Key Points
Chromosomal mimicry occurs when chromosome morphology resembles a known SV but lacks the expected gene-level rearrangement.
Gene-level confirmation unmasks mimicry and ensures appropriate diagnosis and therapeutic management.
Visual Abstract
The detection of structural variants (SVs) represents a critical component in the diagnostic evaluation and treatment of many hematologic malignancies. Although clinical SV testing mainly consists of traditional cytogenetic methodologies, technological innovations have led to alternative approaches with improved resolution. In this study, we sought to characterize the clinical impact of targeted RNA sequencing on the diagnosis of myeloid and immature lymphoid malignancies. Across a cohort (n = 380) of myeloid (87%) and immature lymphoid (13%) tumors, we compared SVs detected by chromosome banding analysis (CBA) and fusion events detected by anchored multiplex polymerase chain reaction (AMP)–targeted RNA sequencing. Variants detected by either assay were categorized using a 5-tier system: tier 1 (established clinical significance); tier 2 (possible clinical significance); tier 3 (unknown significance); tier 4 (known germ line variants), and tier 5 (no variants detected). The combined use of AMP and CBA improved the detection of clinically relevant (tier 1 or 2) findings in 10% of cases. Unexpectedly, in 1% (3/380) of the patients in our study, CBA appeared to detect a defining SV, for example, t(9;22)(q34;q11.2), that was not confirmed by AMP fusion studies. Subsequent evaluation by orthogonal approaches confirmed breakpoints on the expected chromosomes but did not involve the anticipated genes. Our study indicates that “chromosomal mimicry,” a phenomenon in which chromosome morphology resembles a known SV but lacks the expected gene-level rearrangement, is an infrequent but recurrent finding with the potential to confound clinical management. Our study highlights the need for assays with gene-level resolution in the diagnostic evaluation of hematologic malignancies.
Introduction
The detection or exclusion of genomic structural variants (SVs) or transcriptomic oncogenic fusion represents a critical component of the diagnostic evaluation of hematologic malignancies, with a significant impact on clinical management and risk stratification. As such, SVs/fusions are integral to the World Health Organization and International Consensus Classification diagnosis of hematologic malignancies, and include, for example, PML::RARA, RUNX1::RUNX1T1, BCR::ABL1, KMT2A::r, MECOM, and NUP98::r for acute myeloid leukemia (AML), BCR::ABL1, ETV6::RUNX1, DUX4::r, KMT2A::r, and MYC::r in B-cell acute lymphoid leukemia (B-ALL), and various tyrosine kinase fusions (ie, JAK2, PDGFRA, PDGFRB, FGFR1, FLT3, and ETV6::ABL1) across both myeloid and lymphoid neoplasms.1-3 With advancements in targeted therapies, there exists a critical need for the rapid and comprehensive detection of SVs to enable accurate diagnosis and prompt initiation of the appropriate treatment protocol.4
Traditionally, SVs are detected by chromosome banding analysis (CBA).5 The longstanding use of CBA, as a single-cell, genome-wide approach, has created a foundational understanding of both balanced and unbalanced SVs observed in hematologic malignancies, ultimately contributing to the first “cytogenetic/genomic” dissection of broad disease entities, and in turn becoming enshrined in clinical management.6-13 However, CBA necessitates in vitro cell division, which can lead to outgrowth of the nonneoplastic component of a specimen or culture failure.14 Moreover, CBA is generally unable to detect aberrations less than ∼10 Mb in size and may be unable to detect certain alterations involving morphologically similar chromosome loci.15 Existing ancillary methods, such as fluorescence in situ hybridization (FISH), help overcome some of these limitations but require a priori knowledge of the regions/genes of interest and use probes 150 to 1000 kb in size that may span multiple genes.
Recent technological innovations have addressed many limitations associated with CBA and have enabled multiple opportunities for exon-level SV detection. These methods, including but not limited to anchored multiplex polymerase chain reaction (AMP)–targeted RNA fusion assays,16 optical genome mapping (OGM),17,18 Hi-C,19 and whole genome sequencing,20,21 are beginning to challenge the current diagnostic paradigm and may represent more effective modalities for evaluating SVs.
In this study, we sought to evaluate the clinical impact of targeted RNA sequencing on the diagnosis of myeloid and immature lymphoid malignancies by comparing the diagnostic utility of CBA and AMP-targeted RNA sequencing.
Methods
Patient cohort
Between 1 January 2021 and 31 December 2022, we identified 1125 patients seen at Massachusetts General Hospital who had cytogenetic studies (1727 studies) and 756 patients who had AMP studies (1147 studies) performed as part of their evaluation for myeloid and/or lymphoid neoplasms. Patients with a new diagnosis or presentation to our health care system who underwent concurrent cytogenetic studies (ie, CBA supplemented with FISH as necessary) and AMP (n = 476) were included in our initial cohort. Cases were excluded if: (i) there was no active disease (n = 23); (ii) there was no definitive diagnosis established (n = 45); (iii) they were cases of clonal hematopoiesis of indeterminate potential or clonal cytopenia of unknown significance (n = 9); (iv) the disease was outside the scope of this analysis (ie, lymphoma/atypical lymphoid proliferation or carcinoma, n = 16); (v) cytogenetic studies included FISH only (n = 1); (vi) there were no metaphase cells for cytogenetic analysis (n = 1); or (vii) there were no concurrent cytogenetic and AMP studies at the sampled time point (n = 1). After applying these exclusion criteria (see supplemental Figure 1 for a graphical overview), 380 cases were included in the study, which was performed in accordance with the institutional research board approval (Protocol no. 2020P002978). As Massachusetts General Hospital is an agnostic reference center for oncology patients with a population of ∼3 million people, we do not anticipate a profound selection bias in our cohort. However, we acknowledge that the exclusion criteria described above may have unintentionally biased our cohort, and the results reported here should be considered in this context.
CBA
CBA was performed according to standard clinical practices, as previously described.22
AMP–targeted RNA fusion assay
A clinically validated targeted RNA-based next-generation sequencing method using AMP was performed on all study samples during the diagnostic workup. This assay targets 86 commonly rearranged genes in cancer, including those commonly altered in hematologic malignancies.16 Briefly, total nucleic acid was isolated from peripheral blood or bone marrow samples. Random hexamers were used to reverse-transcribe RNA, and second-strand synthesis was performed to create double-stranded complementary DNA (cDNA). The cDNA then underwent end repair, adenylation, and ligation with a half-functional adaptor before 2 hemi-nested polymerase chain reactions (PCR) using ArcherDx Heme Fusion kit primers. Libraries were sequenced on an Illumina NextSeq using paired-end sequencing, which produced 2 × 151 base pair reads. The reads were aligned to the GRCh37 human genome reference using Burrow-Wheeler aligner-maximal exact match. A laboratory-developed algorithm was used to detect and annotate the fusion transcripts. Quality control metrics to assess the integrity of the input nucleic acid and technical performance were obtained using a qualitative reverse transcription quantitative reverse transcription PCR assay for cDNA/RNA content in the sequencing results. The most prevalent fusion variant reported for a particular fusion pair, in terms of read count, was reported. The assay was clinically validated for samples showing ≥5% tumor cellularity.
OGM
Ultra-high-molecular-weight genomic DNA was extracted from selected peripheral blood and bone marrow samples, covalently conjugated with a fluorescent label using an enzyme that recognizes a specific 6 base pair sequence, and loaded onto Bionano Saphyr Chips for imaging. Human genome coverage of ∼300× was achieved for all tested samples. Genome analysis was performed using the rare variant analysis pipeline in Bionano Access 1.6 and Bionano Solve 3.6 to detect SVs. Briefly, SVs were identified based on discrepant alignment with GRCh38, with consensus genome maps confirming the final SV calls.
Hi-C sequencing
Hi-C sample preparation and analysis were performed using Arima Hi-C technology.23 In brief, 2 to 3 smears were crosslinked and scraped, followed by chromatin fragmentation, labeling, proximity ligation, and DNA purification, according to the Arima-Hi-C+ sample preparation kit protocol (P/N: A510008). Subsequently, short-read sequencing libraries were prepared by shearing the proximally ligated DNA, followed by size-selecting DNA fragments using solid-phase reversible immobilization beads. DNA fragments containing ligation junctions were then enriched using Enrichment Beads (provided in the Arima-Hi-C kit) and converted into sequencing libraries using the Swift Accel-NGS 2S Plus kit (Swift Biosciences, P/N: 21024). After adapter ligation, the DNA was PCR-amplified and purified using solid-phase reversible immobilization beads. The purified DNA was subjected to standard quality control (qPCR and Bioanalyzer) and sequenced using a NovaSeq 6000 (Illumina, San Diego, CA) according to the manufacturer’s instructions, to ∼100 M read pairs. Genomic rearrangements were identified using the Arima-SV v1.3 pipeline (Arima SV Pipeline for Mapping, SV Detection and quality control, GitHub repository: Arima Genomics, 2023). The Arima-SV v1.3 pipeline contains several subcomponents. First, raw read-pairs are aligned to GRCh38 and deduplicated using Hi-C user pipeline.24 Genomic rearrangements are called using Hi-C breakfinder.25 For data visualization, deduplicated alignments from Hi-C user pipeline are converted into Hi-C matrices using Juicer software,26 which are then visualized using Juicebox.27
Statistical analysis
All statistical analyses were performed using R, version 4.1.1.
Results
Patient cohort
Over a 2-year period (January 2021 to December 2022), we identified 476 patients who underwent both cytogenetic studies and AMP with a new diagnosis or new presentation to our health care system. After applying the study exclusion criteria (detailed in the “Methods”), 380 patients were eligible for inclusion. Patient characteristics are summarized in supplemental Table 1. The median patient age was 68 (1-92) years. The most common diagnosis was AML (41.3%), followed by myelodysplastic syndrome (MDS) (16.1%), and Philadelphia-negative myeloproliferative neoplasm (Ph-negative MPN) (12.6%). Rare entities that only appeared once within the cohort, including acute undifferentiated leukemia, mixed phenotype acute leukemia, juvenile myelomonocytic leukemia, histiocytic sarcoma, myeloid/lymphoid neoplasm with PDGFRB rearrangement, and systemic mastocytosis, were categorized as “Other” (1.6%). In 226 of 380 (59.5%) cases, SV and fusion were identified by either CBA or AMP studies, respectively.
Clinical risk stratification
All SVs or fusions were reviewed for their clinical significance by 2 investigators (A.M.D. and V.N.). SVs were tiered by consensus based on their established clinical significance in clinical guidelines (ie, European Leukemia Net, International Consensus Classification, and National Comprehensive Cancer Network) and/or literature review (Table 1; supplemental Table 2).
Clinical tiering schema for SVs/fusions
| Tier . | Definition . | AMP examples . | CBA examples . |
|---|---|---|---|
| 1 | SVs/fusions with known diagnostic or prognostic significance supported by clinical guidelines according to the WHO fifth edition, 2022 ELN, ICC, and NCCN | PML::RARA, BCR::ABL1, KMT2A::r | t(15;17)(q24;q21), t(9;22)(q34;q11.2), t(9;11)(p21;q23) |
| 2 | SVs/fusions with possible clinical significance but not incorporated into existing clinical guidelines | KMT2A-PTD, PICALM::MLLT10, STIL::TAL1 | Dic(9;20)(p13;q11.2) Hsr(11)(q23) 1∼26 dmin |
| 3 | SVs/fusions of unknown clinical significance | RUNX1::WASF3, FOXJ2::ETV6 HMGN2::NF1 | Inv(1)(p23q21), t(1;4)(q32;qq25), add(2)(p21) |
| 4 | SVs/fusions known to be population polymorphisms | GPR128::TFG | Inv(9)(p12q13) |
| 5 | No SVs/fusions detected | — | — |
| Tier . | Definition . | AMP examples . | CBA examples . |
|---|---|---|---|
| 1 | SVs/fusions with known diagnostic or prognostic significance supported by clinical guidelines according to the WHO fifth edition, 2022 ELN, ICC, and NCCN | PML::RARA, BCR::ABL1, KMT2A::r | t(15;17)(q24;q21), t(9;22)(q34;q11.2), t(9;11)(p21;q23) |
| 2 | SVs/fusions with possible clinical significance but not incorporated into existing clinical guidelines | KMT2A-PTD, PICALM::MLLT10, STIL::TAL1 | Dic(9;20)(p13;q11.2) Hsr(11)(q23) 1∼26 dmin |
| 3 | SVs/fusions of unknown clinical significance | RUNX1::WASF3, FOXJ2::ETV6 HMGN2::NF1 | Inv(1)(p23q21), t(1;4)(q32;qq25), add(2)(p21) |
| 4 | SVs/fusions known to be population polymorphisms | GPR128::TFG | Inv(9)(p12q13) |
| 5 | No SVs/fusions detected | — | — |
dmin, double minutes; ELN, European Leukemia Net; ICC, International Consensus Classification; NCCN, National Comprehensive Cancer Network; WHO, World Health Organization.
Evaluation of clinical impact
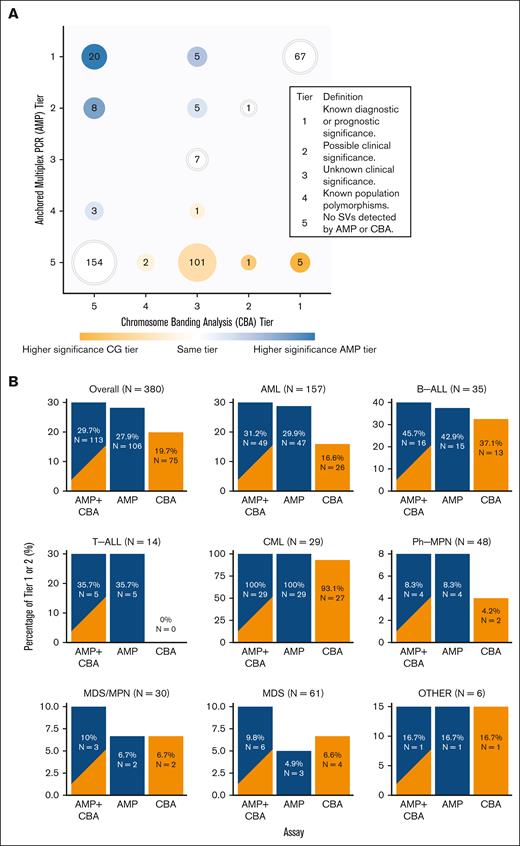
A combination of AMP and CBA led to the detection of a clinically significant (tier 1 or 2) fusion or SV in 29.5% (112/380) of patients, a substantial increase compared with CBA alone (19.5% [74/380]). Clinically significant findings were frequently detected in chronic myeloid leukemia (CML) (100% [29/29]), B-ALL (45.7% [16/35]), and AML (31.2% [49/157]) but uncommonly observed in MDS/MPN (10% [3/30]), MDS (8.2% [5/61]), and Ph-negative MPN (8.3% [4/48]) (Figure 1A-B).
Oncogenic fusion and SVs detected using AMP RNA fusion studies and CBA. (A) Number of SVs detected by AMP (y-axis) and CBA (x-axis) studies and associated clinical significance. CBA tier of higher significance is color-coded as orange circles (eg, tier 1), and the AMP tier of higher significance (eg, tier 1) is color-coded as blue circles. White circles represent SVs detected in the AMP and CBA studies with the same tiering. (B) Bar plots demonstrating the differences in SV detection rates between AMP and CBA, AMP alone, and CBA alone across different types of hematologic malignancies.
Oncogenic fusion and SVs detected using AMP RNA fusion studies and CBA. (A) Number of SVs detected by AMP (y-axis) and CBA (x-axis) studies and associated clinical significance. CBA tier of higher significance is color-coded as orange circles (eg, tier 1), and the AMP tier of higher significance (eg, tier 1) is color-coded as blue circles. White circles represent SVs detected in the AMP and CBA studies with the same tiering. (B) Bar plots demonstrating the differences in SV detection rates between AMP and CBA, AMP alone, and CBA alone across different types of hematologic malignancies.
AMP showed greater detection of clinically significant fusions than SVs detected by CBA (27.9% [106/380]). This difference was most pronounced in T-cell ALL (T-ALL), in which AMP detected a clinically relevant fusion in 35.7% (5/14) of the patients, none of which was noted by concurrent CBA. AML also had a significantly better fusion detection rate using AMP (29.9% by AMP vs 16.6% by CBA). In other entities, including B-ALL (42.9% by AMP and 37.1% by CBA), Ph-negative MPN (8.3% by AMP and 4.2% by CBA), and CML (100% by AMP and 93.1% by CBA), the difference was less pronounced. Overall, AMP improved the detection of clinically relevant (tier 1 or 2) fusions in 10.0% of our cases, particularly T-ALL (35.7% [5/14]), AML (14.6% [23/157]), and B-ALL (8.6% [3/35]).
Comparison of event detection by CBA vs AMP
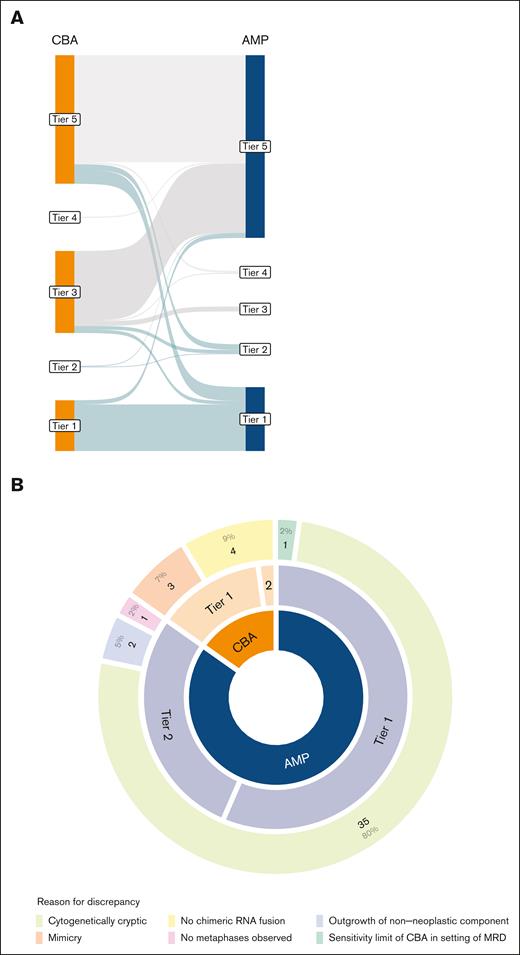
To compare the clinical results achieved by CBA vs AMP, we reviewed all instances in which SV/fusion detection was discrepant between the assays. Across 36.8% (140/380) of our cohort, AMP or CBA detected an SV/fusion (tier 1 through 4) that was not detected by the alternative assay, including 1 case with 2 discordant events (Figure 2A).
Discrepancies in detection of SVs between AMP and CBA studies. (A) Sankey plot showing similarities and differences in tiering by AMP and CBA across cases. Green lines represent clinically significant (Tier 1 and 2) cases. (B) Analysis of the cause of SV discrepancy between AMP and CBA tiering (n = 46). In this figure, “mimicry” is used to describe an identified cytogenetic event that is not as it is reported whereas “cytogenetically cryptic” describes events that were not identified due to limited banding resolution.
Discrepancies in detection of SVs between AMP and CBA studies. (A) Sankey plot showing similarities and differences in tiering by AMP and CBA across cases. Green lines represent clinically significant (Tier 1 and 2) cases. (B) Analysis of the cause of SV discrepancy between AMP and CBA tiering (n = 46). In this figure, “mimicry” is used to describe an identified cytogenetic event that is not as it is reported whereas “cytogenetically cryptic” describes events that were not identified due to limited banding resolution.
Discrepancies in the detection of clinically significant SVs were observed in 11.8% (45/380) of the cohort. These comprised 46 discrepant events, mostly due to the higher frequency of detection by AMP than by CBA (38/46 [84.4%]) (Figure 2B; supplemental Table 3). A detailed evaluation identified alterations that were cryptic to the CBA as the predominant cause of discrepancies (35/45 [80.0%]) (Figure 2B). Other explanations included the outgrowth of nonneoplastic cells in culture (2/45 [4.4%]), failed expansion (ie, absence of metaphases) in culture (1/45 [2.2%]), and the limit of sensitivity of CBA in the setting of minimal residual disease (1/45 [2.2%]).
In the remaining instances (7/46 [15.6%]), CBA detected clinically significant SV that was not detected by AMP studies. Three of these were associated with known enhancer hijacking events, in which a genomic rearrangement occurs in the absence of an RNA chimeric fusion, and is therefore undetectable by AMP. One of these events was due to a dicentric chromosome t(9;20) in B-ALL, resulting in the absence of RNA chimeric fusion. Surprisingly, in the remaining 3 instances (3/46 [6.7%]), chromosome morphology resembled the banding pattern of a pathognomonic SV but lacked the expected gene-level rearrangement when further interrogated, hereafter referred to as “chromosomal mimicry.”
Chromosomal mimicry: a rare but recurrent event
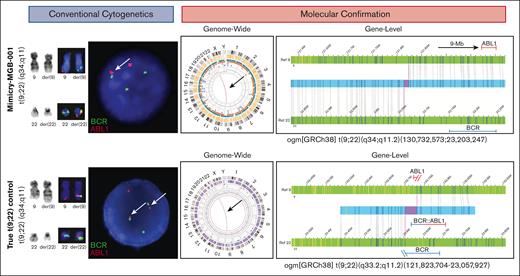
Cases in which chromosomal mimicry was noted are described below. In the first instance (Mimicry-MGB-001), the patient presented with leukocytosis of 65 × 109/L, 6% peripheral myeloblasts, and severe anemia suggestive of CML. CBA identified t(9;22)(q34;q11.2) in all 20 metaphases (Figure 3, top panel). Interphase FISH studies confirmed BCR::ABL1 in 52% of nuclei, whereas metaphase FISH studies demonstrated a BCR::ABL1 signal on derivative chromosome 22, as expected. The presence of cells with a normal FISH pattern suggests that the alteration is likely somatic and not constitutional in origin. However, AMP did not identify BCR::ABL1 but rather unexpectedly identified a partial tandem duplication of KMT2A as the sole oncogenic finding. Chromosome morphology was reviewed by several expert cytogeneticists who were unable to discern differences in the banding pattern and morphology of this tumor and a molecularly confirmed t(9;22)/BCR::ABL1 comparison (Figure 3, bottom panel). OGM was performed to reconcile these findings and confirmed a translocation between chromosomes 9 and 22 but with breakpoints within TTLL11 (9q33) and adjacent to RSPH14 (22q11.2) (Figure 3). The significance of this rearrangement is unknown, and this specific alteration has not been described in the published literature. The breakpoint was 9 Mb upstream of ABL1 (on chromosome 9) but only 122.6 kb upstream of BCR and was thus encompassed by the 1.5 Mb BCR FISH probe region.
CBA and molecular evaluation of chromosomal mimicry in MGB-Mimicry-001 findings. Conventional cytogenetic studies (karyotyping and FISH) are shown on the left. Molecular confirmation with OGM is shown on the right. In the top panel, CBA and FISH studies detect a t(9;22)(q34;q11.2) with BCR::ABL1 fusion by FISH. OGM notes rearrangements on chromosomes 9 and 22 that do not overlap with BCR or ABL1. In the bottom panel, CBA, FISH, and OGM is showing a positive control with a confirmed t(9;22), leading to BCR::ABL1.
CBA and molecular evaluation of chromosomal mimicry in MGB-Mimicry-001 findings. Conventional cytogenetic studies (karyotyping and FISH) are shown on the left. Molecular confirmation with OGM is shown on the right. In the top panel, CBA and FISH studies detect a t(9;22)(q34;q11.2) with BCR::ABL1 fusion by FISH. OGM notes rearrangements on chromosomes 9 and 22 that do not overlap with BCR or ABL1. In the bottom panel, CBA, FISH, and OGM is showing a positive control with a confirmed t(9;22), leading to BCR::ABL1.
In the second case (Mimicry-MGB-002), the patient presented with AML diagnosed at an outside institution. CBA yielded a complex karyotype that included der(9)t(9;22)(q34;q11.2), with concurrent interphase and metaphase FISH studies that were positive for BCR::ABL1 in approximately half of the cells (supplemental Figure 2). However, AMP did not detect any oncogenic fusions. OGM detected t(9;22)(q34.11;q11.21), with breakpoints adjacent to ASS1 (9q34) and MED15 (22q11.2). In this instance, the breakpoints were 8.7 Mb and 2.6 Mb upstream of ABL1 and BCR, respectively (supplemental Figure 2). The extent of chromatic condensation in metaphase chromosomes is believed to have caused a false-positive fusion signal. A retrospective review of interphase FISH studies detected a small gap between the BCR and ABL1 probes in a subset of interphase nuclei; however, this finding was subtle and not clearly distinguishable from true-positive BCR::ABL1 cases.
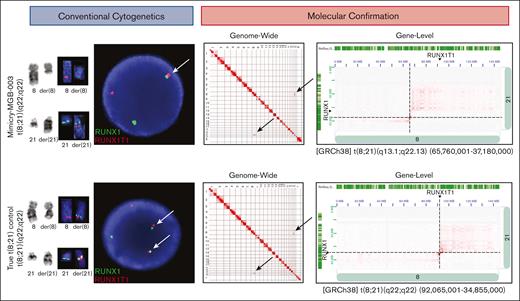
In the third case (Mimicry-MGB-003), the patient presented with gum bleeding and decreased appetite, and was found to have leukocytosis of 76 × 109/L and 72% peripheral blasts with Auer rods, consistent with AML. A t(8;21)(q22;q22.1), presumably leading to a RUNX1::RUNX1T1 gene fusion, was observed by CBA and subsequently confirmed by FISH studies, albeit with a single fused RUNX1::RUNX1T1 signal in 23% of cells. (Figure 4). AMP did not detect RUNX1::RUNX1T1 or other oncogenic fusions. As no viable cells were available for this case to run OGM, Hi-C was performed. This revealed breakpoints at t(8;21)(q13.1;q22.13) within PDE7A and TTC3 (Figure 4). The breakpoint on chromosome 8 was 26.1 Mb upstream of RUNX1T1 but only 2.1 Mb downstream of RUNX1.
CBA and molecular evaluation of chromosomal mimicry in MGB-Mimicry-003 findings. Conventional cytogenetic studies (karyotyping and FISH) are shown on the left. Molecular confirmation with Hi-C is shown on the right. In the top panel, CBA and FISH studies detect a t(8;21)(q22;q22) with RUNX1::RUNX1 fusion by FISH. Hi-C notes rearrangements on chromosomes 8 and 21 that do not overlap with RUNX1 or RUNX1T1. In the bottom panel, CBA, FISH, and OGM is showing a positive control with a confirmed t(8;21) leading to RUNX1::RUNX1T1.
CBA and molecular evaluation of chromosomal mimicry in MGB-Mimicry-003 findings. Conventional cytogenetic studies (karyotyping and FISH) are shown on the left. Molecular confirmation with Hi-C is shown on the right. In the top panel, CBA and FISH studies detect a t(8;21)(q22;q22) with RUNX1::RUNX1 fusion by FISH. Hi-C notes rearrangements on chromosomes 8 and 21 that do not overlap with RUNX1 or RUNX1T1. In the bottom panel, CBA, FISH, and OGM is showing a positive control with a confirmed t(8;21) leading to RUNX1::RUNX1T1.
To evaluate whether SVs identified in cases of chromosomal mimicry represent novel driver alterations, we reviewed concurrent next-generation sequencing studies. In each case, other well-characterized driver alterations were identified, suggesting that these novel SVs represent random passenger alterations, rather than oncogenic drivers (supplemental Table 4). Limited specimen availability precluded germ line analysis of these chromosomal mimics; therefore, a constitutional origin likewise cannot be excluded, although concurrent FISH results are highly suggestive of these events being acquired.
Discussion
In this study, we sought to characterize the clinical impact of AMP RNA sequencing in supporting conventional cytogenetic approaches (ie, CBA) for the detection of SVs/fusions in hematologic malignancies. By analyzing 380 newly diagnosed or presenting cases, our study showed that the combinatorial approach of AMP and CBA increases the detection rate of clinically relevant findings by 10% (1.5× increase) when compared with either analysis in isolation.
The observed clinical utility/advantage appears to be disease dependent. Although both CBA and AMP demonstrated similar performances in their detection rates of t(9;22)(q34;q22.1)/BCR::ABL1 for CML, for other entities, such as AML, B-ALL, or T-ALL, AMP showed a greater fusion detection rate for clinically relevant findings. Our results highlight the limitations of conventional cytogenetic methods, consistent with prior publications.20,21,28 Some of these limitations represent well-described aberrations with diagnostic relevance, including NUP98 or KMT2A rearrangements in AML or rearrangements in Ph-like in B-ALL, which are known to be cytogenetically cryptic by CBA.29,30 In other cases, cytogenetic results are confounded by the outgrowth of nonneoplastic cells or failure of in vitro cultures. The specific rearrangement mechanism also impacts the potential utility of sequence-based approaches. For example, T-ALLs often harbor microdeletions that occur below the resolution limit of the CBA (ie, TAL1::STIL). Our findings suggest the need for higher-resolution methodologies that interrogate gene- or exon-level changes. Fortunately, multiple approaches now exist with the potential to fill these gaps, including but not limited to, the OGM. The continued development of newer methods and/or bioinformatic algorithms that can more reliably characterize smaller structural changes will continue to refine our understanding of the disease and likely expand the spectrum of “cytogenetically cryptic” findings.
Although this study focused on CBA, FISH analysis is often a necessity in the cytogenetic work up of hematologic malignancies. Although its use is highly variable between institutions.31 Here, FISH studies are used as a reflexive test, often after the completion of CBA and/or sequencing. Although cytogenetic methods continue to provide the mainstay of diagnostic yield for many hematologic malignancies, the benefits of concurrent implementation of molecular techniques with gene-/exon-level resolution can be substantial.
Unexpectedly, our study identified 1% (3/380) of cases with chromosomal mimicry, in which chromosome morphology mimicked a disease-defining SV but lacked a functional gene-level chimeric fusion. Chromosomal mimicry is a byproduct of the subjective, visual assessment of CBA, coupled with the limited resolution of FISH studies. In the age of precision medicine and a large genetic classification of hematologic malignancies, misclassification of a patient with a pathognomonic rearrangement has the potential for diverse clinical consequences, such as inappropriate treatment with ABL1 tyrosine kinase inhibitors in cases of t(9;22)(q34;q11) without carrying the concomitant BCR::ABL1 gene fusion driver. Importantly, although chromosomal aberrations identified in each of these mimics were present in 100% of metaphases by CBA, concurrent FISH identified normal nuclei, confirming that these findings are likely somatic alterations.
In each of the cases identified, the presence of chromosomal mimicry interjected clinical uncertainty, which required orthogonal testing outside the standard of care to rectify. Discrepancies between the current gold-standard testing approaches and more modern methodologies are well described in the literature, and the breadth of these diagnostic blind spots continues to evolve.18,32-39 For example, 1 study reported a series of patients with AML that appeared to have an ETV6::PDGFRA rearrangement by FISH and were nonresponsive to imatinib. Subsequent molecular studies, however, were able to demonstrate different partner genes with ETV6 (ie, SCF2, CHIC2, and GSX2).32 In another study, a BCR::ABL1 FISH finding in a patient with B-cell precursor-ALL was resolved as a 3-way fusion between ZNF384, EWSR1, and EHMT1 by RNA sequencing.33 As prior publications have described rare instances in which “mimicry” was driven by a constitutional finding, confirmation of the somatic nature of these findings should also be performed.36 These cases highlight the importance of understanding the limitations and performance characteristics of diagnostic genomic/cytogenetic testing across hematologic entities, and the necessity to comprehensively integrate them in a multimodal manner to achieve the appropriate diagnosis and clinical management. In this study, we used OGM and Hi-C as ancillary methods to confirm the discrepant cytogenetic and molecular findings. Although we do not espouse a singular confirmatory method, our study highlights the importance of gene-/exon-level resolution testing, which may continue to unmask chromosomal mimicry. Future, large cohort, studies should be considered to independently evaluate the frequency of chromosomal mimicry.
To our knowledge, this study is the first to define chromosomal mimicry as a source of false-positive cytogenetic results. Our study suggests that a combinatorial approach of AMP and CBA significantly increases the detection rate of clinically relevant events compared with CBA alone and highlights the critical importance of gene-level resolution to arrive at an accurate diagnosis.
Authorship
Contribution: M.Z., V.N., and A.M.D. created the experimental design; M.Z., G.K.G., S.R., and J.C.K. performed data analysis; S.J.B. performed optical genome mapping experiments with support from A.C.H., K.C., C.B., and T.A.G.; K.S. and A.S. performed data analysis for Hi-C experiments; and M.Z., A.M.D., A.T.F., and S.N.J.S. wrote the manuscript with support from V.N., A.C.S., G.S.H., R.N., P.D.C., A.J.I., and J.C.A.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adrian M. Dubuc, Pathology, Roswell Park Comprehensive Cancer Center, Gratwick Basic Science Building, 5-519, Elm St and Carlton St, Buffalo, NY 14263; email: adrian.dubuc@roswellpark.org.
References
Author notes
M.Z., S.R., V.N., and A.M.D. contributed equally to this work.
Data are available on request from the corresponding author, Adrian M. Dubuc (adrian.dubuc@roswellpark.org). Individual participant data will not be shared.
The full-text version of this article contains a data supplement.