Key Points
FLT3-ITD persists after ICT with and without MIDO in 68% and 87.5% of patients, respectively.
The number of initial FLT3-ITD microclones and macroclones affects the FLT3-ITD persistence rate after MIDO.
Visual Abstract
Despite the use of midostaurin (MIDO) with intensive chemotherapy (ICT) as frontline treatment for Fms-like tyrosine kinase 3 (FLT3)-mutated acute myeloid leukemia (AML), complete remission rates are close to 60% to 70%, and relapses occur in >40% of cases. Here, we studied the molecular mechanisms underlying refractory/relapsed (R/R) disease in patients with FLT3-mutated AML. We conducted a retrospective and multicenter study involving 150 patients with R/R AML harboring FLT3–internal tandem duplication (ITD) (n = 130) and/or FLT3–tyrosine kinase domain mutation (n = 26) at diagnosis assessed by standard methods. Patients were treated with ICT + MIDO (n = 54) or ICT alone (n = 96) according to the diagnosis date and label of MIDO. The evolution of FLT3 clones and comutations was analyzed in paired diagnosis–R/R samples by targeted high-throughput sequencing. Using a dedicated algorithm for FLT3-ITD detection, 189 FLT3-ITD microclones (allelic ratio [AR] of <0.05) and 225 macroclones (AR ≥ 0.05) were detected at both time points. At R/R disease, the rate of FLT3-ITD persistence was lower in patients treated with ICT + MIDO than in patients not receiving MIDO (68% vs 87.5%; P = .011). In patients receiving ICT + MIDO, detection of multiple FLT3-ITD clones was associated with a higher FLT3-ITD persistence rate at R/R disease (multiple clones: 88% vs single clones: 57%; P = .049). If only 24% of FLT3-ITD microclones detected at diagnosis were retained at relapse, 43% became macroclones. Together, these results identify parameters influencing the fitness of FLT3-ITD clones.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of aggressive hematological malignancies defined by the infiltration of bone marrow (BM) and peripheral blood with immature hematopoietic myeloid cells, primarily diagnosed in the elderly.1 The clinical, biological, and prognostic features of AML are driven by underlying genetic abnormalities.2 Among these, Fms-like tyrosine kinase 3 (FLT3) gene mutations are 1 of the most common and play critical role in guiding therapeutic decisions.2,3FLT3 mutations mainly include FLT3–internal tandem duplications (FLT3-ITD) and point mutations in the tyrosine kinase domain (FLT3-TKD) affecting 22% to 30% and 5% to 10% of patients with AML, respectively.3-5 Only FLT3-ITDs are associated with higher rates of chemotherapy failure and relapse, and a poorer overall survival.6 However, both types of mutations lead to constitutive activation of the FLT3 receptor and downstream signaling pathways, making them targets for type 1 FLT3 inhibitors (FLT3i).7 Standard method for FLT3 screening is based on fragment analysis (FA) with polymerase chain reaction amplification followed by capillary electrophoresis detection, providing the allelic ratio (AR) and the length of the mutation.2,8
In the RATIFY trial assessing the combination of the type 1 FLT3i midostaurin (MIDO) with intensive chemotherapy (ICT), an AR threshold of 0.05 was used for MIDO eligibility.9 For several years, this association has become the first-line standard of care in patients diagnosed with FLT3-mutated AML. However, complete remission (CR) rates are close to 60% to 70%, and relapses occur in >40% of cases, demonstrating the ability of leukemic stem cells to resist and evade therapy.9 Such capacity may be related to the acquisition of on-target FLT3 mutations or to the activation of alternative signaling pathways, especially the Rat sarcoma (RAS) pathway.10 The study of paired diagnosis/relapse samples has shown that FLT3-ITD status is still positive in 87.5% and 51% of cases after ICT alone and after ICT + MIDO, respectively.10,11 Moreover, it has been shown that FLT3-ITD mutated AML at relapse can originate from clones with a low AR (<0.05) or clones that were undetectable at diagnosis by routine FA.12,13 However, little is known about the prevalence and overall behavior of such microclones (here defined as clones with an AR of <0.05) under therapy. In this context, high-throughput sequencing (HTS) appears to be a relevant technique for assessing the entire mutational landscape of AML, including the characteristics of FLT3-ITD mutations (ie, nucleotide sequence and gene insertion site) with increased sensitivity. Recently, our team reported a dedicated algorithm for FLT3-ITD detection from HTS data called FiLT3r, allowing the detection of FLT3-ITD clones at an AR threshold of 0.0003.14
In this study, we analyzed a cohort of 150 patients with relapsed or refractory (R/R) AML who had a positive FLT3 screening with FA-based methods at diagnosis and were treated with either ICT alone or ICT combined with MIDO. By using HTS at both diagnosis and R/R stage, we identified the evolution of FLT3-ITD–positive microclones (0.0003 ≤ AR < 0.05) and macroclones (AR ≥ 0.05) as well as their comutations under selective pressure of ICT with or without MIDO.
Methods
Patients and samples
We conducted a retrospective and multicenter study involving patients with R/R AML harboring FLT3 mutation at diagnosis detected by standard method. The presence of FLT3 mutations was defined by a positive FLT3-ITD signal by FA on genomic DNA and/or FLT3-TKD mutation positivity by FA on EcoRV-digested genomic DNA.8 The AR of the mutation was defined as the ratio of the area under the curve for FLT3-mutated signal divided by the area under the curve for FLT3 wild-type (WT) signal (for samples harboring multiple FLT3-ITD mutations, the ARs of each clone were summed to obtain a single value per sample). Patients with unavailable samples at diagnosis and/or R/R disease were excluded.
Paired BM samples obtained at the time of FLT3-mutated AML diagnosis and R/R disease of 150 patients were studied. The patients were diagnosed between 2009 and 2023 and were treated with ICT alone (n = 96, 64%) or ICT + MIDO (n = 54, 36%), according to the applicable recommendations at the date of diagnosis. A total of 103 (69%) patients were in CR with or without complete hematologic recovery after 1 induction course. Overall, 30 patients were classified as refractory (ICT + MIDO, n = 12; ICT, n = 18) and 120 patients were classified as relapsed (ICT + MIDO, n = 42; ICT, n = 78).
Additionally, 68 BM samples from patients at diagnosis of myeloproliferative neoplasms were used as FLT3-ITD–negative controls. This study was approved by the institutional review board of the tumor bank of Lille University Hospital (CSTMT289) and conducted in accordance with the Declaration of Helsinki and French ethics regulations.
HTS
Genomic DNA was extracted following standard procedures. For each sample, an amount of 40 ng of DNA was sequenced using the following methodology. Libraries were prepared according to the Twist next-generation sequencing target enrichment solution (Twist Bioscience), following the manufacturer’s instructions with a custom-designed 48-gene panel (supplemental Table 1) and run on a NovaSeq 6000 platform (Illumina), using paired-end sequencing (2 × 151 base pairs). Regions of interest were successfully sequenced with an average depth of 3000×.
Bioinformatics analysis
The bioinformatics methodology followed a systematic procedure: binary base call sequence files generated by the sequencer were initially transformed into Fastq format using bclconvert (version 3.8.4). Subsequently, Fastq were trimmed by fastp (version 0.20.0). The processed reads were then aligned to the reference genome (hg19) using the bwa-mem aligner (version 0.7.17). To identify genetic variants, we used 2 distinct variant callers: Mutect2 (version 4.1.0.0) and VarDict (version 1.5.8). Copy number analysis was performed according to the Genome Analysis Toolkit best practices (Broad Institute 2022). Simultaneously, Fastq were analyzed using the FiLT3r algorithm (4f569307), a specialized tool for robust detection and quantification of FLT3-ITD mutations.14 For each ITD detected, FiLT3r AR was assessed by the ratio between the mutated reads and the WT reads; FiLT3r variant allele frequency (VAF) was assessed by the ratio between the mutated reads and total reads (mutated reads added to WT reads). Normalized VAFs (norm-VAFs) were defined by the ratio between the FiLT3r VAFs of FLT3-ITDs and the VAFs of NPM1 mutations (considering such mutations are representative of the whole leukemic population), whenever applicable.
Statistical analysis
Continuous data were summarized descriptively including the number of patients (n), mean, standard deviation, median, interquartile range (p25-p75), and minimum and maximum. Categorical data were summarized by frequencies and percentages. Percentages were based on the number of patients with no missing data. For the comparison of continuous variables between 2 groups, either an unpaired 2-sample t test or a paired 2-sample t test was applied, depending on the nature of the data. In case of small sample (n < 30) exhibiting a nonnormal distribution, a nonparametric 2-sided Wilcoxon test was used. The assessment of distribution normality was performed through graphical methods. The analysis of categorical variables was accomplished using χ2 or Fisher exact tests. To predict the outcome of individual FLT3-ITD clones based on their initial characteristics, we performed multivariate logistic regression on variables of interest including ITD AR, clonal interference, ITD length, ITD insertion site, and treatment. Variables were discretized when continuous predictor variables and the logit of the outcome were not linear. The absence of multicollinearity was ascertained using the variance inflation factor method. Statistical analysis was performed using R version 4.3.2, and statistical significance was defined as a P value <.05.
Results
Cohort characteristics
Overall, 150 patients with R/R AML were included and harbored FLT3-ITD (n = 130 with AR ≥ 0.01 including 115 with AR ≥ 0.05) and/or FLT3-TKD (n = 26) mutation at diagnosis, assessed by standard methods; 82 (54.7%) patients had an NPM1 comutation. The median age was 51.0 years (interquartile range, 43.0-57.0), with 134 patients (89.3%) having de novo AML. Biological characteristics were comparable between the 2 treatment groups (Table 1). Most of patients (n = 98, 65.3%) had an intermediate European LeukemiaNet 2022 genetic risk.
Baseline characteristics at diagnosis of 150 patients with FLT3-mutated AML in R/R setting
| . | Total, N = 150 (100%) . | ICT + MIDO, n = 54 (36%) . | ICT, n = 96 (64%) . | P value . |
|---|---|---|---|---|
| Age at diagnosis (y) | .09 | |||
| Median (IQR) | 51.0 (43.0-57.0) | 52.5 (47.0-62.0) | 50.0 (41.5-55.0) | |
| Range | 18.0-76.0 | 23.0-76.0 | 18.0-73.0 | |
| Sex, n (%) | .08 | |||
| Male | 72 (48.0) | 31 (57.4) | 41 (42.7) | |
| Female | 78 (52.0) | 23 (42.6) | 55 (57.3) | |
| AML status, n (%) | .5 | |||
| De novo | 134 (89.3) | 47 (87.0) | 87 (90.6) | |
| Secondary AML | 16 (10.7) | 7 (13.0) | 9 (9.4) | |
| WBC at diagnosis (×109/L) | .5 | |||
| Median (IQR) | 43.6 (12.1-110.0) | 52.0 (15.5-120.9) | 36.5 (12.0-110.0) | |
| Range | 0.9-458.0 | 0.9-458.0 | 1.1-368.8 | |
| Blasts BM (%) | .4 | |||
| Median (IQR) | 81.0 (57.0-89.0) | 82.0 (61.0-91.0) | 76.5 (51.5-89.0) | |
| Range | 15.0-99.0 | 15.0-99.0 | 19.0-98.0 | |
| Blasts PB (%) | .3 | |||
| Median (IQR) | 62.0 (26.0-87.0) | 68.0 (36.0-89.0) | 57.0 (16.0-85.5) | |
| Range | 0.0-98.0 | 3.0-97.0 | 0.0-98.0 | |
| ELN 2022 prognosis, n (%) | .07 | |||
| Favorable | 14 (9.3) | 6 (11.1) | 8 (8.3) | |
| Intermediate | 98 (65.3) | 29 (53.7) | 69 (71.9) | |
| Adverse | 38 (25.3) | 19 (35.2) | 19 (19.8) | |
| FLT3-ITD, n (%)∗ | .08 | |||
| Mutation | 130 (86.7) | 43 (79.6) | 87 (90.6) | |
| No mutation | 20 (13.3) | 11 (20.4) | 9 (9.4) | |
| FLT3-TKD, n (%)∗ | .12 | |||
| Mutation | 26 (17.3) | 13 (24.1) | 13 (13.5) | |
| No mutation | 124 (82.7) | 41 (75.9) | 83 (86.5) | |
| NPM1, n (%)∗ | .4 | |||
| Mutation | 82 (54.7) | 27 (50.0) | 55 (57.3) | |
| No mutation | 68 (45.3) | 27 (50.0) | 41 (42.7) |
| . | Total, N = 150 (100%) . | ICT + MIDO, n = 54 (36%) . | ICT, n = 96 (64%) . | P value . |
|---|---|---|---|---|
| Age at diagnosis (y) | .09 | |||
| Median (IQR) | 51.0 (43.0-57.0) | 52.5 (47.0-62.0) | 50.0 (41.5-55.0) | |
| Range | 18.0-76.0 | 23.0-76.0 | 18.0-73.0 | |
| Sex, n (%) | .08 | |||
| Male | 72 (48.0) | 31 (57.4) | 41 (42.7) | |
| Female | 78 (52.0) | 23 (42.6) | 55 (57.3) | |
| AML status, n (%) | .5 | |||
| De novo | 134 (89.3) | 47 (87.0) | 87 (90.6) | |
| Secondary AML | 16 (10.7) | 7 (13.0) | 9 (9.4) | |
| WBC at diagnosis (×109/L) | .5 | |||
| Median (IQR) | 43.6 (12.1-110.0) | 52.0 (15.5-120.9) | 36.5 (12.0-110.0) | |
| Range | 0.9-458.0 | 0.9-458.0 | 1.1-368.8 | |
| Blasts BM (%) | .4 | |||
| Median (IQR) | 81.0 (57.0-89.0) | 82.0 (61.0-91.0) | 76.5 (51.5-89.0) | |
| Range | 15.0-99.0 | 15.0-99.0 | 19.0-98.0 | |
| Blasts PB (%) | .3 | |||
| Median (IQR) | 62.0 (26.0-87.0) | 68.0 (36.0-89.0) | 57.0 (16.0-85.5) | |
| Range | 0.0-98.0 | 3.0-97.0 | 0.0-98.0 | |
| ELN 2022 prognosis, n (%) | .07 | |||
| Favorable | 14 (9.3) | 6 (11.1) | 8 (8.3) | |
| Intermediate | 98 (65.3) | 29 (53.7) | 69 (71.9) | |
| Adverse | 38 (25.3) | 19 (35.2) | 19 (19.8) | |
| FLT3-ITD, n (%)∗ | .08 | |||
| Mutation | 130 (86.7) | 43 (79.6) | 87 (90.6) | |
| No mutation | 20 (13.3) | 11 (20.4) | 9 (9.4) | |
| FLT3-TKD, n (%)∗ | .12 | |||
| Mutation | 26 (17.3) | 13 (24.1) | 13 (13.5) | |
| No mutation | 124 (82.7) | 41 (75.9) | 83 (86.5) | |
| NPM1, n (%)∗ | .4 | |||
| Mutation | 82 (54.7) | 27 (50.0) | 55 (57.3) | |
| No mutation | 68 (45.3) | 27 (50.0) | 41 (42.7) |
ELN, European LeukemiaNet; IQR, interquartile range; PB, peripheral blood; WBC, white blood cells.
Status assessed by FA.
Evolution of FLT3-ITD clones between diagnosis and R/R disease at the patient level
The evolution of FLT3-ITD mutations was examined by HTS in paired diagnosis and R/R BM samples from 150 patients. Sequencing data were processed with the FiLT3r algorithm allowing to detect small FLT3-ITD clones with an AR of ≥0.0003. Using this approach, we categorized FLT3-ITD mutations as microclones if the AR was <0.05 and as macroclones if the AR was ≥0.05. The specificity of FLT3-ITD detection was confirmed by processing negative controls, and the presence of some FLT3-ITD microclones was validated using droplet digital polymerase chain reaction (see the supplemental Methods). A total of 270 FLT3-ITD clones were detected in diagnostic samples including 141 (52%) microclones and 129 (48%) macroclones (supplemental Figure 1). Among 144 FLT3-ITD clones detected in R/R samples, 48 (33%) were microclones and 96 (67%) were macroclones. Among the patients included on the basis of a FLT3-TKD mutation and classified FLT3-ITD negative at diagnosis by FA (n = 20), 25% (5/20 patients) actually harbored FLT3-ITD microclones at diagnosis unveiled by HTS. Thus, a total of 135 patients were initially positive for FLT3-ITD by HTS, among which 112 patients harbored at least 1 macroclone and the remaining 23 patients harbored only microclones. At the time of diagnosis, the characteristics of the microclones were similar to those of the macroclones in terms of length and insertion site (supplemental Table 2).
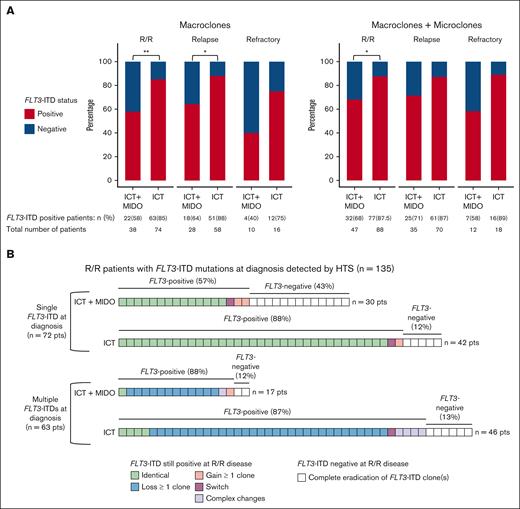
To describe the evolution of FLT3-ITD clones, we focused on patients harboring FLT3-ITD mutations at diagnosis. Considering FLT3-ITD macroclones only, the rate of FLT3-ITD persistence at R/R disease was lower in patients treated with ICT + MIDO than in patients receiving ICT alone (58% [22/38] vs 85% [63/74]; P = .0023). The persistence rates were 64% (18/28) vs 88% (51/58) at relapse (P = .019) and 40% (4/10) vs 75% (12/16) at refractory status in ICT + MIDO–treated patients and ICT-treated patients, respectively (Figure 1A). Considering both micro and macroclones, the rate of FLT3-ITD persistence at R/R disease was also lower in patients treated with ICT + MIDO compared with patients receiving ICT alone (68% [32/47] vs 87.5% [77/88]; P = .011). The persistence rates were 71% (25/35) vs 87% (61/70) at relapse and 58% (7/12) vs 89% (16/18) at refractory status in ICT + MIDO–treated patients and ICT-treated patients, respectively (Figure 1A).
Evolution of FLT3-ITD mutations between diagnosis and R/R disease. (A) FLT3-ITD status at R/R disease assessed by HTS, according to the treatment group and the type of progression. Left panel: only patients harboring FLT3-ITD macroclones at diagnosis are displayed. Only macroclones are taken into account to determine FLT3-ITD status at R/R disease. Right panel: patients harboring FLT3-ITD microclones and/or macroclones at diagnosis are displayed. Both macroclones and microclones are taken into account to determine FLT3-ITD status at R/R disease. (B) Patterns of evolution of FLT3-ITD microclones and macroclones between diagnosis and R/R disease, according to the number of FLT3-ITD clones detected at diagnosis (either a single clone or multiple clones) and the treatment group. Each square represents 1 patient. Patterns of evolution (from diagnosis to R/R disease) were assessed by HTS and defined as follows. Identical: clone A → clone A. Loss: clone A + clone B → clone A. Gain: clone A → clone A + clone B. Switch: clone A → clone B. Complex changes: clone A + clone B → clone B + clone C. Eradication: clone A → ⦰. ∗, <.05; ∗∗, <.01. Pts, patients.
Evolution of FLT3-ITD mutations between diagnosis and R/R disease. (A) FLT3-ITD status at R/R disease assessed by HTS, according to the treatment group and the type of progression. Left panel: only patients harboring FLT3-ITD macroclones at diagnosis are displayed. Only macroclones are taken into account to determine FLT3-ITD status at R/R disease. Right panel: patients harboring FLT3-ITD microclones and/or macroclones at diagnosis are displayed. Both macroclones and microclones are taken into account to determine FLT3-ITD status at R/R disease. (B) Patterns of evolution of FLT3-ITD microclones and macroclones between diagnosis and R/R disease, according to the number of FLT3-ITD clones detected at diagnosis (either a single clone or multiple clones) and the treatment group. Each square represents 1 patient. Patterns of evolution (from diagnosis to R/R disease) were assessed by HTS and defined as follows. Identical: clone A → clone A. Loss: clone A + clone B → clone A. Gain: clone A → clone A + clone B. Switch: clone A → clone B. Complex changes: clone A + clone B → clone B + clone C. Eradication: clone A → ⦰. ∗, <.05; ∗∗, <.01. Pts, patients.
Among 135 patients harboring FLT3-ITD microclones or macroclones at diagnosis, 72 had a single FLT3-ITD clone and 63 had multiple FLT3-ITD clones (henceforth referred to as “clonal interference”), respectively (Figure 1B). Among patients with multiple FLT3-ITD clones, the median number of ITDs per patient was 3. In further detail, 27 patients had 2 ITDs (42.9%), 19 patients had 3 ITDs (30.2%), 8 patients had 4 ITDs (12.7%), 4 patients had 5 ITDs (6.3%), 3 patients had 6 ITDs (4.8%), 1 patient had 8 ITDs (1.6%), and 1 patient had 9 ITDs (1.6%). The majority of patients with multiple FLT3-ITD clones harbored a single macroclone and 1 or several microclone(s) (55.6% [35/63]). In 72 patients with a single FLT3-ITD clone, the addition of MIDO to ICT was associated with a lower rate of FLT3-ITD positivity at R/R disease than in patients receiving ICT alone (57% [17/30] vs 88% [37/42]; P = .005). In the 63 patients who harbored multiple FLT3-ITD clones at diagnosis, the addition of MIDO to ICT did not affect the FLT3-ITD persistence rate at R/R disease (ICT + MIDO: 88% [15/17] vs ICT: 87% [40/46]).
Accordingly, after ICT + MIDO, the FLT3-ITD persistence rate was lower in patients harboring a single FLT3-ITD clone compared with patients harboring multiple clones (57% [17/30] vs 88% [15/17], P = .049) whereas there was no difference in patients treated by ICT alone (88% [37/42] vs 87% [40/46]). The evolution of patients harboring multiple FLT3-ITD clones at diagnosis was similar across the 2 treatment groups, predominantly shaped by the simplification of the FLT3-ITD repertoire upon R/R disease that is the loss of part of the initial clones (ICT + MIDO: 71% [12/17]; ICT: 67% [31/46]; Figure 1B).
Among the 109 patients retaining a positive FLT3-ITD status at R/R disease after either ICT or ICT + MIDO, this simplification of FLT3-ITD repertoire is highlighted by the decrease in the rate of patients harboring multiple FLT3-ITD clones between diagnosis and R/R disease (50% [55/109] vs 23% [25/109]; P = .014; supplemental Figure 2). In those 109 patients, enrichment of the FLT3-ITD repertoire upon R/R disease through the acquisition of new FLT3-ITD clones was a rare phenomenon (ICT + MIDO: 16% [5/32]; ICT: 9% [7/77]; Figure 1B). All those results were consistent, although nonsignificant because of the small number of patients, when refractory and relapsed patients were analyzed separately (supplemental Figure 3A-B).
Evolution of the allele burden between diagnosis and relapse at the patient level
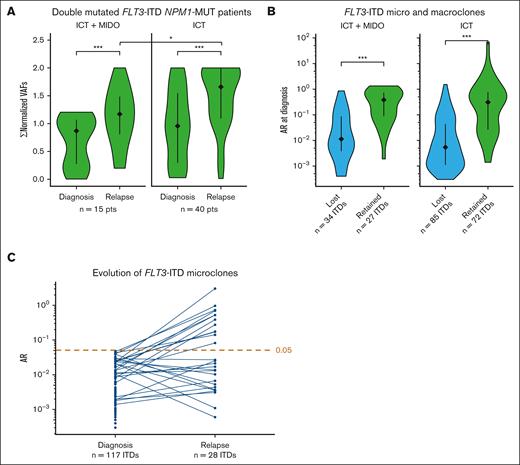
After a qualitative analysis of clonal evolution based on the binary presence or absence of FLT3-ITD clones, we carried out a quantitative analysis (supplemental Table 3). Because the number of patients with refractory disease was limited, we restricted the quantitative analysis to relapsed patients. In patients with mutated NPM1, we calculated norm-VAFs of FLT3-ITD mutations to limit the impact of sample dilution on the allele burden. For samples harboring multiple FLT3-ITD microclones or macroclones, the ARs or the norm-VAFs of each clone were summed to obtain a single value per sample, namely ΣARs or Σnorm-VAFs. We observed that Σnorm-VAFs increased between diagnosis and relapse in each treatment group, strengthening the role of FLT3-ITD clones in the relapse process (median Σnorm-VAFs; ICT + MIDO: 0.87 vs 1.2; P < .001 and ICT: 0.96 vs 1.7; P < .001; Figure 2A; supplemental Table 3), independently of MIDO. Nevertheless, patients who received MIDO displayed lower ΣARs (P = .034) and Σnorm-VAFs (P = .02) at relapse than patients who received ICT alone, indicating that the expansion of FLT3-ITD clones at relapse is probably dampened by MIDO.
Quantification of FLT3-ITD clones at diagnosis and relapse by HTS. (A) Σnorm-VAFs per sample at diagnosis and relapse, according to the treatment group. Only patients with a positive FLT3-ITD status by HTS at both diagnosis and relapse are displayed. (B) AR at diagnosis for lost and retained FLT3-ITD microclones and macroclones upon AML relapse, according to the treatment group. (C) Evolution of the AR of FLT3-ITD microclones detected at AML diagnosis. Each line features the evolution of a FLT3-ITD microclone between diagnosis and relapse. The threshold is positioned at an AR of 0.05. ∗, <.05; ∗∗∗, <.001. MUT, mutated; Pts, patients.
Quantification of FLT3-ITD clones at diagnosis and relapse by HTS. (A) Σnorm-VAFs per sample at diagnosis and relapse, according to the treatment group. Only patients with a positive FLT3-ITD status by HTS at both diagnosis and relapse are displayed. (B) AR at diagnosis for lost and retained FLT3-ITD microclones and macroclones upon AML relapse, according to the treatment group. (C) Evolution of the AR of FLT3-ITD microclones detected at AML diagnosis. Each line features the evolution of a FLT3-ITD microclone between diagnosis and relapse. The threshold is positioned at an AR of 0.05. ∗, <.05; ∗∗∗, <.001. MUT, mutated; Pts, patients.
Evolution of FLT3-ITD clones between diagnosis and relapse at the clone level
After investigating the evolution of FLT3-ITD mutations at the patient level, we examined their evolution at the clonal level. Overall, only the minority of initial FLT3-ITD microclones and macroclones were conserved at relapse, both in the ICT + MIDO group (44% [27/61]) and in the ICT alone group (46% [72/157]; supplemental Table 4). To determine the factors influencing the fate of the FLT3-ITD microclones and macroclones detected at diagnosis, we compared the initial characteristics between FLT3-ITD clones that were retained or lost at relapse in each treatment group (supplemental Table 4). Lost FLT3-ITDs had significantly lower ARs than retained ones, both in the ICT + MIDO group (median AR: 0.011 vs 0.38; P < .001) and in the ICT group (0.0054 vs 0.31; P < .001; Figure 2B; supplemental Table 4). The results were consistent after adjusting for tumor dilution (supplemental Figure 4; supplemental Table 4). Accordingly, only 24% (28/117) of FLT3-ITD microclones were retained at the time of relapse in both treatment groups combined, of which 43% became macroclones (12/28; Figure 2C). In further detail, 26% (23/88) of microclones were retained at the time of relapse in patients treated with ICT only, of which 48% (11/23) became macroclones. In patients receiving ICT + MIDO, only 17% of microclones (5/29) were retained at the time of relapse, of which 20% became macroclones (1/5).
In the whole cohort, the lost clones had significantly lower ITD lengths than retained clones (median length: 42 vs 54 base pairs; P = .037; supplemental Table 4). In a multivariate logistic regression model, AR of ≥0.5 was strongly predictive of clone persistence (odds ratio [OR], 7.89; 95% confidence interval [CI], 2.7-29; P < .001) whereas microclones (OR, 0.31; 95% CI, 0.15-0.64; P = .0014) as well as competing clones (clonal interference; OR, 0.40; 95% CI, 0.17-0.94; P = .038) were more likely to be eradicated at relapse (Table 2). The ITD length, the insertion site, and the treatment had no significant impact on clonal evolution in multivariate analysis (Table 2).
Logistic regression model for the persistence of 218 FLT3-ITD clones at relapse
| Parameter . | OR . | 95% CI . | P value . |
|---|---|---|---|
| AR ≥ 0.5 | 7.89 | 2.7-29 | <.001 |
| Microclones | 0.31 | 0.15-0.64 | .0014 |
| Clonal interference | 0.40 | 0.17-0.94 | .038 |
| ITD length of >51 bp | 1.70 | 0.77-3.8 | .19 |
| Insertion site in TKD1 | 1.54 | 0.68-3.5 | .30 |
| ICT + MIDO | 0.65 | 0.30-1.4 | .26 |
| Parameter . | OR . | 95% CI . | P value . |
|---|---|---|---|
| AR ≥ 0.5 | 7.89 | 2.7-29 | <.001 |
| Microclones | 0.31 | 0.15-0.64 | .0014 |
| Clonal interference | 0.40 | 0.17-0.94 | .038 |
| ITD length of >51 bp | 1.70 | 0.77-3.8 | .19 |
| Insertion site in TKD1 | 1.54 | 0.68-3.5 | .30 |
| ICT + MIDO | 0.65 | 0.30-1.4 | .26 |
bp, base pairs; TKD1, tyrosine kinase domain 1.
Molecular landscape at diagnosis and R/R disease
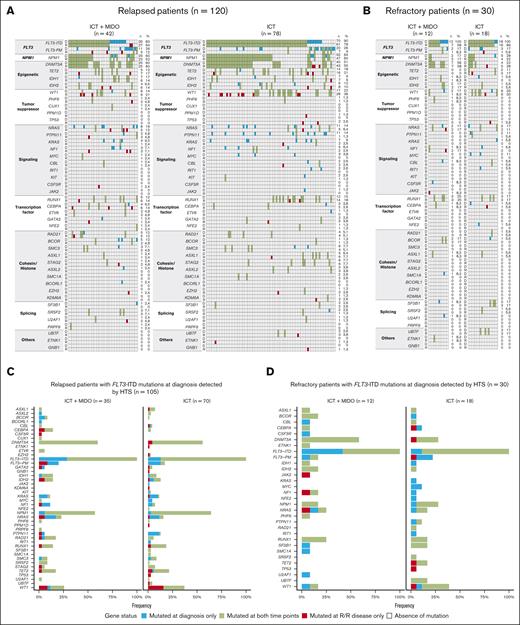
The molecular landscape was examined by HTS at diagnosis and R/R disease using a panel targeting 48 genes (supplemental Table 1). At diagnosis, the landscape of driver mutations was similar between the 2 treatment groups (Figure 3A-B). Genes most frequently mutated at diagnosis were DNMT3A and NPM1.
Evolution of gene mutations between diagnosis and R/R disease assessed by HTS. (A-B) Evolution of gene mutations between diagnosis and R/R disease, according to the type of progression: relapse (A) or refractory disease (B) and the treatment group. Genes are organized according to their functional class. Each column represents a single patient. For each patient, the evolution of the gene status is indicated by the color of the boxes. For each gene, the number (n) and percentage (%) of patients with mutated genes at each time point are displayed on the right of the figures. (C-D) Evolution of gene mutations among patients who experienced relapse (C) or refractory disease (D), according to the treatment group. Only patients harboring FLT3-ITD mutations at diagnosis as assessed by HTS are displayed. D, diagnosis; PM, point mutations; R, relapsed (left side) or refractory disease (right side).
Evolution of gene mutations between diagnosis and R/R disease assessed by HTS. (A-B) Evolution of gene mutations between diagnosis and R/R disease, according to the type of progression: relapse (A) or refractory disease (B) and the treatment group. Genes are organized according to their functional class. Each column represents a single patient. For each patient, the evolution of the gene status is indicated by the color of the boxes. For each gene, the number (n) and percentage (%) of patients with mutated genes at each time point are displayed on the right of the figures. (C-D) Evolution of gene mutations among patients who experienced relapse (C) or refractory disease (D), according to the treatment group. Only patients harboring FLT3-ITD mutations at diagnosis as assessed by HTS are displayed. D, diagnosis; PM, point mutations; R, relapsed (left side) or refractory disease (right side).
We focused on patients harboring FLT3-ITD mutations at diagnosis as assessed by HTS to identify molecular mechanisms of progression. Genes that most frequently became mutated at relapse were FLT3 (regarding point mutations [9%]) and WT1 (9%) in the 35 patients treated with ICT + MIDO vs WT1 (16%) in the 70 patients treated with ICT alone (Figure 3C). Excluding FLT3 mutations, emerging mutations in genes involved in signaling pathways were found in 11% of patients who relapsed after treatment with ICT + MIDO (NRAS: 6% [2/35], PTPN11 + NF1: 3% [1/35], and CSF3R: 3% [1/35]). Several genes became mutated at the time of refractory disease, but each of these genes was involved only once (Figure 3D).
Discussion
We sought to identify the molecular mechanisms underlying R/R disease in patients diagnosed with FLT3-mutated AML and treated with ICT with/without MIDO. To this end, FLT3 mutations and comutations were screened by HTS at diagnosis and R/R disease to unravel clonal evolution. For accurate annotation and quantification of FLT3-ITDs from HTS data, we used the FiLT3r algorithm,14 able to detect small FLT3-ITD clones with an AR of ≥0.0003. In our cohort, a total of 270 FLT3-ITD clones were detected by HTS at diagnosis, including 141 clones (52%) with an AR of <0.05 and 129 clones (48%) with an AR above this limit, respectively designated microclones and macroclones. The average number of FLT3-ITD clones per patient at diagnosis was higher in this study (2.0 [270/135] FLT3-ITDs per patient) than previous studies using FA (1.14,15 1.17,16 and 1.3317FLT3-ITD per patient). Moreover, the rate of patients harboring multiple FLT3-ITD clones at diagnosis was higher in this study (47% [63/135]) than in previous studies based on FA (12%,15 14%,16 23%,6 and 27%17). The presence of multiple FLT3-ITD clones was referred to as clonal interference, a concept from evolutionary biology describing the emergence of several clones harboring independent lesions and competing with each other in the same population.18,19
At R/R disease, the rate of FLT3-ITD persistence was significantly lower in patients treated with MIDO (68%) than in patients not receiving MIDO (87.5%), as previously described.10 More precisely, the persistence rates reached 71% vs 87% at relapse and 58% vs 89% at refractory disease, in ICT + MIDO–treated patients and ICT-treated patients, respectively. Regarding relapsed patients treated with ICT alone, this rate is in line with former data.11,20,21 However, regarding R/R patients treated with the ICT + MIDO combination, there is a discrepancy with a previous study by Schmalbrock et al in which the persistence rate amounted to 54% at R/R disease.10 This discrepancy is probably related to the use of distinct FLT3-ITD screening methods. Indeed, in the former study, FLT3-ITD mutations were screened by FA using an AR threshold of 0.05.9,10 In our study, the screening of FLT3-ITD mutations by HTS revealed that 46% of clones detected at both time points were microclones, defined by an AR of <0.05. Factoring in only the macroclones detected by HTS, the persistence rate after treatment with ICT + MIDO decreased to 58% at R/R disease, in keeping with the previous study by Schmalbrock et al.10
In R/R patients, a key finding of our work was that the addition of MIDO to ICT significantly decreased the FLT3-ITD persistence rate of patients harboring a single FLT3-ITD clone (ICT + MIDO: 57% vs ICT: 88%) but not that of patients harboring multiple FLT3-ITD clones. Accordingly, in patients receiving ICT + MIDO, our data demonstrated that clonal interference was associated with a greater ability to select a FLT3-ITD clone at R/R disease (multiple clones: 88% vs single clone: 57%; P = .049). Further data are warranted to determine whether clonal interference affects the prognosis of patients treated with ICT + MIDO or other FLT3i such as quizartinib.22 Regarding patients treated with ICT exclusively, the presence of multiple FLT3-ITD clones at diagnosis has been correlated with lower CR rates, event-free survival, and overall survival,6,15,23 although these findings have been challenged by other studies.16,17,24
After analyzing the evolution of FLT3-ITD mutations at the patient level, we performed a multivariate logistic regression to predict the outcome of individual FLT3-ITD clones based on their initial characteristics. In this model, an AR of ≥0.5 favored clonal persistence at relapse whereas microclones and clones subjected to clonal interference were more likely to be eradicated, clearing the way to the fittest FLT3-ITD clone. Considering the limited number of persisting microclones in our cohort, additional studies are warranted to uncover the factors underlying the persistence of certain microclones.
Overall, the best predictor of FLT3-ITD persistence in a patient would likely be an indicator that reflects not only the number of FLT3-ITD clones but also the fraction of leukemic cells carrying each FLT3-ITD. However, because the bulk HTS approach cannot determine whether cells are heterozygous or homozygous for a given mutation, it is not suitable to calculate such an indicator.
Other relapse-related changes included the acquisition of additional mutations. In patients harboring FLT3-ITD mutations at diagnosis as assessed by HTS, WT1 gene frequently became mutated at relapse both after ICT + MIDO (9%) and ICT only (16%). Moreover, genes involved in signaling pathways (except FLT3) were newly mutated in 11% of patients who relapsed after treatment with ICT + MIDO, in line with a previous study.10
In summary, MIDO shaped the evolution of patients with a single FLT3-ITD clone in our cohort. In these patients, the addition of MIDO to ICT was associated with a lower rate of FLT3-ITD persistence at R/R disease. These findings emphasize the need to reassess FLT3-ITD status at R/R disease before implementing targeted therapy. Moreover, our study revealed that FLT3-ITD microclones are capable of driving relapse. Prospective studies are warranted to elucidate the impact of FLT3-ITD microclones in patients with AML who tested FLT3-ITD negative at diagnosis using standard FA-based methods.
Acknowledgments
The authors thank Maxime Bucci and Nathalie Helevaut for technical help. The authors also thank Lamya Haddaoui and Krishshanti Sinnadurai (tumor bank for the French Innovative Leukemia Organization [FILO] group, no. BB-033-00073, Hôpital Pitié-Salpêtrière, Paris) as well as Christophe Roumier (tumor bank for the Acute Leukemia French Association [ALFA] group, certification NF 96900-2014/65453-1) for handling, conditioning, and storing patient samples. The work of all investigators from the ALFA-FILO group and clinical research assistants is also acknowledged.
This study was supported by the French National Cancer Institute (PRTK 15-125). R.J. and N.D. were supported by grants from the French League Against Cancer (Appel à Projets [AAP]2021 du Septentrion, Comité du Pas-de-Calais) and Lille University Hospital (AAP 2021 du Fonds Hospitalier d’aide à l'émergence). R.J. received grants from INSERM, Fondation Initiatives-Science-Innovation-Territoires-Economie de l'Université Lille Nord Europe (I-SITE ULNE), and Laurette Fugain association (ALF 2024/13).
Authorship
Contribution: P.-Y.D., N.D., and C.P. designed the study; H.D., C.B., A.P., D.L., P.P., E.T., M.C., L.H., R.I., S. Bertoli, M.H., and C.R. managed patients and provided clinical data; S. Bouzy, P.F.-G., S.T., A. Bidet, and E.D. provided samples and biological data; S.G. performed the experiments; N.D. and L.F. performed the molecular analyses; A. Boudry and M.S. performed the bioinformatics analysis. A. Boudry and R.J. performed the statistical analyses; R.J., N.D. and P.-Y.D. wrote the manuscript; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: M.C. reports honoraria from AbbVie, Astellas, Jazz, and Bristol Myers Squibb. R.I. reports honoraria from Astellas and Daichii-Sankyo, and research support from Novartis. S. Bertoli declares a consulting or advisory role with Abbvie, Astellas, Bristol Myers Squibb-Celgene, Jazz Pharmaceuticals and Novartis; and received travel grants from Abbvie and Pfizer. C.R. reports consulting or advisory role with AbbVie, Amgen, Astellas, Bristol Myers Squibb, Boehringer, Jazz Pharmaceuticals, Johnson & Johnson, and Servier; received research funding from AbbVie, Amgen, Astellas, Bristol Myers Squibb, Iqvia, and Jazz Pharmaceuticals; and received support for attending meetings and/or travel from AbbVie, Novartis, and Servier. P.-Y.D. reports honoraria and research support to institution from Novartis, Servier, Bristol Myers Squibb, Astellas, and Daiichi-Sankyo; honoraria from AbbVie, Jazz Pharmaceuticals, and Janssen; and research support to institution from Roche. The remaining authors declare no competing financial interests.
Correspondence: Pierre-Yves Dumas, Hématologie Clinique et Thérapie Cellulaire, Centre Hospitalier Universitaire Bordeaux, 1 avenue Magellan, Hôpital du Haut-Lévèque, F-33000 Bordeaux, France; email: pierre-yves.dumas@u-bordeaux.fr.
References
Author notes
R.J. and A.B. as well as N.D. and P.-Y.D. contributed equally to this study.
Presented, in part, as an oral presentation (abstract 976) at the 65th annual meeting of the American Society of Hematology, San Diego, CA, 11 December 2023.
Some deidentified data are available on reasonable request from the corresponding author, Pierre-Yves Dumas (pierre-yves.dumas@u-bordeaux.fr); the data sharing will require a detailed proposal to the study investigators, and a data transfer agreement must be signed.
The full-text version of this article contains a data supplement.