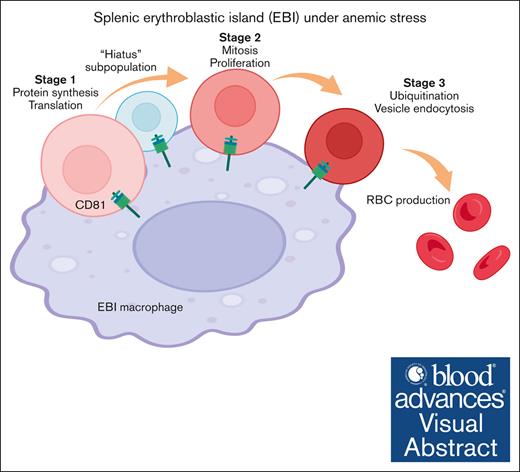
Distinct developmental phases and a “hiatus” quiescent subpopulation were identified in emerging erythroid cells under anemic stress.
CD81, identified as a novel marker for central macrophages in erythroblastic islands, is functionally required for combating anemic stress.
Visual Abstract
Stress erythropoiesis can be influenced by multiple mediators through both intrinsic and extrinsic mechanisms in early erythroid precursors. Single-cell RNA sequencing was conducted on spleen tissue isolated from mice subjected to phenylhydrazine and serial bleeding to explore novel molecular mechanisms of stress erythropoiesis. Our results showed prominent emergence of early erythroblast populations under both modes of anemic stress. Analysis of gene expression revealed distinct phases during the development of emerging erythroid cells. Interestingly, we observed the presence of a "hiatus" subpopulation characterized by relatively low level of transcriptional activities that transitions between early stages of emerging erythroid cells, with moderate protein synthesis activities. Moreover, single-cell analysis conducted on macrophage populations revealed distinct transcriptional programs in Vcam1+ macrophages under stress. Notably, a novel marker, CD81, was identified for labeling central macrophages in erythroblastic islands (EBIs), which is functionally required for EBIs to combat anemic stress. These findings offer fresh insights into the intrinsic and extrinsic pathways of early erythroblasts' response to stress, potentially informing the development of innovative therapeutic approaches for addressing anemic-related conditions.
Introduction
Steady-state erythropoiesis produces mature red blood cells (RBCs) at a constant rate, however, stressors such as hemolysis and bleeding may cause disruption of erythrocytes production, termed as stress erythropoiesis.1 Under these conditions, erythroid output is increased to compensate for the loss of their production, and alternative stress erythropoiesis pathways are activated.2
Spleen has been identified as the primary site for stress erythropoiesis in mice.3 Early studies on how the body responds to acute haemolytic anemia caused by the administration of phenylhydrazine (PHZ) showed that in mice, the steady-state erythroid progenitors originated from bone marrow (BM) migrated to the spleen and differentiated there in response to the increased levels of erythropoietin (EPO) in the blood.4 In adult mice, bone morphogenetic protein 4 (BMP4) and hedgehog signaling have been shown to stimulate the expansion of erythroid progenitor cells in response to acute anemia.5 Despite this knowledge, our understanding of the intrinsic development, expansion, and differentiation of these emerging splenic erythroid populations remains limited. To address this, we conducted a detailed analysis of these erythroid precursors at single-cell resolution.
Furthermore, erythroid precursors reside within a specialized niche known as erythroblastic islands (EBIs), in which they are in close association with central macrophages. EBI macrophages transport iron and secrete cytokines to support erythroblast survival and differentiation, while also acting as phagocytes to degrade nuclei of erythroblasts during enucleation.6,7 Despite decades of research into EBIs, the molecular mechanisms of their function during anemic stress are still not fully understood, including erythroid-macrophage interactions and central macrophage functions.
To fill these important gaps in the understanding of the molecular mechanisms that are both cell-intrinsic and cell-extrinsic in the spleen's response to anemic stress, we mapped the transcriptomic landscape of erythroid precursors/erythroblasts and their EBI macrophage niches in spleen tissue isolated from mice under PHZ and serial bleeding (BLD) stress. Our analysis decoded the trajectory of these erythroid precursors under anemic stress and identified CD81 as a novel surface marker for central EBI macrophages, and more importantly, CD81 blockade impeded erythroid recovery, implying a crucial role of CD81 in overcoming anemic stress.
Collectively, our findings unravel the intricate interplay between cell-intrinsic and -extrinsic pathways in early erythroblasts’ response to stress and may open new avenues for the development of innovative therapeutic approaches.
Methods
Mice
Mice were in a C57/BL6 background and used for experiments at age 8 to 12 weeks. All mice were maintained under experimental protocols approved by institutional animal care and use committees under the National University of Singapore.
PHZ treatment and serial blood withdrawal
As described previously,8 to generate acute hemolytic anemia, mice were intraperitoneally injected with 40 ug/g per mice of PHZ (114715, Sigma-Aldrich). To generate BLD-induced anemic stress, 400 to 500 uL of peripheral blood was obtained from the submental vein of the mice for 3 consecutive days.
Single-cell library construction and sequencing
Spleen tissues were isolated simultaneously from steady state, PHZ- and BLD-treated mice. Cell viability was evaluated using trypan blue and a hemocytometer. Samples were prepared for target capture of 10 000 cells. Single cells were subjected to the Chromium Instrument (10× Genomics) to generate single-cell gel beads in emulsion, followed by complementary DNA synthesis, amplification, and library construction using the Chromium Single Cell 3′ Reagent Kit v3 (10× Genomics, CA) according to the manufacturer’s instructions. The final library was amplified and sequenced using the P5 and P7 primers in a DNA nanoball sequencing at BGI Genomics.
Statistical analysis
Statistical significance was calculated using 2-tailed unpaired t tests as indicated in the figure legends. Statistical significance is represented using the following symbols: P > .05, ns; P ≤ .05∗; P ≤ .01∗∗; P ≤ .001∗∗∗; P ≤ .0001∗∗∗∗.
Results
Dynamics of erythroid recovery from PHZ- and BLD-induced anemic stress
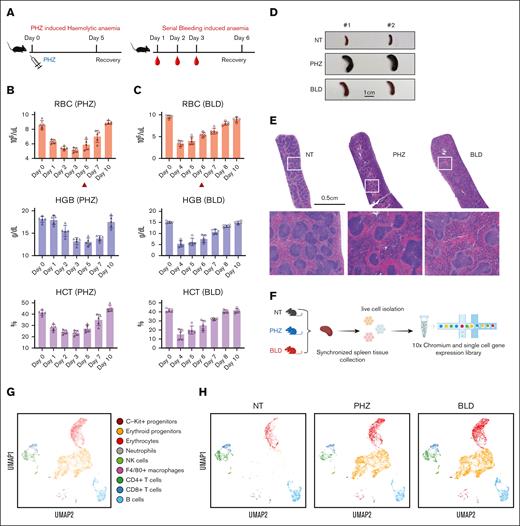
To gain a thorough understanding of the spleen's response to anemic stress, we used 2 mouse models of experimentally induced anemia: PHZ-induced hemolytic anemia and BLD-induced anemia (Figure 1A). To determine the ideal time points for synchronizing the collection of spleen tissue samples for subsequent analysis, we evaluated the dynamics of erythroid recovery under PHZ and bleeding stress conditions, respectively. In mice treatment with PHZ, peripheral blood RBC, hemoglobin, and hematocrit declined to minimum levels ∼3 days after treatment (Figure 1B). In contrast, the BLD model showed that the most severe anemia occurred 1 day after the last blood withdrawal (Figure 1C). As a result, we selected 2 days after the occurrence of the most severe anemia state as the time point for synchronizing the collection of spleen tissue samples for single-cell transcriptomic analysis in both models (ie, day 5 in the case of PHZ-induced anemia and day 6 in the case of BLD-induced anemia; Figure 1B and C).
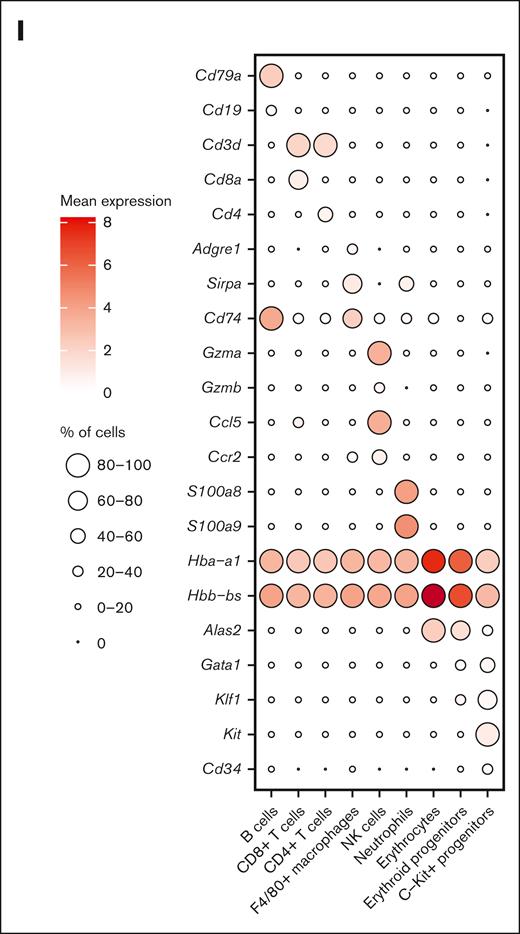
Dynamics of splenic erythroid regeneration in response to PHZ and bleeding stress. (A) Schematic illustration of the experiment to generate PHZ- and BLD-induced stress erythropoiesis models. (B-C) Peripheral blood analysis in mice treated with PHZ (B) or BLD (C) at various time points. Triangle indicates the time point chosen for subsequent analyses. (D) Comparison of spleen morphology under normal conditions, PHZ and bleeding (n = 2). (E) Histological examination (using hematoxylin and eosin staining) of spleen tissue from mice under normal, PHZ, and bleeding conditions. (F) Schematic illustration of experiment to analyze single cell transcriptomes. (G) UMAP visualization of pooled scRNA-seq data from normal, PHZ, and bleeding conditions, with annotations of 9 subpopulations. (H) UMAP visualization of 9 annotated subpopulations from normal, PHZ, and bleeding spleen tissues respectively. (I) Dot plot showing feature genes of each annotated subpopulations. HCT, hematocrit; HGB, hemoglobin; NK, natural killer; NT, no treatment.
Dynamics of splenic erythroid regeneration in response to PHZ and bleeding stress. (A) Schematic illustration of the experiment to generate PHZ- and BLD-induced stress erythropoiesis models. (B-C) Peripheral blood analysis in mice treated with PHZ (B) or BLD (C) at various time points. Triangle indicates the time point chosen for subsequent analyses. (D) Comparison of spleen morphology under normal conditions, PHZ and bleeding (n = 2). (E) Histological examination (using hematoxylin and eosin staining) of spleen tissue from mice under normal, PHZ, and bleeding conditions. (F) Schematic illustration of experiment to analyze single cell transcriptomes. (G) UMAP visualization of pooled scRNA-seq data from normal, PHZ, and bleeding conditions, with annotations of 9 subpopulations. (H) UMAP visualization of 9 annotated subpopulations from normal, PHZ, and bleeding spleen tissues respectively. (I) Dot plot showing feature genes of each annotated subpopulations. HCT, hematocrit; HGB, hemoglobin; NK, natural killer; NT, no treatment.
Under such conditions, both anemia models exhibited varying degrees of splenomegaly, as observed morphologically (Figure 1D). Additionally, both models showed a decrease in reticulocyte/RBC and an increase in early erythroid progenitor populations (gating strategy9; supplemental Figure 1A and B).
Histological staining displayed a reduction in the white pulps and a surge in red pulps in the spleen tissues of mice treated with PHZ and BLD compared with steady state, although the distribution of these changes differed between the 2 models (Figure 1E). With the aim of gaining a deeper understanding of these differences and uncover the underlying molecular mechanisms, we conducted transcriptome analysis of spleen tissue samples subjected to different stress conditions at the single-cell level.
Single-cell transcriptomic profiling of murine spleen erythroid progenitors and their microenvironment
We next conducted single-cell RNA-sequencing (scRNA-seq) on spleen tissue samples collected from mice in steady state, as well as mice subjected to PHZ-induced anemia and BLD (Figure 1F). After normalization and filtering, our data set comprised transcriptomes from a total of 15 819 cells (supplemental Figure 1C). Manifold approximation and projection (UMAP) revealed the diversity of splenic cell populations. We next used curated gene signatures from murine BM cells to annotate the UMAP clusters.10 Cells from clusters 0, 2, 6, and 17 exhibit the most prominent levels of hemoglobin subunit genes (Hba-a1 and Hbb-bs), indicating the presence of late erythroblasts or mature erythrocytes. Subsequently, cells from cluster 1, 3, 4, 5, 8, 10, 14, 16, and 25 shows relatively high expressions of hemoglobin subunit genes. In contrast, the expressions of progenitor genes (Kit, Gata1, and Klf1) are at their peaks among these clusters, indicating the presence of hematopoietic stem and progenitor cells and erythroid precursors. Moreover, it is important to note that the F4/80 (Adgre1) pan-macrophage marker is only expressed in cluster 19 and 22. These clusters also display high levels of Cd74 and Sirpa, indicating the presence of monocyte/macrophage populations (Figure 1G-I; supplemental Figure 1D). Notably, erythroid progenitors are absent in the steady state spleen tissue samples but become prominent during PHZ- and BLD-induced anemia (Figure 1H), hence, from this point on, they will be referred to as “emerging erythroid cells.”
Subpopulations in emerging erythroid cells exhibit distinct transcriptional programs
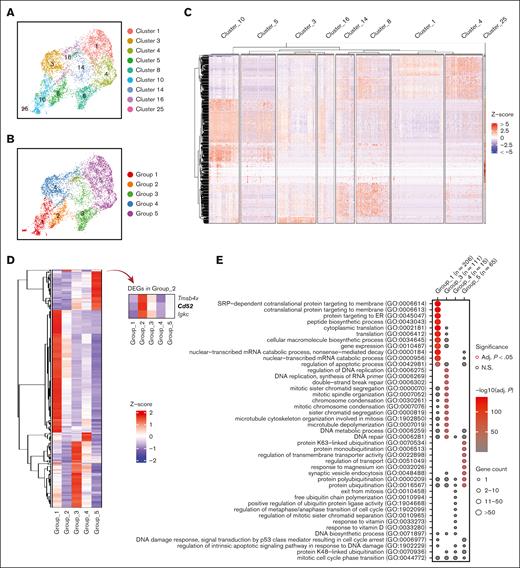
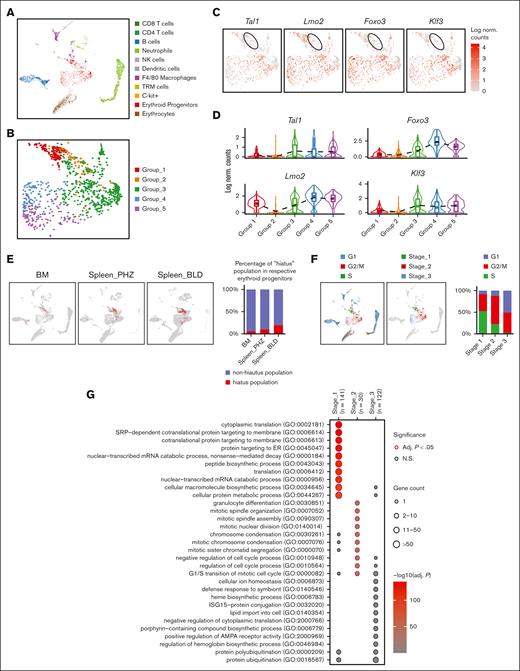
In order to understand the role of each subgroup of emerging erythroid cells during stress and to study their differentiation trajectories, we reclassified the 9 clusters into 5 groups based on their general gene expression patterns as observed from the heat map clustering (Figure 2A-C and Supplemental Figure 2A). The groups (1-5) exhibit distinct gene expression patterns (Figure 2D). Gene ontology (GO) analysis was performed via Enrichr11-13 to investigate the biological processes of the differentially expressed genes (DEGs) in each group. The analysis revealed that Group_1 has a high enrichment of genes involved in protein synthesis and translation, and Group_3 and Group_4 show high enrichment of genes associated with mitotic cell division and the cell cycle related functions, whereas Group_5 has high expression of genes responsible for protein ubiquitination and regulation of transport (Figure 2E).
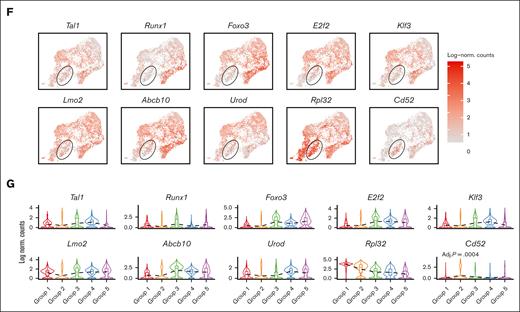
Emerging erythroid cells subpopulations display distinct transcriptional programs. (A) UMAP visualization of the 9 initial clusters of emerging erythroid cells in the pooled scRNA-seq data. (B) UMAP visualization of the 5 reclustered groups in the emerging erythroid cells in the pooled scRNA-seq data. (C) Heat map showing the expression levels of DEGs of single cells of the 9 initial clusters. (D) Heat map showing the average expression levels of DEGs of single cells from group 1 to group 5, as well as 3 individual DEGs from group 2. (E) Dot plot demonstrating the enrichment analysis (GO biological processes) of DEGs from groups_1, 3, 4, and 5, respectively. (F) UMAP visualization of the selected gene expression levels among various groups in emerging erythroid cells, with group 2 highlighted in ovals. (G) Violin plots comparing the differences in selected gene expression levels among various groups in emerging erythroid cells. Adj., adjusted.
Emerging erythroid cells subpopulations display distinct transcriptional programs. (A) UMAP visualization of the 9 initial clusters of emerging erythroid cells in the pooled scRNA-seq data. (B) UMAP visualization of the 5 reclustered groups in the emerging erythroid cells in the pooled scRNA-seq data. (C) Heat map showing the expression levels of DEGs of single cells of the 9 initial clusters. (D) Heat map showing the average expression levels of DEGs of single cells from group 1 to group 5, as well as 3 individual DEGs from group 2. (E) Dot plot demonstrating the enrichment analysis (GO biological processes) of DEGs from groups_1, 3, 4, and 5, respectively. (F) UMAP visualization of the selected gene expression levels among various groups in emerging erythroid cells, with group 2 highlighted in ovals. (G) Violin plots comparing the differences in selected gene expression levels among various groups in emerging erythroid cells. Adj., adjusted.
It is worth mentioning that Group_2 has very few uniquely expressed genes and, thus, did not show up in the GO analysis (Figure 2E). Upon closer examination, it was observed that Group_2 shares a relatively similar gene expression pattern with Group_1, albeit with much lower expression levels. Interestingly, we discovered that several crucial regulators of erythropoiesis, including Tal1, Runx1, Lmo2, Foxo3, and Klf3, are expressed at extremely low levels in Group_2, indicating a temporarily transcriptionally quiescent state (Figure 2F and 2G). The relatively low expression of genes related to cell division and growth, including E2f2, Btg2, and Rb1, implies a possible cessation of cellular proliferation (Figure 2F and 2G and supplemental Figure 2C). In contrast, Group_2 exhibited moderate expression of ribosomal subunit genes and genes involved in protein synthesis after Group_1 (Figure 2F, 2G and Supplemental Figure 2C). In addition, Group_2 exhibited moderate but distinctive expression of CD52 compared with other groups (Figure 2F and 2G). CD52 is widely recognized as a marker of immune cells, but it is also mildly expressed in early erythroid progenitors, as evidenced by Gene Expression Commons data sets14 (supplemental Figure 2B), although its function in this cell population is still unknown.
Of note, Group_2's transcriptional quiescence raises the notion of a "hiatus" concept, which refers to a temporary gap, pause, or interruption in a sequence or process. The appearance of this subgroup during stress erythropoiesis prompts the need to functionally characterize the concept of “hiatus” in subsequent sessions.
Uncovering the differentiation trajectory of emerging erythroid cells
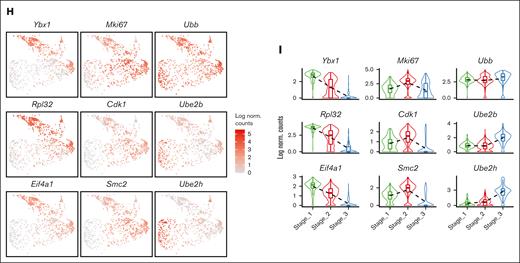
Then we investigated the differentiation trajectory of erythroid populations and identified a developmental pathway that starts with Group_1/2, progresses to Group_3/4, then to Group 5, and finally culminates in the generation of mature erythrocytes (Figure 3A-C). Given the overall similarity of gene signatures between Group_1/2, as well as Group_3/4, we reorganized the 5 groups into 3 distinct stages (Stage_1-3) (Figure 3D). GO analysis on the DEGs in each stage uncovered that these erythroblasts initiate protein synthesis during Stage_1, subsequently progress to mitotic cell division during Stage_2, and finally reach Stage_3 in which protein ubiquitination takes place (Figure 3G; supplemental Figure 4A-C). Interestingly, the distribution of cells across Stage_1 to Stage_3 is correlated with their cell cycle status. Specifically, a majority of S phase cells were found in Stage_1, whereas Stage_2 had the highest proportion of M phase cells, and G1 phase cells were predominant in Stage_3 (Figure 3E-F; supplemental Figure 2D). Hence, there is an interplay between the cells’ biological functions and their distribution across different cell cycle stages.
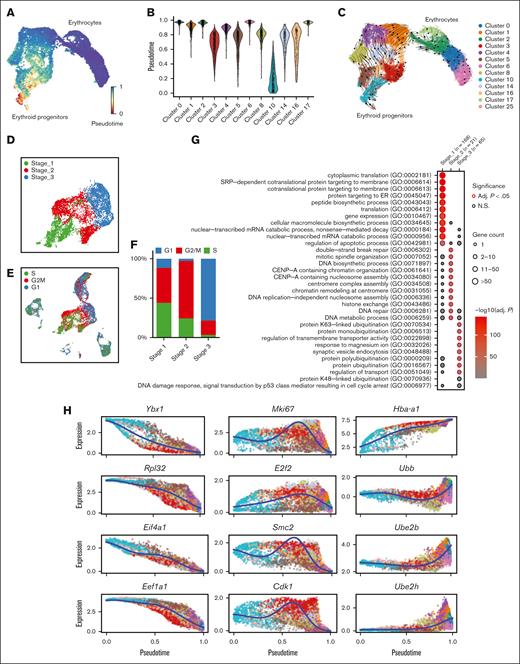
Emerging erythroid cells exhibit functionally distinct stages along the differentiation trajectory. (A) UMAP plot of erythroid cell populations colored by pseudotime. (B) Violin plot showing the distribution of pseudotime in all initial erythroid cell population clusters. (C) Pseudotime trajectory analysis demonstrating the directions of erythroid differentiation as indicated by the arrows along various erythroid populations. (D) UMAP visualization of the 3 regrouped stages in the emerging erythroid cells in the pooled scRNA-seq data. (E) UMAP visualization of the various stages of cell cycle in the pooled scRNA-seq data. (F) Bar graph showing the proportions of different cell-cycle phases (G1, S, and G2M) of pooled spleen cells from different stages. (G) Dot plot demonstrating the enrichment analysis (GO biological processes) of DEGs from stages 1, 2, and 3, respectively. (H) Expressions of selected genes in all erythroid populations along the pseudotime axis.
Emerging erythroid cells exhibit functionally distinct stages along the differentiation trajectory. (A) UMAP plot of erythroid cell populations colored by pseudotime. (B) Violin plot showing the distribution of pseudotime in all initial erythroid cell population clusters. (C) Pseudotime trajectory analysis demonstrating the directions of erythroid differentiation as indicated by the arrows along various erythroid populations. (D) UMAP visualization of the 3 regrouped stages in the emerging erythroid cells in the pooled scRNA-seq data. (E) UMAP visualization of the various stages of cell cycle in the pooled scRNA-seq data. (F) Bar graph showing the proportions of different cell-cycle phases (G1, S, and G2M) of pooled spleen cells from different stages. (G) Dot plot demonstrating the enrichment analysis (GO biological processes) of DEGs from stages 1, 2, and 3, respectively. (H) Expressions of selected genes in all erythroid populations along the pseudotime axis.
To verify this finding, we analyzed specific genes across the pseudotime axis. We observed that genes responsible for protein synthesis and translation (Ybx1, Rpl32, Eif4a1, and Eef1a1) show a gradual increase along the pseudotime trajectory, whereas genes involved in proliferation and mitotic cell division (Mki67, E2f2, Smc2, and Cdk1) have peak expressions in the middle of the axis. Additionally, we found that genes associated with ubiquitination (Ubb, Ube2h, and Ube2b) display upregulation toward the end of the trajectory axis (Figure 3H and supplemental Figure 4A-C).
These results collectively suggest that splenic emerging erythroblasts exhibit a stepwise progression pattern in preparation for erythrocyte production to combat anemic stress.
A temporary “hiatus” population resides between early stages of emerging erythroid cells
The discovery of the “hiatus” subpopulation during stress erythropoiesis highlights the importance of functionally characterizing this concept. One notable finding is that the weight percentage of different groups differs significantly between PHZ and BLD conditions, with the “hiatus” Group_2 population being more prevalent in the BLD stress condition than in the PHZ condition (Figure 4A and 4B).
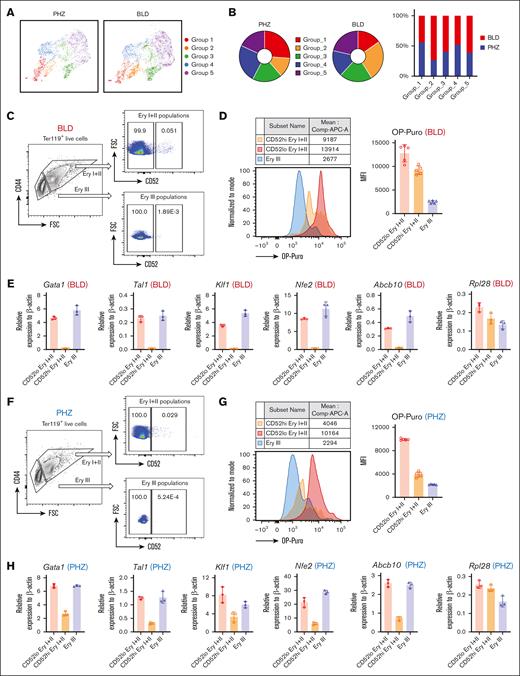
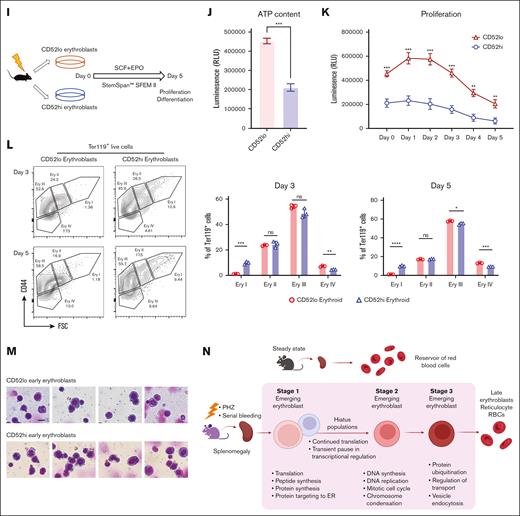
“Hiatus” subpopulation resides between early stages of emerging erythroid cells. (A) UMAP visualization of Group_1 to Group_5 in emerging erythroid cells under PHZ and bleeding conditions, respectively. (B) Pie chart (left) and bar graph (right) showing the proportions of each group in PHZ and BLD conditions, respectively. (C) Representative gating strategy of CD52lo high and CD52lo Ery I+II and Ery III fractions within splenic early erythroblasts during bleeding stress. (D) Protein synthesis rate, as determined by OP-Puro incorporation, of the indicated erythroblast populations (CD52hi and CD52lo Ery I+II and Ery III) under bleeding stress. (E) mRNA expression of Gata1, Tal1, Klf1, Nfe2, Abcb10, and Rpl28 isolated from CD52lo and CD52hi Ery I+II and Ery III populations under bleeding stress. Error bars indicate the standard error of the mean (SEM) for 3 independent experiments. (F) Representative gating strategy of CD52lo high and CD52lo Ery I+II and Ery III fractions within splenic early erythroblasts during PHZ stress. (G) Protein synthesis rate, as determined by OP-Puro incorporation, of the indicated erythroblast populations (CD52hi and CD52lo Ery I+II and Ery III) under PHZ stress. (H) mRNA expression of Gata1, Tal1, Klf1, Nfe2, Abcb10, Rpl28 isolated from CD52lo and CD52hi Ery I+II and Ery III populations under PHZ stress. (I) Schematic illustration of the experiment to isolate CD52lo and CD52hi early erythroblasts (Ery I) from mice subjected to bleeding stress and culture in vitro. (J) Adenosine triphosphate content of CD52lo and CD52hi early erythroblasts measured by CellTiter-Glo Luminescent Cell Viability Assay. (K) Proliferation of CD52lo and CD52hi early erythroblasts in the in vitro culture from day 0 to day 5. (L) Erythroid differentiation of CD52lo and CD52hi early erythroblasts in the in vitro culture at day 3 and day 5, measured by flow cytometry analysis. Representative flow cytometric plots were shown on the left. (M) Morphology of CD52lo and CD52hi early erythroblasts after cytospin. Scale bar, 10 μm. (N) Illustration of the proposed model for cell intrinsic differentiation trajectory of emerging erythroid cells during anemic stress. Under steady state, murine spleen contains a reservoir of mature erythrocytes, whereas during anemic stress, mice undergo splenomegaly, and early erythroblasts emerge in the spleen, displaying transition of various biological processes during differentiation, with a “hiatus” subpopulation residing between early stages. Illustration was created via biorender.com. For all quantification, means ± SEMs; ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001; ∗∗∗∗ P < .0001 by t test. mRNA, messenger RNA; OP-Puro, O-Propargyl-puromycin.
“Hiatus” subpopulation resides between early stages of emerging erythroid cells. (A) UMAP visualization of Group_1 to Group_5 in emerging erythroid cells under PHZ and bleeding conditions, respectively. (B) Pie chart (left) and bar graph (right) showing the proportions of each group in PHZ and BLD conditions, respectively. (C) Representative gating strategy of CD52lo high and CD52lo Ery I+II and Ery III fractions within splenic early erythroblasts during bleeding stress. (D) Protein synthesis rate, as determined by OP-Puro incorporation, of the indicated erythroblast populations (CD52hi and CD52lo Ery I+II and Ery III) under bleeding stress. (E) mRNA expression of Gata1, Tal1, Klf1, Nfe2, Abcb10, and Rpl28 isolated from CD52lo and CD52hi Ery I+II and Ery III populations under bleeding stress. Error bars indicate the standard error of the mean (SEM) for 3 independent experiments. (F) Representative gating strategy of CD52lo high and CD52lo Ery I+II and Ery III fractions within splenic early erythroblasts during PHZ stress. (G) Protein synthesis rate, as determined by OP-Puro incorporation, of the indicated erythroblast populations (CD52hi and CD52lo Ery I+II and Ery III) under PHZ stress. (H) mRNA expression of Gata1, Tal1, Klf1, Nfe2, Abcb10, Rpl28 isolated from CD52lo and CD52hi Ery I+II and Ery III populations under PHZ stress. (I) Schematic illustration of the experiment to isolate CD52lo and CD52hi early erythroblasts (Ery I) from mice subjected to bleeding stress and culture in vitro. (J) Adenosine triphosphate content of CD52lo and CD52hi early erythroblasts measured by CellTiter-Glo Luminescent Cell Viability Assay. (K) Proliferation of CD52lo and CD52hi early erythroblasts in the in vitro culture from day 0 to day 5. (L) Erythroid differentiation of CD52lo and CD52hi early erythroblasts in the in vitro culture at day 3 and day 5, measured by flow cytometry analysis. Representative flow cytometric plots were shown on the left. (M) Morphology of CD52lo and CD52hi early erythroblasts after cytospin. Scale bar, 10 μm. (N) Illustration of the proposed model for cell intrinsic differentiation trajectory of emerging erythroid cells during anemic stress. Under steady state, murine spleen contains a reservoir of mature erythrocytes, whereas during anemic stress, mice undergo splenomegaly, and early erythroblasts emerge in the spleen, displaying transition of various biological processes during differentiation, with a “hiatus” subpopulation residing between early stages. Illustration was created via biorender.com. For all quantification, means ± SEMs; ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001; ∗∗∗∗ P < .0001 by t test. mRNA, messenger RNA; OP-Puro, O-Propargyl-puromycin.
More importantly, the detection of CD52 as a marker for the “hiatus” subpopulation allows for the functional characterization of this concept. Our flow cytometry analysis indicated that CD52 is predominantly expressed in the Ery I and II population, while being almost undetectable in the Ery III population, under both PHZ and BLD conditions. Subsequently, based on CD52 expression levels, we further subdivided the Ery I+II population into CD52hi and CD52lo erythroblasts (Figure 4C and 4F). By using the OP-Puro protein synthesis assay, we determined that CD52hi cells within the Ery I+II population function as intermediates between the CD52lo Ery I+II and Ery III populations (Figure 4D and 4G). This discovery is in line with the expression patterns observed for protein synthesis/translation signatures in scRNA-seq and quantitative polymerase chain reaction validations for the “hiatus” populations (Figure 2F and 2G; Figure 4E and 4H; and supplemental Figure 2C).
To gain a comprehensive understanding of the characteristics associated with the “hiatus” subpopulation, we isolated 2 distinct groups of early erythroblasts: CD52hi and CD52lo Ery I populations. These isolated populations were subsequently cultured in vitro (Figure 4I). Notably, the CD52hi labelled early erythroblasts exhibited significantly reduced levels of adenosine triphosphate content and proliferative capacity, as assessed by the CellTiter-Glo Luminescence assay (Figure 4J and 4K). This finding suggests that the CD52hi subpopulation operates in a relatively lower energy state than their CD52lo counterparts. Furthermore, as a consequence, the CD52hi early erythroblasts displayed compromised differentiation capability (Figure 4L), although no evident morphological distinctions were observed between the 2 groups of early erythroblasts (Figure 4M). These findings strongly suggest that the “hiatus” subpopulation is characterized by a lower energy state, which in turn leads to reduced proliferative and differentiation capabilities. It is plausible that this phenomenon might be related to an adaptive strategy adopted by early erythroblasts aimed at conserving energy during emergency situations.
Collectively, our analysis revealed a step-wise functionally distinct differentiation trajectory of splenic early erythroblasts under stress, that they progress from protein synthesis to mitotic cell cycle, then ubiquitination, and eventually maturation into erythrocytes to overcome stress. Of particular importance, we have identified a “hiatus” subpopulation that undergoes a temporary pause in major transcriptional processes, while maintaining moderate protein biosynthesis activities (Figure 4N).
Single-cell transcriptomic profiling of steady-state BM corroborates splenic responses to stress
To expand on our findings regarding splenic response to anemic stress, we conducted a single cell transcriptomic analysis of the BM under steady state and performed a comprehensive analysis by integrating these data with the stressed spleen data set. (Figure 5A; supplemental Figure 3A and 3B).
Transcriptomic analysis of steady state BM single cells. (A) UMAP visualization of scRNA-seq data from steady state BM samples, with annotations of 11 subpopulations. (B) UMAP visualization of the 5 reclustered groups in the erythroid progenitor populations in steady state bone marrow scRNA-seq data. (C) UMAP visualization of the selected gene expression levels among various groups in steady state BM erythroid progenitors, with Group_2 highlighted in ovals. (D) Violin plots comparing the differences in selected gene expression levels among various groups in steady state bone marrow erythroid progenitors. (E) UMAP visualization and quantification of the distribution of “hiatus” subpopulation in steady state BM, PHZ-treated spleen and bleeding-treated spleen, respectively. (F) UMAP visualization and bar graph showing the proportions of different cell-cycle phases (G1, S, and G2M) of steady state BM cells from different stages (stages_1 to _3). (G) Dot plot demonstrating the enrichment analysis (GO biological processes) of DEGs from stages_1, 2, and 3 in steady state BM erythroid progenitors. (H) UMAP visualization of the selected gene expression levels among various stages in steady state BM erythroid progenitors. (I) Violin plots comparing the differences in selected gene expression levels among various stages in steady state BM erythroid progenitors. NK cell, natural killer cell; SRP, signal-recognition particle.
Transcriptomic analysis of steady state BM single cells. (A) UMAP visualization of scRNA-seq data from steady state BM samples, with annotations of 11 subpopulations. (B) UMAP visualization of the 5 reclustered groups in the erythroid progenitor populations in steady state bone marrow scRNA-seq data. (C) UMAP visualization of the selected gene expression levels among various groups in steady state BM erythroid progenitors, with Group_2 highlighted in ovals. (D) Violin plots comparing the differences in selected gene expression levels among various groups in steady state bone marrow erythroid progenitors. (E) UMAP visualization and quantification of the distribution of “hiatus” subpopulation in steady state BM, PHZ-treated spleen and bleeding-treated spleen, respectively. (F) UMAP visualization and bar graph showing the proportions of different cell-cycle phases (G1, S, and G2M) of steady state BM cells from different stages (stages_1 to _3). (G) Dot plot demonstrating the enrichment analysis (GO biological processes) of DEGs from stages_1, 2, and 3 in steady state BM erythroid progenitors. (H) UMAP visualization of the selected gene expression levels among various stages in steady state BM erythroid progenitors. (I) Violin plots comparing the differences in selected gene expression levels among various stages in steady state BM erythroid progenitors. NK cell, natural killer cell; SRP, signal-recognition particle.
Through this investigation, we also identified a distinct subpopulation of erythroid progenitors enriched with Cd52, which we previously termed as “hiatus” subpopulation. Cells in this subpopulation exhibit low messenger RNA expression levels of key erythropoiesis regulators (Figure 5B-D and supplemental Figure 3C). However, we observed that the proportion of the “hiatus” subpopulation in steady state BM was lower than that in spleen erythroid progenitors under stress conditions (Figure 5E). This finding suggests that the “hiatus” phenomenon may be more enriched under stress conditions in the splenic erythroblasts, possibly due to the increased demand for RBC production. It is plausible that stressed erythroblasts within the "hiatus" subpopulation adopt this strategy to conserve energy while fulfilling the surge in the requirement for protein synthesis during early stages to overcome stress erythropoiesis.
In a similar manner to the spleen data sets, we reclassified the distinct groups of BM erythroblasts into 3 stages: the transition from translation to mitotic cell division and subsequent posttranslational modifications, including protein ubiquitination, which generally aligns with findings from previously reported human bulk-transcriptomic data sets.15 Notably, we found that the distinct functions exhibited by these 3 stages are strongly correlated with the cell cycle status (Figure 5F-I). These findings suggest that erythroblasts undergo a stepwise transition of distinct functions in both steady state BM erythropoiesis and stress erythropoiesis in spleen cells, indicating a parallel pattern of functional changes during erythropoiesis across different physiological conditions.
Single-cell transcriptomic profiling of splenic macrophages
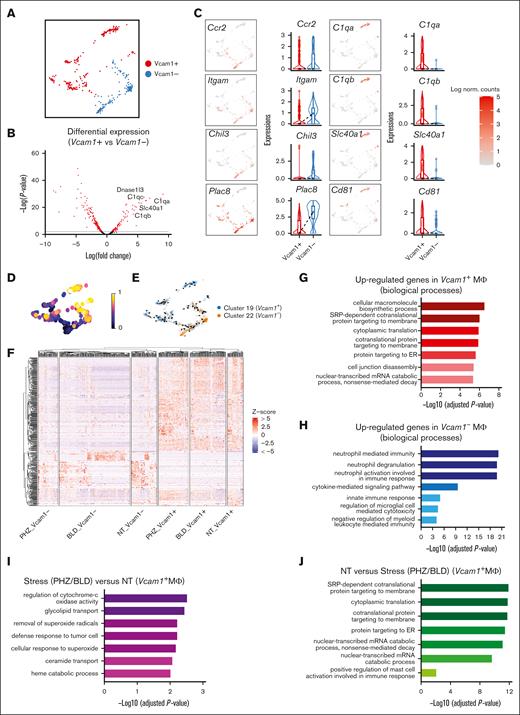
Next, we proceeded to investigate the cell-extrinsic mechanisms related to the erythroid niche, that is, EBI macrophages. To this end, we identified macrophage populations expressing F4/80 belonging to 2 subclusters (clusters 19 and 22), and Vcam1 gene is found to be significantly upregulated in Cluster 19 (supplemental Figure 1D). Because Vcam1 is a crucial marker for EBI macrophages, we designated the 2 subclusters as the Vcam1+ and Vcam1– subpopulations to facilitate further analysis (Figure 6A).
Splenic macrophage populations display distinct gene expression signatures. (A) UMAP visualization of Vcam1– and Vcam1+ macrophages in the pooled scRNA-seq data of F4/80+ macrophage populations. (B) Volcano plots showing the DEGs between Vcam1+ vs Vcam1– macrophages. (C) UMAP visualization and Violin plots comparing the differences in selected gene expression levels between Vcam1+ vs Vcam1– macrophages. (D) UMAP plot of macrophage populations coloured by pseudotime. (E) Pseudotime trajectory analysis demonstrating the directions of macrophage differentiation as indicated by the arrows. (F) Heat map showing the expression levels of DEGs of single cells in Vcam1– and Vcam1+ macrophages from NT, PHZ, and BLD conditions, respectively. Enrichment analysis (GO biological processes) of genes upregulated in Vcam1+ macrophages (G), genes upregulated in Vcam1– macrophages (H), genes upregulated in stress (PHZ/BLD) vs NT Vcam1+ macrophages (I), and genes upregulated in NT vs stress (PHZ/BLD) Vcam1+ macrophages (J). ER, endoplasmic reticulum; NT, no treatment.
Splenic macrophage populations display distinct gene expression signatures. (A) UMAP visualization of Vcam1– and Vcam1+ macrophages in the pooled scRNA-seq data of F4/80+ macrophage populations. (B) Volcano plots showing the DEGs between Vcam1+ vs Vcam1– macrophages. (C) UMAP visualization and Violin plots comparing the differences in selected gene expression levels between Vcam1+ vs Vcam1– macrophages. (D) UMAP plot of macrophage populations coloured by pseudotime. (E) Pseudotime trajectory analysis demonstrating the directions of macrophage differentiation as indicated by the arrows. (F) Heat map showing the expression levels of DEGs of single cells in Vcam1– and Vcam1+ macrophages from NT, PHZ, and BLD conditions, respectively. Enrichment analysis (GO biological processes) of genes upregulated in Vcam1+ macrophages (G), genes upregulated in Vcam1– macrophages (H), genes upregulated in stress (PHZ/BLD) vs NT Vcam1+ macrophages (I), and genes upregulated in NT vs stress (PHZ/BLD) Vcam1+ macrophages (J). ER, endoplasmic reticulum; NT, no treatment.
Notably, we observed differential expression of infiltrating macrophage markers in Vcam1– macrophages, including Ccr216 and CD11b (Itgam)17 as well as expression of genes previously identified in Ly6chi infiltrating macrophages in the kidney, such as Chil3 and Plac8.18 In contrast, genes that have been identified as resident macrophage markers 18 (eg, C1qa, C1qb, and Cd81) were found to be highly expressed on Vcam1+ macrophages in both spleen and BM (Figure 6B-C and Supplemental Figure 3D-E). Furthermore, trajectory analysis demonstrated the differentiation pathway of macrophage subpopulations, from a less mature Vcam1– state to a more developed Vcam1+ state (Figure 6D and 6E). Functionally, GO analysis of biological processes revealed that the genes upregulated in Vcam1+ macrophages are highly enriched in macromolecule/protein biosynthesis (Figure 6G), whereas genes upregulated in Vcam1– subpopulations are predominantly associated with immune response and signaling pathways (Figure 6H). This suggests that different subpopulations of macrophages in the spleen have distinct biological roles.
We next investigate the transcriptomes of Vcam1+ macrophages upon stress. Heat map shows distinct transcriptional programs in Vcam1+ and Vcam1– macrophages, under steady state, PHZ and bleeding treatment, respectively (Figure 6F and supplemental Figure 4D). GO analysis demonstrated that Vcam1+ macrophages undergoing stress (PHZ and BLD) are strongly associated with pathways involved in combating superoxide, whereas Vcam1+ macrophages in a steady state exhibit enriched biological processes related to macromolecule biosynthesis (Figure 6I and 6J), which are typically linked to the functions of resident macrophages. This suggests that Vcam1+ macrophages have a specialized role under stress conditions.
CD81 marks EBI macrophages and is required for erythroid regeneration from anemic stress
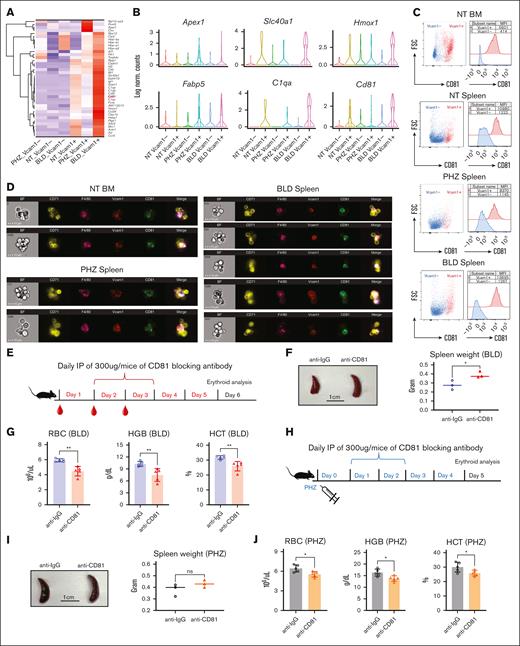
To identify novel markers and functional regulators for EBI macrophage under both steady state and stress conditions, we examined the DEGs closely. In contrast to bleeding, hemolysis caused by PHZ is a result of cellular breakdown. Therefore, the significant expression of genes encoding responses to DNA damage caused by oxidative stress, including Prmt5, Apex1, and Ctrc,19-21 under PHZ treatment (Figure 7A-B and Supplemental Figure 4E) is intriguing. These molecules may elicit distinct cellular responses compared with bleeding, as evidenced by the observed differences in spleen morphology (Figure 1E).
CD81 is expressed on EBI macrophages and functionally required to overcome anemic stress. (A) Heat map showing the average expression levels of DEGs of cells from Vcam1+ macrophages in PHZ and BLD conditions, respectively. (B) Violin plots comparing the differences in selected gene expression levels in Vcam1– and Vcam1+ macrophages from NT, PHZ, and BLD conditions, respectively. (C) Flow cytometry analysis showing the expression of CD81 in Vcam1+ and Vcam1– macrophages, respectively. (D) Coexpression of CD81 with F4/80 and Vcam1 on EBI macrophages in steady state BM, as well as spleen under PHZ and BLD conditions, as demonstrated by Amnis ImageStream. (E) Schematic illustration of experiment to validate the function of CD81 in erythroid recovery from BLD stress. CD81 blocking antibody (anti-CD81) was administered into BLD-treated mice. (F) Comparison of spleen morphology with or without CD81 blockade under BLD conditions (n = 3). (G) Peripheral blood analysis in mice subjected to BLD stress with and without CD81 blockade (n = 5). (H) Schematic illustration of experiment to validate the function of CD81 in erythroid recovery from PHZ stress. CD81 blocking antibody (anti-CD81) was administered into PHZ-treated mice. (I) Comparison of spleen morphology with or without CD81 blockade under PHZ conditions (n = 3). (J) Peripheral blood analysis in mice subjected to PHZ stress with and without CD81 blockade (n = 5). For all quantification, means ± SEMs; ∗ P < .05; ∗∗ P < .01 by t test. HCT, hematocrit; HGB, hemoglobin; NT, no treatment.
CD81 is expressed on EBI macrophages and functionally required to overcome anemic stress. (A) Heat map showing the average expression levels of DEGs of cells from Vcam1+ macrophages in PHZ and BLD conditions, respectively. (B) Violin plots comparing the differences in selected gene expression levels in Vcam1– and Vcam1+ macrophages from NT, PHZ, and BLD conditions, respectively. (C) Flow cytometry analysis showing the expression of CD81 in Vcam1+ and Vcam1– macrophages, respectively. (D) Coexpression of CD81 with F4/80 and Vcam1 on EBI macrophages in steady state BM, as well as spleen under PHZ and BLD conditions, as demonstrated by Amnis ImageStream. (E) Schematic illustration of experiment to validate the function of CD81 in erythroid recovery from BLD stress. CD81 blocking antibody (anti-CD81) was administered into BLD-treated mice. (F) Comparison of spleen morphology with or without CD81 blockade under BLD conditions (n = 3). (G) Peripheral blood analysis in mice subjected to BLD stress with and without CD81 blockade (n = 5). (H) Schematic illustration of experiment to validate the function of CD81 in erythroid recovery from PHZ stress. CD81 blocking antibody (anti-CD81) was administered into PHZ-treated mice. (I) Comparison of spleen morphology with or without CD81 blockade under PHZ conditions (n = 3). (J) Peripheral blood analysis in mice subjected to PHZ stress with and without CD81 blockade (n = 5). For all quantification, means ± SEMs; ∗ P < .05; ∗∗ P < .01 by t test. HCT, hematocrit; HGB, hemoglobin; NT, no treatment.
Moreover, genes upregulated in Vcam1+ macrophages during bleeding stress are involved in multiple biological processes related to response to various stimuli or oxidative stress. Notably, these genes are enriched in functions such as response to oxidative stress (Fth1, Hmox1, Aif1, Sdc3, Ptgs1, Actn1, Axl, and Ccr3), regulation of inflammatory response (Cd5l, Clec1b, C1qa, C1qb, C1qc, and Ccl24), response to oxygen levels (Hbb-bs, Hbb-bt, Hba-a1, Hba-a2, and Epb41l3), as well as response to cytokine stimulus (Hmox1, Ckb, Vcam1, and Cd81) (Figure 7A-B and supplemental Figure 4E). These finding suggests that EBI macrophages may have an important role in combating the stimuli induced by inflammation and oxidative stress in response to stressors.
Notably, we observed prominent CD81 expression on Vcam1+ macrophages before stress in both spleen and BM cells, and its expression is significantly increased under bleeding conditions (Figure 7A-B; supplemental Figure 3D-E). CD81, also known as TAPA-1, belongs to tetraspanin superfamily and is expressed on numerous immune cell types. Importantly, evidence from previous studies suggests that CD81 is involved in various cellular processes, including cell signaling and cell-cell contact.22 This suggests that CD81 may serve as a promising surface marker for identifying EBI macrophages.
To verify our hypothesis, we performed flow cytometry analysis on F4/80+ macrophages isolated from BM and spleen under various conditions, and our results showed that CD81 expression is strongly associated with Vcam1 expression under steady state conditions in both BM and spleen, as well as in spleen under PHZ and BLD treatment (Figure 7C). The coexpression of CD81 on F4/80+Vcam1+ macrophages was confirmed by multispectral imaging flow cytometry (supplemental Figure 5A). Next, to confirm the presence of CD81 on EBI macrophages, we performed imaging flow cytometry that enables the characterization the structural and morphological details of the EBI macrophages and associated erythroblasts. Our analysis revealed that CD81 was highly expressed on EBI macrophages under BM steady state conditions and remained elevated in spleen during stress (Figure 7D; supplemental Figure 5B-C). These findings suggest that CD81 could be a significant factor in the function of EBI macrophages under both steady state and stress conditions.
To investigate the functional role of CD81 in EBI macrophages' response to stress, we administered a CD81 blocking antibody in vivo after treatment with either BLD or PHZ (Figure 7E and 7H). We found that under BLD conditions, CD81 blockade resulted in more severe splenomegaly (Figure 7F) and significant impairment of erythroid recovery (Figure 7G), along with a significant reduction in the association between erythroid cells and EBI macrophages (Supplemental Figure 5D). Although the effects were less pronounced under PHZ treatment, they were still observed (Figure 7I and 7J). These data suggest that CD81 plays an important functional role in the ability of EBI macrophages to mediate erythroid recovery under stress conditions. The underlying mechanisms may involve CD81's roles in immune response, cell-cell fusion, and cell signaling.
Discussion
Although several single-cell analyses of stress erythropoiesis were documented, ranging from investigations involving fetal liver/EPO-stimulated BM23 to studies focusing on anemia-activated cis elements,24 our study distinguishes itself by concentrating on exploring the broader landscape of the splenic response to PHZ and bleeding stress, particularly emphasizing insights into EBI dynamics. Specifically, we delve into scrutinizing the differentiation dynamics of erythroblasts during the recovery phase from anemic stress, as well as identifying the cellular diversity and surface marker profiles of resident EBI macrophage populations (supplemental Figure 5E).
More specifically, the identification of the “hiatus” population of erythroblasts during stress is intriguing. The term “hiatus” is commonly used in the field of immunology to refer to a group of cells that have undergone a sudden transition from an immature state to a mature state. This phenomenon was first described in the context of leukemia, in which it is known as “hiatus leukemicus,” and refers to a sudden jump in the development of leukemic cells from an early to a late stage with no intermediate stages.25 In the context of stress erythropoiesis, the discovery of a subpopulation of Stage_1 erythroblast displaying a transient pause in most transcriptional activities, except for moderate level of protein synthesis, is captivating. One possible reason for the existence of such transcriptionally quiescent erythroid progenitor cells under stress is that they may be conserving energy and resources by temporarily halting major transcriptional activities until conditions become more favourable for them to proliferate and differentiate. To gain a comprehensive understanding of the mechanisms underlying the “hiatus” population, it is necessary to conduct more extensive analysis, including the identification of more potent surface markers and additional functional studies.
Another important finding is the distinct transcriptional programs in various macrophage populations under different stressors (Figure 7A). The differential gene expressions related to DNA damage repair, oxidative stress, and inflammation responses may explain the observed differences in spleen histology. Furthermore, we identified a novel surface marker labeling EBI macrophages and functionally required to combat stress. This marker, CD81, was previously found to facilitate cell-cell fusion in various contexts, including between gametes, myoblasts, and virus-infected cells.26 In addition, CD81 is known to participate in signal transduction and cell adhesion within the immune system.27 Studies also provided evidences for the downstream signaling of CD81. More specifically, CD81 was shown to interact with the tetraspanin CD19 in B cells, leading to activation of the B-cell receptor signaling pathway. This results in the activation of the MAPK pathway and phosphoinositide 3-kinase pathway, ultimately leading to B-cell proliferation and survival.28 Hence, the discovery of CD81 as a novel surface marker on EBI macrophages has shed light on the investigation of comprehensive downstream signaling pathways of CD81 under both steady state and anemic stress conditions. Therefore, targeting CD81 could potentially modulate erythroblastic island function and offer therapeutic opportunities in various hematological disorders.
Collectively, our study expands upon previous analyses of murine erythroid response to stress mediated by EBI interactions8 and identifies novel cell-intrinsic and -extrinsic responses of EBIs to anemic stress. Such advancements could potentially inform the development of novel therapies for individuals suffering from anemia and related diseases.
Acknowledgment
The study is supported by the Singapore Translational Research Investigator (STaR) Award from the National Medical Research Council in Singapore (NMRC/STaR 18 may-0004; T. Suda); and the Open Fund-Young Individual Research Grant (OF-YIRG) from the National Medical Research Council in Singapore (OFYIRG19may-0009; C.Y.).
Authorship
Contribution: T. Suda and C.Y. conceived the study, designed the experiments, and wrote the manuscript; R.Y. performed scRNA-seq analyses. L.H.C., S.H.T., and M.Y.K. assisted in the experiments; H.T., T. Sanda participated in the writing of manuscript, discussion, and analysis of experiments; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Toshio Suda, Cancer Science Institute of Singapore, National University of Singapore, 14 Medical Dr, #12-01 Centre for Translational Medicine, Singapore 117599; email: csits@nus.edu.sg; and Chong Yang, Cancer Science Institute of Singapore, National University of Singapore, 14 Medical Dr, #12-01 Centre for Translational Medicine, Singapore 117599; email: csiyc@nus.edu.sg.
References
Author notes
The scRNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (accession number GSE230331).
All data are available upon reasonable request from the corresponding authors, Toshio Suda (csits@nus.edu.sg) and Chong Yang (csiyc@nus.edu.sg).
The full-text version of this article contains a data supplement.