Human hematopoietic stem cells can be corrected to restore endogenous RAG2 gene expression while preserving durable engraftment potential.
Gene-corrected RAG2 locus restores V(D)J recombination in stem cells of patients with RAG2-SCID, promoting TCR and B-cell receptor formation.
Visual Abstract
Recombination-activating genes (RAG1 and RAG2) are critical for lymphoid cell development and function by initiating the variable (V), diversity (D), and joining (J) (V(D)J)-recombination process to generate polyclonal lymphocytes with broad antigen specificity. The clinical manifestations of defective RAG1/2 genes range from immune dysregulation to severe combined immunodeficiencies (SCIDs), causing life-threatening infections and death early in life without hematopoietic cell transplantation (HCT). Despite improvements, haploidentical HCT without myeloablative conditioning carries a high risk of graft failure and incomplete immune reconstitution. The RAG complex is only expressed during the G0-G1 phase of the cell cycle in the early stages of T- and B-cell development, underscoring that a direct gene correction might capture the precise temporal expression of the endogenous gene. Here, we report a feasibility study using the CRISPR/Cas9-based “universal gene-correction” approach for the RAG2 locus in human hematopoietic stem/progenitor cells (HSPCs) from healthy donors and RAG2-SCID patient. V(D)J-recombinase activity was restored after gene correction of RAG2-SCID-derived HSPCs, resulting in the development of T-cell receptor (TCR) αβ and γδ CD3+ cells and single-positive CD4+ and CD8+ lymphocytes. TCR repertoire analysis indicated a normal distribution of CDR3 length and preserved usage of the distal TRAV genes. We confirmed the in vivo rescue of B-cell development with normal immunoglobulin M surface expression and a significant decrease in CD56bright natural killer cells. Together, we provide specificity, toxicity, and efficacy data supporting the development of a gene-correction therapy to benefit RAG2-deficient patients.
Introduction
Severe combined immunodeficiencies (SCIDs) are genetic diseases caused by inherited mutations in genes required for T-, B-, and, sometimes, natural killer (NK)-cell development and function.1 Defects in the recombination-activating genes 1 and 2 (RAG1/2) are the second most common cause of SCID in the United States. RAG1 and RAG2 form a heterotetrameric complex (with 2 copies of each protein) that initiates the variable (V), diversity (D), joining (J) (V(D)J) recombination process and mediates DNA cleavage at recombination signal sequences flanking the V(D)J coding elements of the T-cell receptor (TCR) and immunoglobulin (Ig) receptor loci. The nonhomologous end-joining (NHEJ)–mediated DNA repair of RAG-induced DNA breaks joins coding elements to generate functional TCR and Ig molecules.
Over 200 unique pathogenic variants have been identified that mutate the RAG1/2 genes, causing the T–B–NK+ SCID phenotype2,3 and a spectrum of immune dysregulation4,5: Omenn syndrome (OS), leaky SCID (LS),6-8 and delayed-onset combined immunodeficiency with granulomas and/or autoimmunity (CID-G/AI).9-11 Patients with severe forms of RAG deficiency, including SCID, OS, and LS, achieve immune reconstitution with high survival rates (>90%) following allogeneic hematopoietic cell transplantation (allo-HCT) from human leucocyte antigen (HLA)-matched donors.12-15 In contrast, patients with RAG mutations who rely on haploidentical HCT without myeloablative conditioning experience high graft failure rates (>75%) or poor lymphoid reconstitution. This is because the host’s genetically defective common lymphoid progenitors (CLPs) occupy the bone marrow (BM) and thymus niches16,17 and prevent the donor’s CLPs from establishing lymphopoiesis. In addition, the host’s NK cells have heightened cytotoxic activity, which may mediate graft rejection.18
To identify therapeutic options beneficial to all patients with RAG deficiency, feasibility studies using lentivirus (LV)-based gene therapy were performed in Rag1/2 disease mouse models.19-21 These studies showed variable therapeutic outcomes, suggesting that overexpressing Rag genes might not robustly and faithfully recapitulate the levels of regulation and expression necessary to support immune system development and function, thus raising concerns about potential clinical efficacy.
Advances in genome-editing technology22-24 have allowed for gene correction of human hematopoietic stem/progenitor cells (HSPCs). This strategy offers significant therapeutic benefits for diseases such as RAG1/2 deficiency, where the underlying gene relies on strict spatiotemporal gene regulation and expression.25 The RAG protein complex is only expressed in the G0/G1 during early T- and B-cell development,3 thus demonstrating precise cell cycle and developmental specificity. Cluster regulatory interspaced short palindromic repeats–associated Cas9 nuclease (CRISPR/Cas9) is a genome engineering platform23-25 that has been successfully applied to primary human cells,26-28 including HSPCs,28-32 to introduce the desired genomic modification through homologous recombination–mediate genome targeting (HR-GTed).25 We have previously demonstrated the in vitro rescue of T-cell developmental blockage using CRISPR/Cas9–adeno-associated virus of serotype 6 (AAV6)–based correction of a patient’s RAG2 homozygous null mutation (c.831T>A; p.Y277∗).33 The efficacy of in vitro CRISPR-based correction of the RAG2-SCID phenotype was further validated in a recent study34 in healthy donors and patient-derived HSPCs. However, the in vivo efficacy of this gene-correction approach remains to be determined. Here, we showed functional correction of RAG2-SCID in vivo using immunodeficient NSG-SGM3 mice that received transplantation with HSPCs derived from patients with HR-GTed RAG2-SCID. We repaired the DNA sequence of HSPCs from patients with RAG2-SCID carrying 2 null compound heterozygous variants causing RAG2-SCID, which restored V(D)J activity and rescued both T- and B-cell developmental blocks. We further showed the in vivo rescue of the immature NK cell phenotype derived from HR-GTed RAG2-SCID HSPCs.
Methods
Human CD34+ HSPCs
Peripheral blood (PB)-CD34+ cells from a RAG2-SCID subject were collected after receiving informed consent (protocol NCT00001405, approved by the National Institutes of Health institutional review board as protocol 94-I-0073). Additional information is detailed in supplemental Methods.
On- and off-target activity assay
The top 48 putative off-target (OT) sites for RAG2 were identified using CRISPR OT sites with mismatches, insertions, and deletions (COSMID)35 and were tested using rhAmpSeq36,37 (Integrated DNA Technologies). On-target analysis was performed on polymerase chain reaction–amplified GTed and non-GTed alleles from a bulk codon-optimized RAG2 (coRAG2) GTed cell population. The amplicons were sequenced using the PacBio Sequel IIe platform and analyzed using CRISPAltRations.38 A detailed description is included in supplemental Methods.
Colony-forming unit assay
Single-live HSPCs were sorted and analyzed as previously described.29 Colony genotyping is detailed in supplemental Methods.
Transplantation of human gene-modified CD34+ HSPCs and engraftment assessment
Rescue experiments, detailed in supplemental Methods, were performed using frozen peripheral blood (PB)-HSPCs from 1 patient with RAG2-SCID and compound heterozygous functionally null variants (c.296C>A, c.1324C>A; p.P99Q, A442T).
T- and B-cell receptors repertoire analysis
High-throughput sequencing (HTS) of TCRα and TCRβ and immunoglobulin M (IgM) heavy chain (Vh) analysis was performed as described in the supplemental Methods. Polymerase chain reaction assays for the 7 Vh chains were performed as described.39
Karyotype analysis
RAG2-SCID patient-derived PB-HSPCs mock (control) or ribonucleoprotein (RNP)-treated were subjected to G-band karyotyping analysis of 20 cells derived from each condition (WiCell Cytogenetics, Madison, WI).
Statistical analysis
Statistical tests were performed using GraphPad Prism (version 9.3.0 software), as detailed in the figure legends and supplemental Methods.
Ethics and animal approval statement
All work described in this study was carried out in compliance with all relevant ethical regulations. The animal studies were reviewed, approved, and monitored by the Stanford University Institutional Animal Care and Use Committee.
Results
Efficient universal gene-targeting strategy of RAG2 to the endogenous locus in human HSPCs
Two central concepts of the corrective therapeutic approach for RAG2 deficiency are that hematopoietic stem cells (HSCs) and their hematopoietic progenitor cells will preserve the physiological gene regulation necessary to achieve temporal and lineage-specific RAG2 protein expression and activity (Figure 1A-B) and that most of the RAG2-causing mutations, including deletions, will be corrected. We screened and selected a RAG2-specific guide RNA (sgRNA-3; supplemental Figure 1A-B) that, when complexed with HiFi Cas9 nuclease, achieved a median insertion and deletions (INDEL) frequency (gene-editing–mediated NHEJ; GE-NHEJ) of 52.6% (range, 27.1%-91.0%) in cord blood (CB)-HSPCs and 68.0% (range, 31.0%-81.0%) in PB-HSPCs. The INDEL profile of sgRNA-3 is characterized by 2-17 bp deletions (supplemental Figure 1C-F), which abrogated RAG2 protein expression in vivo (supplemental Figure 1D-E).
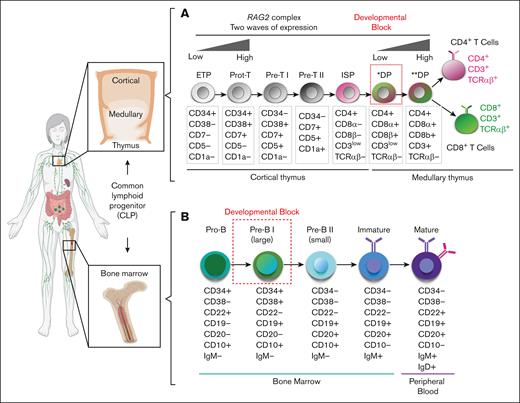
Schematics of hematopoietic developmental defect for patients with RAG2-SCID. (A) Overview of human T-cell (top) and (B) B-cell (bottom) developmental stages in the thymus and BM from cCLP. Dotted red squares mark developmental block for patients with RAG2-SCID. ∗DP, preselection double-positive stage; ∗∗DP, postselection double-positive stage; ETP, early thymic progenitor; ISP, immature single-positive stage.
Schematics of hematopoietic developmental defect for patients with RAG2-SCID. (A) Overview of human T-cell (top) and (B) B-cell (bottom) developmental stages in the thymus and BM from cCLP. Dotted red squares mark developmental block for patients with RAG2-SCID. ∗DP, preselection double-positive stage; ∗∗DP, postselection double-positive stage; ETP, early thymic progenitor; ISP, immature single-positive stage.
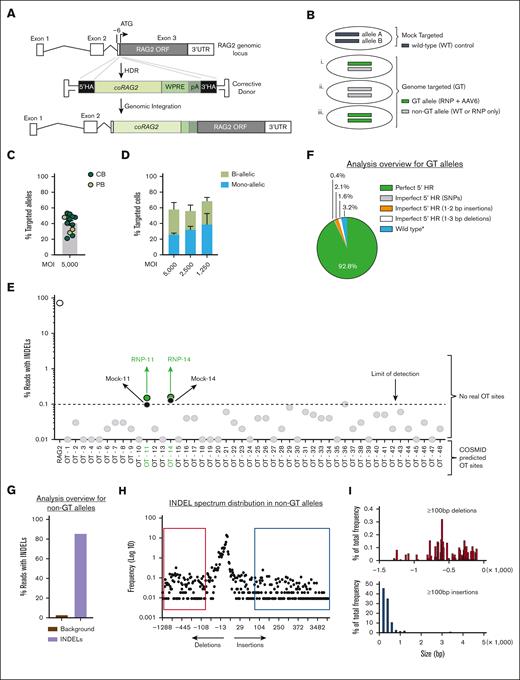
We used AAV6 to deliver coRAG2 transgene–bearing homology arms to the cut sites of the sgRNA (Figure 2A), generating various genome-editing outcomes in the bulk cell population (Figure 2Bi-iii). Absolute quantification showed an average of 40.6 ± 3 (standard error of the mean [SEM]; n = 13 unique HSPCs donors) alleles carrying the coRAG2 (Figure 2C) or higher (1.4- to 1.7-fold increase) when total alleles were quantified at single-cell resolution (Figure 2D, green and hashed bars combined; supplemental Figures 2A-B and 3A-B): 58.0% (5000 multiplicity of infection [MOI]; n = 5 donors); 56.0% (2500 MOI; n = 4 donors); and 68.7% (1250 MOI; n = 2 donors). We estimated the distribution of cells with 1 (monoallelic) or 2 (biallelic) alleles targeted as a function of the viral MOI. We observed an increase in monoallelic HR-GTed with decreased MOI (Figure 2C): 5000 MOI 25.6 ± 2.0 (SEM) monoallelic and 32.4 ± 8.7 (SEM) biallelic; 2500 MOI 31.7 ± 4.5 (SEM) monoallelic and 24.5 ± 7.4 (SEM) biallelic; and 1250 MOI 38.6 ± 14.2 (SEM) monoallelic and 30.1 ± 4.6 (SEM) biallelic. Correcting a single allele is sufficient to revert the disease phenotype because the disease is autosomal recessive, and heterozygous parents have normal immune systems.
Efficacy and specificity of genome editing at the RAG2 locus using universal correction strategy. (A) Schematics of gene-targeted integration of codon- coRAG2 and an expression cassette. The coRAG2 sequence is under the control of an endogenous promoter. (B) Overview of genome-editing outcomes. (C) Percent coRAG2-GTed in healthy donor (HD)purified HSPCs from fresh CB and frozen PB. Each circle represents a unique human HSPC donor. Genome-targeted (GTed) integration of coRAG2 was quantified by digital-droplet polymerase chain reaction (ddPCR). (D) Frequency of cells with 1 (monoallelic) or 2 (biallelic) alleles targeted as a function of the virus’ MOI. Analysis was performed on single cells sorted on methylcellulose plates (n = 5 at 5000 MOI; n = 4 at 2500 MOI; and n = 2 at 1250 MOI). Bars ± SEM (E) OT analysis using RAG2-SCID (c.296C>A and c.1342C>A) patient-derived HSPCs. Next-generation sequencing of 48 COSMID predicted OT sites in edited-only (RNP-sgRNA guide no. 3 and HiFi Cas9 nuclease) or electroporated-only (mock, nucleofected without RNP) cells. Shown INDELs reads for on-target (RAG2 gene, white circle) and OT sites below the limit of detection (gray circles) and above the limit of detection (green circles). PacBio-based analysis overview of (F) quantification of perfect and imperfect homologous recombination events using PacBio sequencing. Marked as “wild-type∗” are events that retained the wild-type protospacer adjacent motif sequence even after perfect HR repair. (G) A total of 85.5% of the bulk non-GT alleles have INDELs and 14.5% are wild-type (WT) alleles. The brown bar represents the background signal from sequencing error. (H) An overview of the INDEL spectrum in non-GT alleles identified short deletions (∼13 bp) as the predominant events. Each dot represents the frequency of a unique PacBio long-read size bin. Red box shows ≥100 bp deletions in (I) top histogram. Blue box shows ≥100 bp insertions in (I) bottom histogram. Percent total frequency of ≥100 bp deletions is 4.2 and of ≥100 bp insertions is 4.0 (supplemental Figure 4D). SNP, single nucleotide polymorphism.
Efficacy and specificity of genome editing at the RAG2 locus using universal correction strategy. (A) Schematics of gene-targeted integration of codon- coRAG2 and an expression cassette. The coRAG2 sequence is under the control of an endogenous promoter. (B) Overview of genome-editing outcomes. (C) Percent coRAG2-GTed in healthy donor (HD)purified HSPCs from fresh CB and frozen PB. Each circle represents a unique human HSPC donor. Genome-targeted (GTed) integration of coRAG2 was quantified by digital-droplet polymerase chain reaction (ddPCR). (D) Frequency of cells with 1 (monoallelic) or 2 (biallelic) alleles targeted as a function of the virus’ MOI. Analysis was performed on single cells sorted on methylcellulose plates (n = 5 at 5000 MOI; n = 4 at 2500 MOI; and n = 2 at 1250 MOI). Bars ± SEM (E) OT analysis using RAG2-SCID (c.296C>A and c.1342C>A) patient-derived HSPCs. Next-generation sequencing of 48 COSMID predicted OT sites in edited-only (RNP-sgRNA guide no. 3 and HiFi Cas9 nuclease) or electroporated-only (mock, nucleofected without RNP) cells. Shown INDELs reads for on-target (RAG2 gene, white circle) and OT sites below the limit of detection (gray circles) and above the limit of detection (green circles). PacBio-based analysis overview of (F) quantification of perfect and imperfect homologous recombination events using PacBio sequencing. Marked as “wild-type∗” are events that retained the wild-type protospacer adjacent motif sequence even after perfect HR repair. (G) A total of 85.5% of the bulk non-GT alleles have INDELs and 14.5% are wild-type (WT) alleles. The brown bar represents the background signal from sequencing error. (H) An overview of the INDEL spectrum in non-GT alleles identified short deletions (∼13 bp) as the predominant events. Each dot represents the frequency of a unique PacBio long-read size bin. Red box shows ≥100 bp deletions in (I) top histogram. Blue box shows ≥100 bp insertions in (I) bottom histogram. Percent total frequency of ≥100 bp deletions is 4.2 and of ≥100 bp insertions is 4.0 (supplemental Figure 4D). SNP, single nucleotide polymorphism.
We report no significant bias in the ability of coRAG2-modified HSPCs (coRAG2-HSPCs) to generate all 4 early myeloid and erythroid hematopoietic progenitor cells compared with control conditions, demonstrating an absence of unintended perturbations in the nonlymphoid lineages (supplemental Figure 2A-C). To address virus-related toxicity at higher MOIs, we tested p53 binding protein 1 (53BP1) inhibitor (i53) with a lower AAV6 concentration. i53 treatment blocks the NHEJ pathway and tips the balance toward the homology repair (HR) pathway.40 We observed a 1.5-fold higher HR-GTed in the i53-treated cells than in the untreated cells without diminishing the modified cells’ differentiation potential41 (supplemental Figure 2B). Overall, we showed that coRAG2-HSPCs supported early hematopoietic progenitor development.
Safety and specificity of the RAG2 gene-correction approach
To evaluate the OT profile of the RAG2 lead guide (sgRNA-3), we used COSMID,35 a bioinformatic tool, to predict the putative guide’s binding sites, and rhAMP-Seq36,37 to confirm them. OT activity at a total of 48 predicted loci was quantified38 in the genome of PB-derived HSPCs from a patient with RAG2null carrying compound heterozygous missense mutations (c.[296C>A; 1342C>A]; Figure 2E). We compared the percent INDELs (GE-NHEJ) detected in cells electroporated with RNP to mock-treated (control) cells. The sites considered true OT were those with GE-NHEJ > 0.1% in the RNP-treated condition relative to the control sample. Using this criterion, none of the sites were identified as true OTs (Figures 2E; Table 1). OT-11 and OT-14 were not bona fide OT sites because there was no increase in the frequency of INDELs compared with that in the mock (Table 1). To characterize the genome-editing events at the sgRNA cut site (on-target), we used the PacBio Sequel IIe sequencing platform38 on non-GTed (Figure 2Bi-iii, gray bars; supplemental Figure 4A,C) and GTed alleles (Figure 2Bi-iii, green bars; supplemental Figure 4B-C). We confirmed perfect HR in >90% of GTed alleles (Figure 2F), with the remaining 7.2% of GTed alleles harboring small-range INDELs (Table 2). In the non-GTed allele population, 85.5% carried predominantly small deletions (Figure 2G-H). INDELs ≥ 100 bp were detected at low frequencies relative to the total INDELs (Figure 2I; supplemental Figure 4D). These larger deletions would convert a hypomorphic allele into a null allele with no functional consequence on the null genotypes. These results and the absence of karyotype abnormalities (supplemental Figure 5A-B) suggest a high degree of specificity for the lead RAG2 guide (sgRNA-3) when complexed with HiFi S.p.Cas924 (RNP).
Description of the OT sites above limit of detection (<0.1)
 |
 |
Mismatched bases from RAG2 guide sequence are shown in red and the PAM sequences are shown in bold. When normalized to mock, all % INDELs are < 0.1 using high fidelity (HiFi) Cas9 protein. Distance from transcription start site is shown in base pairs. ncRNA: non-coding RNA.
Description of on-target sites above limit of detection (>0.2)
 |
 |
coRAG2 treated cells were characterized for possible unintended genome-editing induced mutations at the site of genomic integration. Allele frequencies were normalized∗ to remove background signal (>0.2%), using a normalization factor of 0.96. Color coded bars match the legend in Figure 2F. To prevent re-cutting, the PAM sequence was changed from C∗C∗N to GGN. SNP: single nucleotide polymorphism; PAM: protospacer adjacent motif.
Cumulatively, we subjected 63 mice to transplantation with 28 million gene-modified HSPCs (3.5 million modified cells derived from healthy donors and 24.5 million modified cells derived from patients with RAG2-SCID) and analyzed them from 18 to 22 weeks after transplantation. No hematopoietic skewing (supplemental Figures 6 and 7) or detectable gross tumors were observed. In summary, these data offer strong genetic and functional evidence for the safety of our gene-correction strategy for RAG2 deficiency.
Human hematopoietic system is derived from coRAG2-HSPCs
To determine if gene-modified HSPCs can support long-term normal hematopoiesis, we performed xenotransplantation studies of immunodeficient NSG and NSG-SGM3 mice (Figure 3A). We first tested the hematopoietic reconstitution of coRAG2-HSPCs in fresh cord blood (CB)-HSPCs using intrahepatic transplantation (Tx) into NSG neonates because this system has been shown to best support T-cell lymphopoiesis.42 Primary human engraftment was measured 18 weeks posttransplant by fluorescence-activated cell sorting (FACS) analysis for human chimerism (% hCD45+ HLA A-B-C+ cells) in the BM (Figure 3B, left panel; supplemental Figure 6A) and spleen (Figure 3B, right panel; supplemental Figure 6B) as follows: unmodified 28.6% (range, 10.3%-39.4%), RNP 15.4% (range, 4.4%-41.8%), and 15.8% (range, 7.0%-40.3%). We observed no difference in the repopulation capacity of HSPCs that underwent HR-GTed or GE-NHEJ compared with cells that did not (mock or wild-type). Within the hCD45+HLA+ cells, the mean frequency of cells expressing coRAG2 purified from the BM was 19.5 ± 6.5 (SEM; n = 10) and from the spleen was 20.7 ± 5.3 (SEM; n = 11), as compared with 21.0% bulk targeted alleles before Tx; Figure 3D –Tx vs +Tx). coRAG2 cDNA was also detected in sorted lymphocytes (CD19+ B cells and CD3+ T cells) from BM (Figure 3D, yellow squares and triangles) and spleen (Figure 3D, red squares and triangles). The quantification only showed a proliferative advantage in T cells sorted from the spleen [53.5 ± 6.1 (SEM; n = 4)], with the remaining sorted cells expressing the transgene at levels comparable to bulk (not sorted) human hCD45+HLA+ cells: 16.6 (n = 1; sorted CD19+ from BM); 27.9 ± 9.9 (SEM; n = 3 sorted CD19+ from spleen); 53.6 ± 6.2 (SEM; n = 4; sorted CD3+ from BM); 34.4 ± 11.5 (SEM; n = 5; sorted CD3+ from the spleen; Figure 3D). The enrichment of HR-GTed cells in T cells is indirect evidence of the developmental selective advantage cells containing the cDNA knock-in have over cells with biallelic knockouts of RAG2.
In vivo lymphoid lineage development from coRAG2-targeted healthy donor-derived HSPCs. (A) Schematic of engraftment protocol of human HSPCs into NSG immunodeficient mice and secondary analysis. (B) Percent human cell chimerism (CD45+ HLA A-B-C+ double-positive cells) in bone marrow (BM) and spleen (SP) of mice 22 weeks posttransplant with CB (intra-hepatic [IH] transplant [Tx]) or (C) PB-derived HSPCs (intra-femoral [IF] transplant [Tx]) edited with RAG2-sgRNA guide 3 (RNP, light grey circles in panel B) or targeted with coRAG2 cassette (GTed, yellow and red circles in panel B, dark grey and red circles in panel C). Each dot represents an individual mouse; panel B: WT (n = 10), RNP (n = 5), GTed (n = 11); panel C: WT (n = 5), GTed-i53 (n = 5), GTed+i53 (n = 6). Bars, median (D) percent coRAG2-GTed by ddPCR before (-Tx) and after (+Tx) transplant of CB and (E) PB-derived CD34+ HSPCs into NSG mice and FACS sort from BM, as marked. Bars, mean ± s.e.m.; -i53, without p53 inhibitor; +i53, with p53 inhibitor; stats. One-way ANOVA, nonparametric test, Kruskal-Wallis test, Dunn’s multiple comparisons test.
In vivo lymphoid lineage development from coRAG2-targeted healthy donor-derived HSPCs. (A) Schematic of engraftment protocol of human HSPCs into NSG immunodeficient mice and secondary analysis. (B) Percent human cell chimerism (CD45+ HLA A-B-C+ double-positive cells) in bone marrow (BM) and spleen (SP) of mice 22 weeks posttransplant with CB (intra-hepatic [IH] transplant [Tx]) or (C) PB-derived HSPCs (intra-femoral [IF] transplant [Tx]) edited with RAG2-sgRNA guide 3 (RNP, light grey circles in panel B) or targeted with coRAG2 cassette (GTed, yellow and red circles in panel B, dark grey and red circles in panel C). Each dot represents an individual mouse; panel B: WT (n = 10), RNP (n = 5), GTed (n = 11); panel C: WT (n = 5), GTed-i53 (n = 5), GTed+i53 (n = 6). Bars, median (D) percent coRAG2-GTed by ddPCR before (-Tx) and after (+Tx) transplant of CB and (E) PB-derived CD34+ HSPCs into NSG mice and FACS sort from BM, as marked. Bars, mean ± s.e.m.; -i53, without p53 inhibitor; +i53, with p53 inhibitor; stats. One-way ANOVA, nonparametric test, Kruskal-Wallis test, Dunn’s multiple comparisons test.
To assess the efficacy of the gene-correction strategy in a cell source of similar derivation as that of patients with RAG2-deficiency, we repeated xenotransplantation studies using frozen PB-HSPCs derived from healthy donors. An equal number of unmodified (wild-type and mock) and HR-GTed PB-HSPCs, treated (+i53) or untreated (–i53), were Tx intrafemoral into sublethally irradiated 8-week-old NSG-SGM3 mice (Figure 3A,C; supplemental Figure 7). Twenty-two weeks posttransplant, the median frequencies of hCD45+HLA+ cells in BM were as follows: unmodified, 6.8% (range, 4.0%-14.2%); HR-GTed (–i53) 1.12% (range: 0.2%-25.0%); and HR-GTed (+i53) 5.9% (range: 3.6%-11.4%). We observed no statistical difference in the repopulation capacity of cells that underwent HR-GTed compared with cells that did not, nor among HR-GTed cells treated or not with i53 in the BM (Figure 3C left panel) or spleen (Figure 3C, right panel). GTed-coRAG2 cells were detected among sorted mature B cells: CD19+ (–i53: 11.5 ± 4.1 [SEM; n = 4]; +i53: 18.8 ± 9.6 [SEM; n = 5]) and CD20+IgM+ (–i53: 10.9 ± 0.4 [SEM; n = 2]; +i53: 9.9 ± 5.2 [SEM; n = 4]), providing further evidence that coRAG2 cDNA enables productive V(D)J recombination (Figure 3E).
To assess the HR-GTed cells’ ability to differentiate into multiple hematopoietic lineages, we used human CD3, CD19, CD14, and CD235a markers to identify lymphoid (B and T), myeloid (monocytes), and erythroid populations, respectively. Compared with the unmodified (mock) cells, GTed-coRAG2–derived cells showed no skewing toward a particular lineage in the BM (supplemental Figure 6C, left panel) and in circulating cells in the PB (supplemental Figure 6C, right panel) or spleen (supplemental Figure 6D). These data demonstrated that the coRAG2 transgene can support durable in vivo hematopoiesis and drive lymphopoiesis without lineage skewing.
In vivo B-cell lineage reconstitution from gene-corrected, patient-derived RAG2null HSPCs
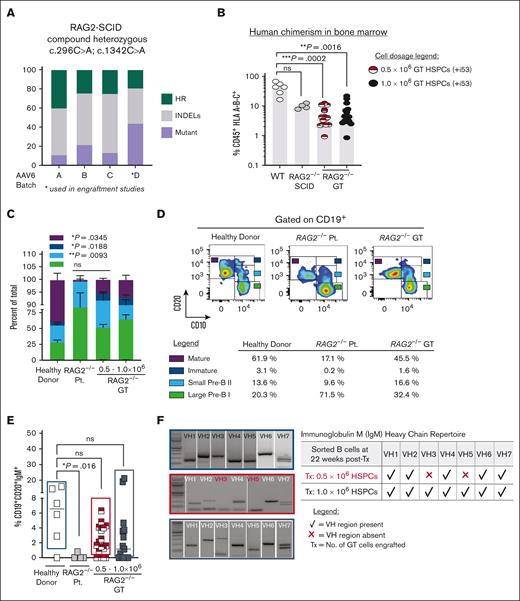
We performed xenotransplantation experiments in NSG-SGM3 immunodeficient mice to assess phenotypic correction of B-cell developmental defect in RAG2null patient-derived HSPCs (RAG2null HSPCs). Four different AAV6 virus batches carrying the coRAG2 transgene (AAV6coRAG2) were purified and tested (5000 MOI) on frozen RAG2null HSPCs (Figure 4A). Total HR-GTed (green bars) or GE-NHEJ (gray bars) generated by 3 different virus batches in the absence of i53 treatment showed a mean percent INDEL of 54.9 ± 3.7 (SEM), mean percent HR of 29.9 ± 5.1 (SEM), respectively, with 15.2% alleles (± 5.6 SEM) remaining unmodified. These viruses, however, decreased cellular viability and diminished colony-forming units and long-term engraftment potential of gene-corrected RAG2null HSPCs. We purified a fourth AAV6coRAG2 batch to transduce patient cells at a lower MOI (2500) in the presence of a p53 inhibitor (+i53). Under these optimized conditions, we achieved 19.2% HR-GTed and 37.0% GE-NHEJ with no observed toxicity.
Ex vivo gene-targeted RAG2null HSPCs correct in vivo B cell developmental block. (A) Percent of total genome editing (INDELs and HR) in RAG2null patient-derived HSPCs, using 4 different AAV6 production lots. AAV6 lot D (asterisk marked) was used for all subsequent engraftment studies. (B) Percent human cells (CD45+ HLA A-B-C+) engrafted in BM after transplanting (22 weeks post-Tx) 0.5 x106 uncorrected RAG2null patient-derived HSPCs (n=4 mice, grey circles), 0.5 x106 coRAG2-GT HSPCs (n=15 mice, half red circles) or 1.0 x106 coRAG2-GT HSPCs (n=15 mice, black circles). Healthy donor (HD) HSPCs were used as control (n=6, white circles). RAG2null patient genotype: c.296C>A; c.1342C>A. (C) FACS-based quantification is shown in (D) of large Pre-B I, small Pre-B II, immature, and mature B cells derived from coRAG2-GT HSPCs. Each population is graphed as a percent of total B cells. Bars: mean ± s.e.m. (D) Representative FACS plots of B-cell developmental stages derived from a healthy donor (left panel), RAG2null patient (middle panel), and coRAG2-GT RAG2null HSPCs. FACS-based quantification of percent cells in each developmental stage is shown. (E) FACS-based quantification of CD19+CD20+IgM+ triple-positive B cells derived from each condition tested. (F) PCR-based sequencing of immunoglobulin M (IgM) heavy chain (Vh) families from sorted triple-positive B-cells.
Ex vivo gene-targeted RAG2null HSPCs correct in vivo B cell developmental block. (A) Percent of total genome editing (INDELs and HR) in RAG2null patient-derived HSPCs, using 4 different AAV6 production lots. AAV6 lot D (asterisk marked) was used for all subsequent engraftment studies. (B) Percent human cells (CD45+ HLA A-B-C+) engrafted in BM after transplanting (22 weeks post-Tx) 0.5 x106 uncorrected RAG2null patient-derived HSPCs (n=4 mice, grey circles), 0.5 x106 coRAG2-GT HSPCs (n=15 mice, half red circles) or 1.0 x106 coRAG2-GT HSPCs (n=15 mice, black circles). Healthy donor (HD) HSPCs were used as control (n=6, white circles). RAG2null patient genotype: c.296C>A; c.1342C>A. (C) FACS-based quantification is shown in (D) of large Pre-B I, small Pre-B II, immature, and mature B cells derived from coRAG2-GT HSPCs. Each population is graphed as a percent of total B cells. Bars: mean ± s.e.m. (D) Representative FACS plots of B-cell developmental stages derived from a healthy donor (left panel), RAG2null patient (middle panel), and coRAG2-GT RAG2null HSPCs. FACS-based quantification of percent cells in each developmental stage is shown. (E) FACS-based quantification of CD19+CD20+IgM+ triple-positive B cells derived from each condition tested. (F) PCR-based sequencing of immunoglobulin M (IgM) heavy chain (Vh) families from sorted triple-positive B-cells.
Two different cell doses (0.5 × 106 and 1.0 × 106) of HR-GTed RAG2null HSPCs were Tx intrafemoral into NSG-SGM3 mice and unmodified controls (mutant and wild-type cells). Human chimerism (hCD45+HLA+) was quantified at 22 weeks after transplantation in the BM and showed the following results: 4.4% (range, 0.9-21.7) in mice Tx with 1.0 × 106 HR-GTed; 4.1% (range, 0.9-12.3) in mice Tx with 0.5 × 106 HR-GTed; 9.6% (range, 7.78-12.5) in mice Tx with 1.0 × 106RAG2null HSPCs; and 43.5% (range, 16.4-67.3) in mice Tx with 1.0 × 106 healthy donor HSPCs (Figure 4B). A twofold decrease in human chimerism was observed between the healthy donor and HR-GTed RAG2null HSPCs, regardless of the cell dose transplanted. We observed no difference in the level of engraftment between the healthy donor and unmodified RAG2null HSPCs (Figure 4B, white and gray circles), suggesting an increased sensitivity of RAG2null HSPCs to the gene-correction reagents. Transplanting NSG-SGM3 mice with a lower dose of HR-GTed RAG2null HSPCs generated a statistically significant increase in the fractions of small pre–B II (light blue bar) cells, immature (white bar), and mature (burgundy bar) cells when compared with uncorrected mutant cells (Figure 4C-D). Triple-positive CD19+CD20+IgM+ mature B cells derived from HR-GTed RAG2null HSPCs (Figure 4E, red/white, and black squares) developed at comparable levels as healthy donor-engrafted cells (Figure 4E, white squares). When sorted (supplemental Figure 9) and analyzed for IgM Vh repertoire, the HR-GTed RAG2null HSPCs engrafted with a higher cell dose (1.0 × 106) showed expression of all 7 Ig Vh variable gene families (Figure 4F, black bottom panel). The lower cell dose (0.5 × 106) expressed 5 out of 7 Vh variable regions (Figure 4F, middle red panel). Together, these data show that up to 84.8% of RAG2null alleles are gene-modified, of which up to 29.9% are corrected and up to 54.9% are disrupted. HR-GTed RAG2null HSPCs can restore B-cell development blocks from patient-derived cells.
Transplantation of RAG2null gene-corrected HSPCs rescues T-cell development in NSG-SGM3 mice
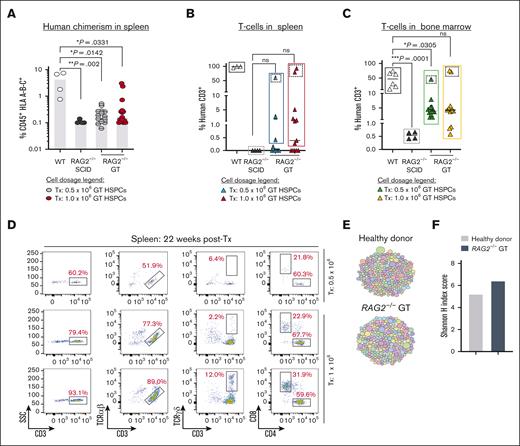
We previously demonstrated in vitro restoration of lymphopoiesis following HR-GTed of RAG2-SCID patient-derived induced pluripotent stem cells carrying a homozygous null mutation (c.831T>A).33 Here, we assessed the in vivo potential of HR-GTed to re-establish lymphopoiesis using HSPCs derived from a patient with RAG2-SCID with compound heterozygous null mutations (c.296C>A; c.1324C>A). Corrected RAG2null HSPCs Tx into NSG-SGM3 mice gave rise to CD3+ T cells at levels comparable to those of healthy donor–derived T cells (in 3 mice; Figure 5A-C). The derived CD3+ T cells detected in the spleen of 3 mice express αβ TCR and γδ TCR and gave rise to single-positive CD4+ and CD8+ T cells (Figure 5D), demonstrating that CLPs derived from corrected RAG2null HSPCs undergo normal lymphopoiesis and achieve maturation.
Correction of RAG2 gene function in RAG2null HSPCs restores V(D)J activity and normal T cells development. (A) Percent human cells (CD45+ HLA A-B-C+) detected in spleen (SP) (22 weeks post-Tx) with coRAG2-GTed RAG2null HSPC (0.5 x 106, grey circles or 1.0 x 106, red circles). HD (white circles) and uncorrected RAG2-/- (black circles) HSPCs-derived human cells were engrafted and analyzed in parallel. (B) Human CD3+ T-cells detected in the spleen (SP) and (C) bone marrow (BM) derived from coRAG2-GTed RAG2null HSPCs. (D) FACS plots showing T-cells analysis derived from 3 mice with the highest level of human CD3+ cells (dotted squares). Functional V(D)J rearrangement is demonstrated by the presence of CD3+TCR α/β, CD3+TCR γ/δ, and single-positive CD4+ and CD8+ derived from coRAG2-GTed RAG2null HSPCs. (E) Treemap diversity analysis for TCRA/TCRD CDR3 sequences from sorted CD3+ cells from (C). Each circle is a unique CDR3 sequence, and the size of the circle represents the frequency out of the total number of reads. (F) Shannon H index score quantification of CDR3 sequence from (E-F), showing oligoclonal repertoire. Shannon index score of ≥8 indicates polyclonal repertoire. Stats. One-way ANOVA, nonparametric, Kruskal-Wallis test. Median plotted in A-C.
Correction of RAG2 gene function in RAG2null HSPCs restores V(D)J activity and normal T cells development. (A) Percent human cells (CD45+ HLA A-B-C+) detected in spleen (SP) (22 weeks post-Tx) with coRAG2-GTed RAG2null HSPC (0.5 x 106, grey circles or 1.0 x 106, red circles). HD (white circles) and uncorrected RAG2-/- (black circles) HSPCs-derived human cells were engrafted and analyzed in parallel. (B) Human CD3+ T-cells detected in the spleen (SP) and (C) bone marrow (BM) derived from coRAG2-GTed RAG2null HSPCs. (D) FACS plots showing T-cells analysis derived from 3 mice with the highest level of human CD3+ cells (dotted squares). Functional V(D)J rearrangement is demonstrated by the presence of CD3+TCR α/β, CD3+TCR γ/δ, and single-positive CD4+ and CD8+ derived from coRAG2-GTed RAG2null HSPCs. (E) Treemap diversity analysis for TCRA/TCRD CDR3 sequences from sorted CD3+ cells from (C). Each circle is a unique CDR3 sequence, and the size of the circle represents the frequency out of the total number of reads. (F) Shannon H index score quantification of CDR3 sequence from (E-F), showing oligoclonal repertoire. Shannon index score of ≥8 indicates polyclonal repertoire. Stats. One-way ANOVA, nonparametric, Kruskal-Wallis test. Median plotted in A-C.
We used HTS to quantify the diversity and composition of the TCR repertoire and determine the clonality at the complementary-determining region 3 (CDR3) level. CDR3 is the region that promotes TCR-peptide major histocompatibility complex formation. Using a treemap profile analysis of the CDR3 of sorted HR-GTed RAG2null–derived CD3+ T cells and healthy donor control cells, we determined a similar pattern of TCR CDR3 specificity, with Shannon H entropy index of 6.4 and 5.2, respectively (Figure 5E-F). Virtual spectratyping analysis revealed similar CDR3 lengths of productive rearrangements in bulk T cells from HR-GTed RAG2null and healthy donor control (supplemental Figure 10).
It has been reported that mature T cells derived from patients with RAG2 carrying hypomorphic mutations have an abnormal TRA repertoire composition defined by markedly reduced usage of most of the 5′ TRAV and 3′ TRAJ genes.43 HTS of TCR rearrangements at the TCRα/TCRδ (TRA/TRD) loci were found. HR-GTed showed a similar rearrangement pattern of TRAV and TRAJ gene usage in RAG2null–derived and healthy donor–derived CD3+ T cells (supplemental Figure 11A-B). Analysis of TRB rearrangements showed comparable patterns between HR-GTed RAG2null–derived CD3+ T cells and healthy donor control CD3+ T cells (supplemental Figure 11C-D). Together, these data demonstrate restoration of the RAG complex after genomic integration of coRAG2, promoting T-cell development with normal VJ pairing in gene-edited cells, supporting previously published in vitro results.33
Correction of the immature NK CD56bright phenotype for patients with RAG2null
It has been reported that Rag-deficient mice display an increased proportion of NK cells with an immature phenotype, heightened cytotoxic activity, and limited survival potential.44 These observations have also been reported for patients with SCID caused by defects in RAG and NHEJ genes.45 The FACS analysis of human chimerism in mouse BM confirmed that NK cells derived from RAG2null HSPCs expressed significantly higher CD56brightCD16– cell surface markers than those from healthy donors (supplemental Figure 12A). After gene correction, the CD56brightCD16– population was reduced, although not to the same levels as those observed in the healthy donors (supplemental Figure 12B). These results show that gene-corrected HSPCs rescue both adaptive and innate immune defects.
Discussion
We describe a proof-of-concept study of a novel therapy for RAG2 deficiencies. Through a broad and diverse set of preclinical studies performed in vitro and in vivo in immunodeficient mice, using healthy donor and HSPCs derived from patients with RAG2nul, we have demonstrated the application of the CRISPR/Cas9-AAV6 platform in correcting HSPCs of patients with RAG2null to restore V(D)J activity in support of lymphopoiesis. For primary immunodeficiencies, such as RAG2-SCID, in which a strict spatiotemporal level of gene expression and regulation is required for normal immune development and function,3,46 a gene-correction approach has several safety and therapeutic benefits when compared with the semirandom gene addition strategy: (1) it restricts the gene activity to the lymphoid lineage and the G0-G1 cell-cycle phase [safety]; (2) it circumvents the risk of genotoxicity from ubiquitous RAG activity [safety]; (3) it allows for physiological gene expression [efficacy]; and (4) it is easily adaptable to correct other forms of RAG2 deficiencies, such as OS, AS, and CID-G/AI, for which disrupting and correcting the mutant alleles both alleviates disease burden and achieves cure. In this regard, although in this manuscript we provided pre-clinical evidence for the correction of only 1 form of RAG deficiency (ie, SCID), developing 1 gene correction platform (with CRISPR/Cas9-AAV6 inserting a full-length coRAG2 cDNA at the endogenous locus) holds the highest flexibility to achieve the full therapeutic scope for the entire spectrum of RAG2-deficiency phenotypes.
Although a phase 1 to 2 LV gene therapy clinical trial is underway for infants with RAG1-SCID47 (closely but genetically distinct from RAG2-SCID), and the safety and efficacy of delivering the RAG1 nuclease in HSPCs are being tested (NCT04797260); no alternative therapies, other than allo-hematopoietic stem cell transplantation (HSCT), are currently available for patients with RAG2 deficiencies. To our knowledge, we report the first feasibility study of gene correction at the RAG2 locus in human HSPCs derived from healthy donors and patients with RAG2null to assess the safety and efficacy of this strategy. Allo-HSCT for patients with RAG1/2-deficiency shows clinical improvement if a minimum of 20% donor chimerism is achieved,12 an allele correction threshold we have achieved and surpassed. We demonstrated that coRAG2-HSPCs can support durable hematopoiesis (18-20 weeks in immunodeficient mice), with only a twofold decrease in the engraftment potential of corrected cells of patients with RAG2null. A heightened sensitivity to the non–good manufacturing process grade purified virus could explain this decrease, an observation confirmed by the colony-forming unit assay, or a reduced regenerative potential of patients' cells. In support of the latter, we show that the unmodified RAG2null HSPCs have a 4.5-fold lower engraftment capacity than healthy donor HSPCs controls. Alternatively, in the absence of RAG2 expression, patient cells with lower DNA repair capacity are not selected against, resulting in an overall decrease in the fitness of hematopoietic progenitor cells, necessary for supporting HSCs engraftment after conditioning.45
We demonstrated in vivo rescue of peripheral T- and B-cell compartments, as demonstrated in the spleen and BM of mice transplanted with HR-GTed RAG2null HSPCs. These data are consistent with the notion that the correction of RAG2 deficiency is expected to provide a selective advantage during T-cell development, allowing the generation of spleen CD4+ and CD8+ thymocytes. Correction of the peripheral T cells was especially evident when a higher cell dose was transplanted into mice. It is possible that with lower dose cells, fewer corrected early thymic progenitors survive the mouse thymic selection in support of lymphopoiesis. Delivering a higher dose of corrected cells could compensate for this limitation. We show that the early thymic progenitors that survived the thymic selection develop into mature spleen CD4+ and CD8+ T lymphocytes expressing αβ and γδ TCRs that can propagate developmental signals, supporting single-positive CD4+ and CD8+ T cells. The receptors derived from the gene-targeted condition and healthy controls had comparable rearrangement patterns at the TRA/TRD locus with similar TRAV and TRAJ gene usage. This supports previous findings, in which we demonstrated a polyclonal TCR repertoire in T cells derived in vitro from gene-corrected RAG2null-induced pluripotent stem cells.33 It is reasonable to assume that of the gene-corrected and transplanted HSPCs clones, only a limited number repopulated the mouse thymus, thus restricting the number of stem cells that can contribute to T-cell lineage development. This is supported by a recent study that used LV cellular barcoding and purified human BM- or CB-derived HSPCs transplanted into NSG mice, <10 HSC clones contributed to the repopulation of the mouse thymus microenvironment.48 Although the study further showed that even with a limited clonal contribution to the T-cell lineage, a diverse and polyclonal TCR repertoire can be generated,48 we reason that the fresh CB- or BM-derived HSPCs used in their study holds greater lymphoid potential than the frozen healthy PB– or patient-derived HSPCs used in this study.
The limited absolute numbers of genome-edited derived lymphoid cells recovered from transplanted mice reflect the limitation of the xenograft model and precluded us from performing functional assays to test T- and B-cell immune responses. Nonetheless, we demonstrated that gene editing of RAG2null HSPCs rescued the generation of T and B cells and TCR and BCR diversity.
Without myeloablative conditioning, patients with RAG deficiency show a high graft failure rate, and those who engraft often have poor lymphoid reconstitution.12 It has been speculated that genetically defective early CLPs occupy the BM and thymic niches.16 Without niche clearing, donor functional CLPs must compete with endogenous genetically defective progenitors for space to engraft and develop. Moreover, we and others have recently shown that CD34+ cells from patients with RAGnull can differentiate into CD4+CD8+ double-positive cells in vitro17,49,50 (albeit with reduced efficiency), suggesting that post-HCT competition between host and donor-derived cells might exist up to the double-positive stage. Our data suggest that RAG2 gene editing should overcome this issue because of the selective advantage during T-cell development conferred to progenitor cells with an intact V(D)J recombination machinery. Preliminary data from the RAG1 gene therapy lentiviral trial further support this notion.
Additionally, it has been reported that the RAG-patient-derived NK cells have higher perforin expression and increased degranulation potential despite their more immature (CD56bright) phenotype.18,44 These cytotoxic NK cells can attack the graft in the absence of conditioning. Following gene correction, we observed a reduction in the proportion of CD56bright NK cells associated with an increase in the proportion of more mature CD56dim CD16+ cells. We, thus, propose that autologous HCT using gene-corrected HSPCs with reduced-intensity myelosuppressive conditioning alone or in combination with antibody-based depletion, such as anti-CD45,51,52 would conceivably provide therapeutic benefits by eliminating the competitive environment in the hematopoietic niches and assuring graft survival through correction of the cytotoxic function of the derived NK cells.
Finally, using i53 treatment resulted in ∼1.5-fold higher HR-GTed without altering engraftment potential. We anticipate that a good manufacturing process-grade virus would decrease cellular toxicity and eliminate the use of i53, simplifying the path to clinical translation. Our study provides a rationale for further testing this strategy to correct immune dysregulations in patients with RAG2 deficiency to advance it to the clinic.53
Acknowledgments
The authors thank Stanford Binns Program for Cord Blood Research and the patients for providing an invaluable source of CD34+ HSPCs for research. The authors thank the Stanford fluorescence-activated cell sorting Core facility for providing technical support. The authors acknowledge the BioRender graphic software for helping with high-quality schematics. The authors thank the members of Porteus Laboratory for their valuable discussions and support.
M.P.-D. gratefully acknowledges funding support from the Primary Immune Deficiency Treatment Consortium (U54 AI 082973), part of the National Center for Advancing Translational Sciences (NCATS) Rare Diseases Clinical Research Network and an initiative of the Office of Rare Diseases Research, NCATS. The Primary Immunodeficiency Disease Treatment Consortium is funded by a collaboration between NCATS and the National Institute of Allergy and Infectious Diseases (NIAID). M.H.P. gratefully acknowledges support from the Amon Carter Foundation and Lacob Endowment. L.D.N. gratefully acknowledges the funding support from the Division of Intramural Research (National Institutes of Health/NIAID).
Products and tools supplied by IDT are for research use only and are not intended for diagnostic or therapeutic purposes (purchaser and/or user are solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations).
Authorship
Contribution: M.P.-D. and C.L.G. designed and performed most experiments; Y.N. carried out the intra-femoral engraftments with cells from patients with RAG2-SCID; Y.N. and R.M. provided thoughtful comments on the manuscript; T.K. carried out the B-cell receptor analysis; O.M.D., B.P., M.B., and F.P. carried out TCR analysis; S.V. assisted with mouse studies and provided thoughtful comments on the manuscript; H.Y.G. assisted with AAV6 virus production and testing; N.M.B., G.L.K., and C.A.V. carried out OT analysis and designed and purified the i53 protein; A.M.D. prepared and sequenced the PacBio libraries and acquired data for on-target analysis; N.F.N. is the CZ Biohub sequencing platform director and supervised sequencing and data acquisition; G.L.K. analyzed the on-target analysis; A.S. and S.T.M. obtained and purified CD34+ HSPCs from donated CB; J.C. and E.M.H.G. assisted with colony-forming unit assays; H.L.M. obtained patient cells and provided thoughtful comments on the manuscript; L.D.N. and M.H.P. supervised the work, assisted with the experimental design, and provided thoughtful feedback and manuscript discussion; and M.P.-D., L.D.N., and M.H.P. were responsible for all data.
Conflict-of-interest disclosure: M.H.P. holds equity and is on the board of directors of Graphite Bio; is on the scientific advisory board and holds equity in Allogene Therapeutics; and is a founder and holds equity in Kamau Therapeutics. R.M. is on the board of directors of Beyond Spring Inc, and the scientific advisory boards of Zenshine Pharmaceuticals, and Kodikaz Therapeutics Solutions Inc; none of these companies had any input in the design, execution, interpretation, or publication of the work in this manuscript. G.L.K., N.M.B., and C.A.V. are employees of integrated DNA technologies, which offer reagents for sale that are identical or similar to some of the compounds described in the manuscript. The remaining authors declare no competing financial interests.
Correspondence: Matthew H. Porteus, Pediatrics, Stanford University, Lokey Stem Cell Research Bldg MC 5462, 1291 Welch Rd, Stanford, CA 94305; email: mporteus@stanford.edu; and Luigi D. Notarangelo, Immune Deficiency Genetics Section, Laboratory of Clinical Immunology and Microbiology, DIR, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 10 Center Dr, Bldg 10 CRC, Room 5-3950, Bethesda, MD 20892-1456; email: luigi.notarangelo2@nih.gov.
References
Author notes
C.L.G. and M.P.-D. contributed equally to this work.
Off-target sequencing data have been deposited in BioProject (ID PRJNA817273; accession code SAMN26753110) at https://www.ncbi.nlm.nih.gov/biosample/26753110.
The data supporting this study are available within the article and its supplemental Files or upon reasonable request from the corresponding authors, Matthew H. Porteus (mporteus@stanford.edu) and Luigi D. Notarangelo (luigi.notarangelo2@nih.gov).
The full-text version of this article contains a data supplement.


![In vivo lymphoid lineage development from coRAG2-targeted healthy donor-derived HSPCs. (A) Schematic of engraftment protocol of human HSPCs into NSG immunodeficient mice and secondary analysis. (B) Percent human cell chimerism (CD45+ HLA A-B-C+ double-positive cells) in bone marrow (BM) and spleen (SP) of mice 22 weeks posttransplant with CB (intra-hepatic [IH] transplant [Tx]) or (C) PB-derived HSPCs (intra-femoral [IF] transplant [Tx]) edited with RAG2-sgRNA guide 3 (RNP, light grey circles in panel B) or targeted with coRAG2 cassette (GTed, yellow and red circles in panel B, dark grey and red circles in panel C). Each dot represents an individual mouse; panel B: WT (n = 10), RNP (n = 5), GTed (n = 11); panel C: WT (n = 5), GTed-i53 (n = 5), GTed+i53 (n = 6). Bars, median (D) percent coRAG2-GTed by ddPCR before (-Tx) and after (+Tx) transplant of CB and (E) PB-derived CD34+ HSPCs into NSG mice and FACS sort from BM, as marked. Bars, mean ± s.e.m.; -i53, without p53 inhibitor; +i53, with p53 inhibitor; stats. One-way ANOVA, nonparametric test, Kruskal-Wallis test, Dunn’s multiple comparisons test.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/7/10.1182_bloodadvances.2023011766/1/m_blooda_adv-2023-011766-gr3.jpeg?Expires=1769095807&Signature=gSnbmoq4pJ7KGIuhPwDonPFjgORaZK0Of9610LdTo-rEh7Lqn34CyjplZj2ajdairg1H78lQVeYXsWRbLtV-qqFlQc7UBalKeMN3phhks5gIWJVqStTWNqfIJh3mvBKlAoWT7THujr1bjqgMIBYu3iNqxhE1KAq0ZIcBLK9nZPvkoOChfLD-rtoophdU-9N7hmKJ4RU1db95WA~AKYDsZzbu-unY~iUv-TcUMK-v4oTyO6CEJBTgYtY0nu1jz2fprQs6chjT~NMw82UjcAlrDZ0qm4HD-6DT0zc~kGPVjw-ElKKGGVdaRxfU-atDpW3yRylbYa-PTGSldc2sHf0ADQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)