results.
adj, risk adjusted; Dauno, daunorubicin; Etop, etoposide; I, inert; R, resistant; RNA-seq, RNA sequencing; S, sensitive.
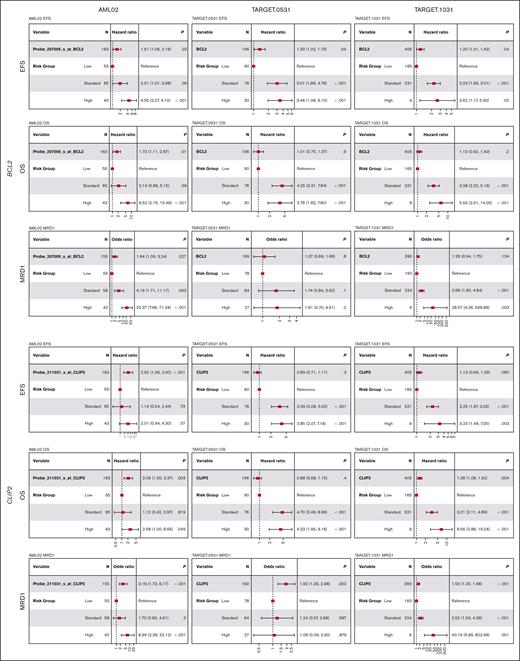
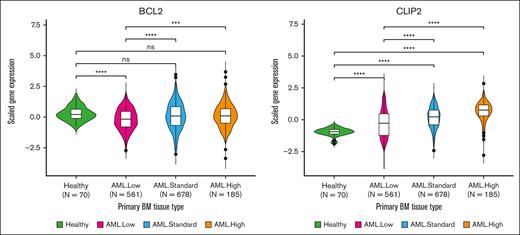
BCL2 was defined as a significant resistant marker in AraC and daunorubicin CRISPR screens, and its elevated expression in pAML was associated with worse outcomes, even after adjusting for the risk group (Table 2; Figure 5), with significantly worse EFS (risk-adjusted HR, 1.51 [95% CI, 1.06-2.16]; P = .02), OS (risk-adjusted HR, 1.73 [1.11-2.67]; P = .01), and MRD1 (risk-adjusted OR, 1.84 [1.07-3.16]; P = .03) in AML02; worse EFS (risk-adjusted HR, 1.33 [1.02-1.73]; P = .04) in TARGET-0531; and worse EFS (risk-adjusted HR, 1.2 [1.01-1.42]; P = .04) in TARGET-1031. BCL2 expression was much lower in low-risk AML (n = 561) than healthy bone marrow (n = 70), but no difference was seen for standard (n = 678) and high-risk (n = 185) AMLs; however, BCL2 gene expression was shown to increase significantly as pAML’s risk increased (Figure 6). CLIP2, another resistant marker in AraC and daunorubicin CRISPR screens, was associated with detrimental outcomes (Table 2; Figure 5); worse EFS (risk-adjusted HR, 2.02 [1.36-2.99]; P < .001), OS (risk-adjusted HR, 2.09 [1.29-3.37]; P < .01), and MRD1 [risk-adjusted OR, 3.16 [1.68-5.94]; P < .001) in AML02; worse MRD1 (risk-adjusted OR, 1.92 [1.27-2.91]; P < .01) in TARGET-0531; and worse OS (risk-adjusted HR, 1.28 [1.08-1.52]; P < .01) and MRD1 (risk-adjusted OR, 1.53 [1.19-1.97]; P < .001) in TARGET-1031. Compared with bone marrow specimens from the healthy pediatric population, patients with pAML had a significantly increased CLIP2 gene expression. Furthermore, patients with pAML within the high-risk group had higher CLIP2 expression than other groups (Figure 6). VAV3 gene is another drug-resistant marker identified in daunorubicin and etoposide CRISPR screens and was shown to be associated with detrimental outcomes at higher expression levels in pAML (Table 2; supplemental Figure 2A). The VAV3 gene expression level in all patients with pAML showed significantly higher expression than healthy individuals, but the expression varied among different risk groups (supplemental Figure 3).
BCL2 and CLIP2 expression association analysis across 3 pediatric patients with AML treated with ADE-containing regimens. High expression of BCL2 and CLIP2 showed unfavorable outcomes in at least one of the end points (EFS, OS, and MRD1) in AML02, TARGET.0531, and TARGET.1031. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
BCL2 and CLIP2 expression association analysis across 3 pediatric patients with AML treated with ADE-containing regimens. High expression of BCL2 and CLIP2 showed unfavorable outcomes in at least one of the end points (EFS, OS, and MRD1) in AML02, TARGET.0531, and TARGET.1031. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
BCL2 and CLIP2 expression in pediatric healthy vs pAML bone marrow. Diagnostic gene expression of healthy and pAML from the whole TARGET data set was evaluated, regardless of treatment protocols. All patients with AML were subdivided into low, standard, and high-risk groups. The Wilcoxon test was used to compare each AML risk group with healthy individuals, and logistic regression with the risk additive model was used to estimate the OR of gene expression, which increases with risk. BM, bone marrow; ns, not significant.
BCL2 and CLIP2 expression in pediatric healthy vs pAML bone marrow. Diagnostic gene expression of healthy and pAML from the whole TARGET data set was evaluated, regardless of treatment protocols. All patients with AML were subdivided into low, standard, and high-risk groups. The Wilcoxon test was used to compare each AML risk group with healthy individuals, and logistic regression with the risk additive model was used to estimate the OR of gene expression, which increases with risk. BM, bone marrow; ns, not significant.
On the contrary, GRPEL1, HCFC1, and TAF10 from CRISPR results were identified as significant sensitive markers to AraC and etoposide, and their increased expression levels in pAML were associated with favorable outcomes (Table 2). Particularly, increased GRPEL1 expression in pAML was associated with better EFS, OS, and MRD1 in AML02 and favorable MRD1 in both TARGET-0531 and TARGET-1031 after risk adjustment (supplemental Figure 2A). Increased HCFC1 expression in pAML was associated with better EFS in both AML02 and TARGET-0531 and favorable MRD1 in TARGET-1031; and increased TAF10 expression in pAML was associated with better OS in both AML02, favorable MRD1 in TARGET-0531, and favorable EFS and MRD1 in TARGET-1031, after risk adjustment (supplemental Figure 2B). Among these 3 significant sensitive markers from CRISPR screens with beneficial outcomes at high expression levels in pAML, GRPEL1 showed a significant decrease in gene expression level when comparing patients with AML vs healthy patients, and its expression level significantly decreased as patients' risks increased; whereas TAF10 gene expression showed a significantly elevated expression compared with healthy individuals, and it also significantly increased as patients' risks increased (supplemental Figure 3).
siRNA-mediated knockdown of BLC2, CLIP2, and VAV3 confirmed increased cellular sensitivity to ADE components in AML cell lines
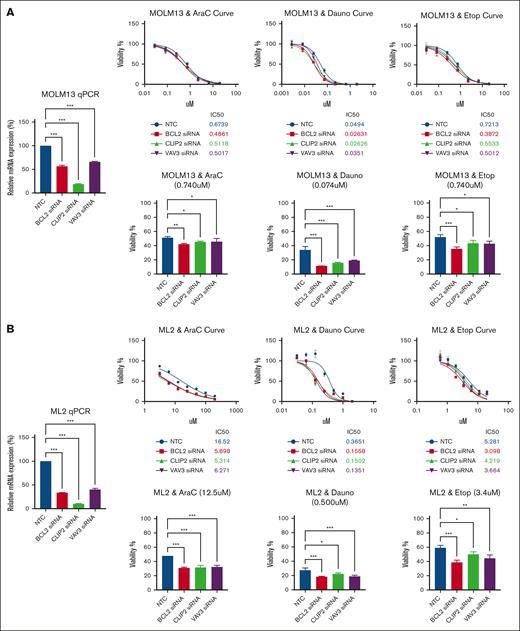
siRNA-mediated knockdown of BCL2, CLIP2, or VAV3 expression increased cellular sensitivity to ADE components in both MOLM-13 and ML-2 AML cells (Figure 7). siRNA knockdown on marker gene of interests showed increased sensitivity with lower 50% inhibitory concentration (IC50) than NTC across all 3 drugs; the IC50 for MOLM-13 treated with AraC was 0.675 μM for NTC and 0.487 μM, 0.514 μM, and 0.504 μM for BCL2, CLIP2, or VAV3, respectively; for daunorubicin, it was 0.0494 μM for NTC and 0.0263 μM, 0.0263 μM, and 0.0351 μM for BCL2, CLIP2, or VAV3 siRNA-mediated knockdown, respectively; and for etoposide, it was 0.721 μM for NTC and 0.387 μM, 0.553 μM, and 0.501 μM for BCL2, CLIP2, or VAV3 siRNA-mediated knockdown, respectively (Figure 7A). Similar results were seen for ML-2, in which siRNA-mediated knockdown of BCL2, CLIP2, or VAV3 expression increased drug sensitivity to AraC, daunorubicin, and etoposide compared with NTC. The IC50 for ML-2 treated with AraC was 16.52 μM for NTC and 5.989 μM, 5.314 μM, and 6.271 μM for BCL2, CLIP2, or VAV3, respectively; for daunorubicin, it was 0.365 μM for NTC and 0.156 μM, 0.150 μM, and 0.135 μM for BCL2, CLIP2, or VAV3 siRNA-mediated knockdown, respectively; and for etoposide, it was 5.281 μM for NTC and 3.098 μM, 4.219 μM, and 3.664 μM for BCL2, CLIP2, or VAV3 siRNA-mediated knockdown, respectively (Figure 7B). We were interested in evaluating the cosilencing of BCL2 and CLIP2 effects with dual siRNA knockdown, in addition to each gene alone, in the MOLM-13 AML cell line (supplemental Figure 4). Although the siRNA-mediated knockdown of BCL2, CLIP2, and the combination of BCL2 and CLIP2 increased the sensitivity to the ADE components, the result showed a minimal additive response effect when simultaneously knocking down 2 genes compared with individual genes for each ADE component in viability assays.
siRNA-mediated BCL2, CLIP2, and VAV3 knockdown increased ADE components’ sensitivity. (A) Results of MOLM-13 cell line, with bar plot (left) showing the effect of siRNA-mediated knockdown for each gene in relative messenger RNA expression compared with NTC. The viability inhibition curves for each ADE component (top); an siRNA-mediated knockdown confirmation quantitative polymerase chain reaction (qPCR) plot (left) for each gene interest compared with NTC, and IC50 values were calculated for each cell condition per drug plot. Bar plots (bottom) showing specific drug concentrations with cell viability (%) of siRNA in the gene of interest compared with NTC for each ADE component. (B) Results of ML-2. Data are presented as mean ± SD. Cell viability and qPCR experiments included 3 technical replicates for each sample condition. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Dauno, daunorubicin; Etop, etoposide.
siRNA-mediated BCL2, CLIP2, and VAV3 knockdown increased ADE components’ sensitivity. (A) Results of MOLM-13 cell line, with bar plot (left) showing the effect of siRNA-mediated knockdown for each gene in relative messenger RNA expression compared with NTC. The viability inhibition curves for each ADE component (top); an siRNA-mediated knockdown confirmation quantitative polymerase chain reaction (qPCR) plot (left) for each gene interest compared with NTC, and IC50 values were calculated for each cell condition per drug plot. Bar plots (bottom) showing specific drug concentrations with cell viability (%) of siRNA in the gene of interest compared with NTC for each ADE component. (B) Results of ML-2. Data are presented as mean ± SD. Cell viability and qPCR experiments included 3 technical replicates for each sample condition. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Dauno, daunorubicin; Etop, etoposide.
Discussion
The application of CRISPR/Cas9 knockout synthetic lethal screens with pooled sgRNA at the genome-wide scale in conjunction with drug treatment has significantly transformed the mechanistic investigation of chemotherapy, and it has uncovered mediators of drug resistance profiles in many malignancies, including AML.15-23 Thus, CRISPR has been proven to be a powerful tool for functional genomic studies to identify biomarkers of significant biological and prognostic relevance. We previously reported novel mediators of etoposide resistance with prognostic potential via integrating etoposide CRISPR lethal screening with AML outcomes.4 Given that ADE is administered in combination and continues to be the backbone of remission induction treatment in pAML, in this study, we expanded the CRISPR synthetic lethal screens to all 3 drugs: cytarabine (AraC), daunorubicin, and etoposide to identify their drug-sensitive and -resistant genes. We chose to use MAGeCK-MLE to provide more comprehensive coverage in multiple conditions, as opposed to our previous study with etoposide CRISPR synthetic lethal screens using the MAGeCK-RRA pipeline with strict cutoff criteria. MAGeCK-MLE also captured the majority of the significant genes from etoposide at early drug treatments (day 4) from the previous analysis and an additional 1549 gene markers that were not previously identified for downstream analysis (supplemental Figure 5).4 These significant markers identified from each drug’s CRISPR screen were then further integrated with clinical outcomes from 3 distinct pediatric cohorts treated with ADE, unveiling prognostically significant biological mediators. CRISPR screen results enabled us to profile a list of sensitive and resistant markers for each ADE component and confirm the involvement of well-characterized pharmacology marker genes for each drug from our CRISPR screens with ADE components. For example, higher expression of DCK is a maker for increased sensitivity to AraC because it is the rate-limiting enzyme for AraC activation; TOP2A is the drug target and a sensitive marker for both daunorubicin and etoposide; and ABCC1 is a resistant marker for etoposide because it is involved in the etoposide efflux transporter. Well-characterized pharmacology gene makers involved in ADE’s pathways confirm the validity of our synthetic lethal CRISPR screen as a tool to profile sensitive and resistant genes for the 3 drugs. Through a thorough evaluation of functional pathways from the Reactome database via GSEA with ranked genome-wide genes and ORA with significant CRISPR hits for each ADE component, we showed that sensitive markers for all 3 drugs had many functional pathways; however, resistant markers mapped to distinct biological processes. In particular, the involvement of all sensitive markers shared common functions, such as rRNA processing and cell translational pathways, across all 3 drugs, validating the mechanism of action of these drugs as inhibition of DNA and RNA synthesis and cell replication. Distinct enriched pathway profiles from resistant markers suggest that separate unique pathways cause drug resistance.
To our knowledge, this is the first study integrating significant hits from synthetic lethal CRISPR screens of the standard-of-care treatment with ADE in 3 independent pAML clinical cohorts of patients treated with ADE-containing regimens. By profiling mediators that met significance with at least 2 of the 3 drugs from CRISPR screens and significance with at least 1 clinical end point, either MRD1, EFS, or OS (with or without risk adjustments), in 3 clinical cohorts, our results showed BCL2, CLIP2, and VAV3 as resistant markers with high expression showing detrimental outcomes and GRPEL1, HCFC1, and TAF10 as sensitive markers with high expression showing beneficial outcomes in pAML treated with ADE. Among the 3 sensitive mediators to ADE components, knowledge of the roles of GRPEL1, HCFC1, and TAF10 in AML is extremely limited. GRPEL1 encoded for GrpE Like 1 mitochondrial protein, with functions in enabling and unfolding protein binding activity, and the pathology atlas of the human cancer transcriptome shows that GRPEL1 is a favorable prognostic marker in renal cancer.24GRPEL1 high expression showed an association with favorable outcomes in this study. HCFC1 encodes for host cell factor 1 that involves transcriptional coregulators. Although high expression of HCFC1 shows favorable outcomes in our study for pAML treated with ADE, upregulation of HCFC1 has been reported to promote hepatocellular carcinoma growth by preventing cell cycle arrest and is connected with immune infiltration. Furthermore, the pathology atlas of the human cancer transcriptome shows that HCFC1 high expression is associated with unfavorable outcomes in liver cancer.24-26TAF10 encodes for Taf10 TATA-box binding protein associated factor 10 involved in the enabling of DNA binding activity and promoter-specific chromatin binding activity. TAF10 expression was reported to be upregulated in AML with isolated trisomy 8 (+8) to cytogenetically normal AML cases,27,28 which is consistent with our result of upregulation of TAF10 gene expression in pAML compared with pediatric healthy individuals. We were particularly interested in resistant mediators because they could be potential drug targets to be used in combination with ADE to overcome drug resistance; therefore, for orthogonal functional validation with siRNA-mediated knockdown, we selected BCL2, CLIP2, and VAV3 genes. Interestingly, venetoclax is the first selective BCL2 inhibitor approved to treat adults with newly diagnosed AML. However, venetoclax use in pAML remained limited,29-31 and its use in combination with chemotherapy in pediatric patients has only been studied in relapsed or refractory AML settings.32 Our study demonstrated that high BCL2 is a marker that contributes to the chemoresistance of ADE, and its high diagnostic expression in pAML led to detrimental outcomes. Our results showed that siRNA-mediated knockdown of BCL2 gene expression made the AML cell lines more sensitive to ADE, suggesting that the use of BCL2 inhibitors such as venetoclax in combination with ADE could overcome drug resistance and potentially improve outcomes in de novo pAML. The ORA findings in supplemental Table 4 revealed that BCL2 remains significantly associated with several genes across multiple pathways, including the PI3K-Akt signaling pathway, the JAK-STAT signaling route, and apoptosis, consistent with its established biological role. In contrast, there remained a minimal pathway interaction between CLIP2 and VAV3 and other marker genes. In literature, CLIP2 was shown to be an unfavorable prognostic marker in endometrial cancer and is highly expressed in glioma compared with other cancer types.24 The results of our study showed that CLIP2 is a resistant marker for ADE from the CRISPR screens, and its high expression is associated with poor outcomes in all 3 pAML cohorts investigated. Furthermore, CLIP2 gene expression level was significantly elevated in pAML compared with healthy individuals, and its expression increased with patient risk, which is also consistent with our proteomics pilot study, in which CLIP2 protein levels were significantly enriched in MRD1 positive and were highly expressed in higher-risk pAML treated with ADE.33CLIP2 siRNA-mediated knockdown sensitized cells to ADE, implying it as a potential prognostic marker as well as a druggable target to be pursued in future studies. Lastly, VAV3, a member of the VAV gene family that encodes for the Vav guanine nucleotide exchange factor 3 protein, is involved in signal transmission between cell membrane receptors and intracellular mediators. The accumulation of VAV3 has been linked to progression of cancer, and increased VAV3 levels in human cancers could play an oncogenic role in cancer progression, as well as serve as a potential prognostic biomarker and a chemoresistance mediator in various cancer pathogenesis.34 However, there are limited studies of VAV3 within hematological malignancies, especially pAML. Our study showed that the VAV3 gene is a resistant marker for ADE, and its high expression showed unfavorable outcomes in pAML treated with ADE; thus, knocking its expression down via siRNA confirmed the increase in sensitivity of these drugs. All together, we showed that GRPEL1, HCFC1, TAF10, BCL2, CLIP2, and VAV3 are significant prognostic markers in pAML treated with ADE-containing regimens and mediators involved in the response of ADE chemotherapy.
In summary, we combined synthetic lethal CRISPR/Cas9 screens to profile resistant and sensitive response mediators for the ADE regimen components. After integrating CRISPR screen results with 3 independent clinical cohorts of pAML treated with ADE-containing regimens, we were able to identify several mediators that would represent clinically and biologically important biomarkers for ADE treatment. Notably, BCL2, CLIP2, and VAV3 orthogonal validation with siRNA gene knockdowns sensitize AML cell lines to the 3 drugs, opening up opportunities to investigate these as possible therapeutic targets in AML. This may also encourage studies examining the use of BCL2 inhibitors in comparison with standard chemotherapy to treat pAML. Some of the limitations of our study include restricting the study to only 4 days of drug treatment and using US pAML cohorts. Because we were interested in capturing early response modulators, there was limited information on the genes and pathways modulated at longer treatments. The CRISPR screen identified key genes predominantly influenced by single-gene perturbations, but the functional impacts of many genes, their interactions, and other patients’ mutation profiles and genetic abnormalities can significantly affect clinical outcomes. Nevertheless, our findings emphasize the pivotal role that CRISPR/Cas9-based synthetic lethal screens play, when combined with clinical data, as a powerful instrument to collect meaningful data about prognostic markers, novel therapeutic targets, drug response mediators, and development approaches for novel drug combinations.
Acknowledgments
The authors thank Dario Campana and Coustan-Smith for the minimal residual disease data.
The authors gratefully acknowledge funding from National Institutes of Health (NIH) grants R01-CA132946 and R01-CA270120 (J.K.L. and S.B.P.), University of Florida (UF)-Research Opportunity Seed funds (J.K.L. and C.D.V.), UF Health Cancer Center, supported in part by state appropriations provided in Florida Statutes §381.915, and the National Cancer Institute of the NIH under award number P30CA247796. N.H.K.N. is supported by the NIH under grant T32HG008958, NIH Loan Repayment Program, American Foundation for Pharmaceutical Education Predoctoral Research Fellowship, and UF Health Cancer Center Predoctoral Award.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the State of Florida.
Authorship
Contribution: N.H.K.N. performed all formal analysis, siRNA functional validation, analyzed and interpreted all formal results, and contributed to manuscript writing; R. Rafiee performed in vitro CRISPR drug screening and prepared samples for next generation sequencing; P.K.P. performed siRNA-mediated knockdown experiment; A.T. generated CRISPR-sgRNA library pool and performed the transductions and associated quality control; J.R., R. Ribeiro, X.C., S.B.P., and C.D.V. contributed to AML patient data and reviewed the manuscript; J.K.L. conceptualized the project, analyzed data, acquired funding, supervised studies, and wrote, reviewed, and edited the manuscript; and all authors have reviewed and contributed to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jatinder K. Lamba, Department of Pharmacotherapy and Translational Research, College of Pharmacy, University of Florida, 1225 Center Dr, Gainesville, FL 32610; email: jlamba@cop.ufl.edu.
References
Author notes
Raw sequencing data are available on request from the corresponding author, Jatinder K. Lamba (jlamba@cop.ufl.edu).
The full-text version of this article contains a data supplement.