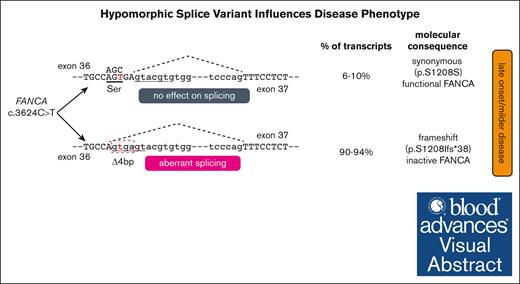
An exonic FANCA variant, c.3624C>T, predicted to be synonymous (p.Ser1208=), induces a pathogenic splicing defect.
The pathogenic variant is hypomorphic, resulting in low level of normal splicing, which delays the onset of Fanconi anemia.
Visual Abstract
Fanconi anemia (FA) is a hereditary, DNA repair deficiency disorder caused by pathogenic variants in any 1 of 22 known genes (FANCA-FANCW). Variants in FANCA account for nearly two-thirds of all patients with FA. Clinical presentation of FA can be heterogeneous and include congenital abnormalities, progressive bone marrow failure, and predisposition to cancer. Here, we describe a relatively mild disease manifestation among 6 individuals diagnosed with FA, each compound heterozygous for 1 established pathogenic FANCA variant and 1 FANCA exon 36 variant, c.3624C>T. These individuals had delayed onset of hematological abnormalities, increased survival, reduced incidence of cancer, and improved fertility. Although predicted to encode a synonymous change (p.Ser1208=), the c.3624C>T variant causes a splicing error resulting in a FANCA transcript missing the last 4 base pairs of exon 36. Deep sequencing and quantitative reverse transcription polymerase chain reaction analysis revealed that 6% to 10% of the FANCA transcripts included the canonical splice product, which generated wild-type FANCA protein. Consistently, functional analysis of cell lines from the studied individuals revealed presence of residual FANCD2 ubiquitination and FANCD2 foci formation, better cell survival, and decreased late S/G2 accumulation in response to DNA interstrand cross-linking agent, indicating presence of residual activity of the FA repair pathway. Thus, the c.3624C>T variant is a hypomorphic allele, which contributes to delayed manifestation of FA disease phenotypes in individuals with at least 1 c.3624C>T allele.
Introduction
Fanconi anemia (FA) is a rare, inherited, genomic instability disorder that affects ∼1 in 100 000 births and is not confined to any specific geographical region.1 FA is primarily characterized by congenital defects and progressive bone marrow failure, resulting in pancytopenia, reduced fertility, and predisposition to cancer.2 Inactivating variants in 1 of 22 known FANC genes (FANCA-FANCW) can cause FA, with FANCA pathogenic variants alone accounting for 64% of patients with FA.3,4 The FANC genes encode proteins that participate in the FA pathway and orchestrate the repair of DNA interstrand cross-links.2,3 In the absence of a functional FA pathway, as in individuals with FA, DNA damage accumulates, leading to genomic instability.
The clinical presentation of FA is heterogeneous, with at least 1 physical abnormality observed in 80% of individuals with FA.5 The reported median age for the onset of hematological disease is 7 years,6,7 and nearly 90% of patients with FA present with progressive depletion of ≥1 blood cell lineages resulting in bone marrow failure by age 40 years,8 requiring allogeneic hematopoietic cell transplantation as definitive therapy. Another manifestation of the disease can be myelodysplastic syndrome (MDS), and acute myeloid leukemia (AML), with patients with FA having ∼500-fold increased relative risk for AML.9-11 Patients with FA are also at higher risk of developing solid tumors, with a 700-fold increased relative risk of developing squamous cell carcinoma of the head and neck.12-14 A common feature of FA is reduced fertility, for males are rarely reported to be fertile and approximately half of female patients with FA are infertile.15
The heterogeneity of FA phenotypes has presented a challenge for genotype–phenotype correlation studies. Pathogenic variants in BRCA2/FANCD1 and PALB2/FANCN lead to embryonal tumors and AML in early childhood16-19 and represent severe disease manifestations, although, even among patients with BRCA2 and PALB2 variants, a milder disease course may result from hypomorphic variants and the encoded protein retaining some functional activity.20,21 We recently found that individuals with FANCB loss-of-function variants develop a more severe disease presentation than individuals with FANCB missense variants.22 Among those with missense variants, individuals who retained increased residual FANCB function exhibited a considerably reduced disease severity.
Characterizing distinct genotype–phenotype correlations is integral to understanding the clinical consequence of FA gene variation and can inform FA disease management, decisions on the timing and the choice of treatments, and the development of novel therapeutic options. This is especially true for the largest FA group with FANCA variants, which is highly heterogeneous, with large deletions accounting for 25% of the variants.23,24
Here, we present 6 individuals who share FANCA c.3624C>T as 1 of their biallelic pathogenic variants and displayed a milder phenotype than other individuals with biallelic FANCA variants. The c.3624C>T variant caused aberrant splicing of most messages, but cells retain between 6% and 10% of wild-type (WT) transcript, resulting in partial activation of the FA pathway and, most likely, delaying disease manifestation.
Methods
Study participants
The study participants included 6 individuals diagnosed with FA and enrolled in the International Fanconi Anemia Registry. The institutional review board of The Rockefeller University (New York, NY) approved these studies. The Office of Human Subjects Research at the National Institutes of Health and institutional review board of the National Human Genome Research Institute approved the receipt of deidentified cell lines and DNA samples from The Rockefeller University. Written informed consent was obtained.
Identification of disease-causing variants
Targeted next-generation sequencing, Sanger sequencing, and array comparative genomic hybridization were used to identify sequence variants and large deletions, as previously described.23 TruSeq custom amplicon kit (Illumina, San Diego, CA) was used for targeted next-generation sequencing of entire genomic regions of all known FA genes.25 The sequencing data was collected using 101–base pair (bp) paired-end reads to a depth of ∼500× using the HiSeq 2000 system (Illumina).
Cell culture
Lymphoblastoid cell lines were cultured in RPMI 1640 (Gibco) plus 20% fetal bovine serum, 1% penicillin/streptomycin (Gibco), 1% amphotericin B (Gibco), and 1% GlutaMAX (Gibco). Fibroblasts were cultured in Dulbecco modified Eagle medium (Gibco) plus 15% fetal bovine serum (Atlanta Biologicals), 1% penicillin/streptomycin, and 1% GlutaMAX. Fibroblast cell lines were transformed and/or immortalized by expression of HPV16 E6E7 and the catalytic subunit of human telomerase, respectively. All cell lines used in this study were established prior to bone marrow transplantation (BMT).
Western blot and antibodies
Whole-cell lysates were prepared by lysing in Laemmli sample buffer (Bio-Rad) and sonication. Samples were boiled and proteins were separated on NuPAGE gradient gels (3%-8% Tris-Acetate gel for FANCD2, and 4%-12% Bis-Tris gel for FANCA; Invitrogen) and transferred onto polyvinylidene difluoride membrane (Millipore). Immunoblotting was performed using the following antibodies: FANCA (Bethyl Antibody, A301-980A) at 1:500 dilution; FANCD2 (Novus NB100-182) at 1:2000 dilution.
Immunofluorescence
Cells were preextracted with 0.5% TritonX-100 in phosphate-buffered saline (PBS) for 5 minutes at room temperature before fixation with 3.7% formaldehyde. Subsequently, cells were permeabilized with 0.5% NP-40 in PBS, blocked in 5% (volume per volume) fetal bovine serum in PBS, and incubated with indicated antibodies (1:1000) in blocking buffer. Cells were incubated with Alexa Fluor secondary antibodies to visualize FANCD2 foci. The cells were washed, and the coverslips were mounted with 4′,6-diamidino-2-phenylindole Fluoromount-G (SouthernBiotech, Birmingham, AL). Four independent coverslips were assayed. They were scored by an individual blinded to the conditions.
Survival assay
For survival assay, 3.5 × 104 cells were seeded per well of a 6-well dish, in triplicate. The next day, mitomycin C (MMC) was added at final concentrations ranging from 0 to 75 nM. After culturing for 4 days without media change, the cells were split at appropriate dilutions, and cultured for another 5 days. Viable cell count was obtained using NucleoCounter NC-3000 (ChemoMetec, Denmark) as per the “cell vitality assay” protocol of the manufacturer. The cell numbers at each dose of drug were divided by the cell number in the untreated sample to calculate percent survival. Two independent experiments were performed. Survival of RA2087 E6E7 transformed and hTERT immortalized (EH)+WT, RA2349, and RA2565 cells were compared to survival of RA3087EH+empty vector (EV) cell lines. Adjusted P values were derived from a 2-way analysis of variance corrected for multiple comparisons.
Cell cycle analysis
Cells were cultured with or without 25 ng/mL MMC for 48 hours. The harvested cells were fixed in ice-cold 70% ethanol and stored overnight at 4°C. Staining of DNA and removal of RNA was performed by incubating cells in Solution 2 (ChemoMetec) supplemented with 200 mg/mL RNase A at 37°C for 30 minutes. The quantification of DNA content to determine cell cycle phases was done using NucleoCounter NC-3000 (ChemoMetec) as per the “cell cycle analysis of fixed cells” protocol of the manufacturer.
Quantitative fluorescence polymerase chain reaction (qfPCR)
The relative levels of FANCA transcripts with or without 4-bp deletion at the end of exon 36 were measured using a qfPCR method as previously described.22 PCR products were amplified using fPCR primers listed in supplemental Table 1. The expected product size from a WT and 4-bp deletion transcript are 319 bp and 315 bp, respectively. The average peak amplitude from 3 fPCR reactions was taken to calculate average percent splice products.
Illumina MiSeq library preparation and next-generation sequencing and analysis
Amplicon libraries were prepared using a 2-step PCR approach, as previously described.22 Briefly, in the first amplification step, the complementary DNA template was used to amplify the exon 36 region of FANCA gene along with universal overhangs on both ends using FAA-ex35-MiSeq-F and FAA-ex37-MiSeq-R primers (supplemental Table 1). In the second amplification step, the product from first-step PCR was used as the template for amplification using i5 and i7 adapter primers with unique barcode sequences (supplemental Table 1). The amplicon library products are quantified using Qubit and checked for any nonspecific products by gel electrophoresis and a Bioanalyzer. Cluster generation and sequencing of PCR products were performed with the MiSeq Reagent Kit version 2 (300 cycles) with 2 × 300 paired-end reads on an Illumina MiSeq instrument (Illumina). The MiSeq data analysis was performed using Burrows-Wheeler Aligner, Samtools, and Integrative Genomics Viewer. The Miseq reads were aligned, indexed, and quantitated comparing the WT sequence and the variant sequence.
Results
The FANCA c.3624C>T variant, predicted to be synonymous (p.Ser1208=), causes aberrant splicing
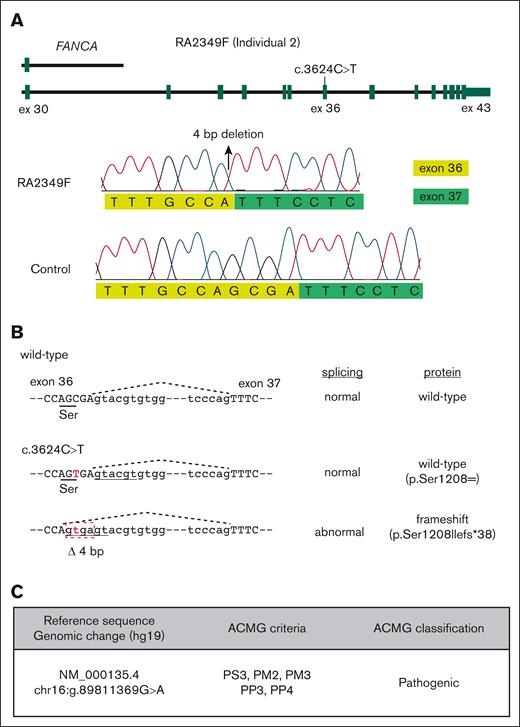
In our search for the affected gene and the disease-causing variants for individuals enrolled in the International Fanconi Anemia Registry, we identified 6 individuals who were compound heterozygotes for a synonymous FANCA variant, c.3624C>T (p.Ser1208=), and a distinct second FANCA pathogenic variant (Table 1). The second FANCA variant in individuals 1 to 5 was a loss-of-function variant, either an indel (2), a stop-gain (2), or a large deletion (1),23 whereas individual 6 carried a missense variant, c.2534T>C; p.Leu845Pro, that has been shown to be pathogenic by functional studies.26 The c.3624C>T variant, although predicted to be synonymous, is located near the exon 36 splice donor site, indicating a potential effect on splicing. Reverse transcription PCR analysis was performed on the region around FANCA exon 36 using RNA from a cell line derived from individual 2, whose other allele carries a >20-kilobase deletion, encompassing exons 31 to 43 and extending beyond the 3′ end of the gene (Figure 1A). The c.3624C>T variant caused a shift in the splice donor site, generating an aberrant splice product missing 4 bp from the end of exon 36, c.3623_3626delGCGA; p.Ser1208Ilefs∗38 (Figure 1). The frequency of this variant, rs149797103, in gnomAD (https://gnomad.broadinstitute.org/variant/16-89811369-G-A) is 0.0001075 (30 of 279 094 alleles), of which all are heterozygous carriers, and mostly from the European population (28 of 30). According to the American College of Medical Genetics and Genomics guidelines,27 we classify the c.3624C>T variant as “pathogenic” based on the all the available evidence included in this study (Figure 1C).
Clinical presentation of individuals with FANCA variant c.3624C>T
| FANCA variant (p.Ser1208=) . | Individual 1 . | Individual 2 . | Individual 3 . | Individual 4 . | Individual 5 . | Individual 6 . |
|---|---|---|---|---|---|---|
| c.3624C>T . | c.3624C>T . | c.3624C>T . | c.3624C>T . | c.3624C>T . | c.3624C>T . | |
| Second FANCA variant | c.1073_1074delTG (p.Tyr357Profs∗49) | Large deletion Exons 31-43∗ | c.1027C>T (p.Gln343∗) | c.2021C>A (p.Ser674∗) | c.1115_1118delTTGG (p.Val372Alafs∗42) | c.2534T>C (p.Leu845Pro) |
| Sex | M | F | F | F | M | M |
| Age (y) at FA diagnosis | 23 | 11 | 18 | 42 | 33 | 63 |
| Chromosome breaks per metaphase at baseline | 0.2 | 0.14 | 0.24 | 0.2 | 0.2 | 0 |
| Chromosome breaks per metaphase after DEB/MMC | 3.6 | 2.2 | 2.1 | 2.3 | 1.06 | 0.1† |
| Clinical presentation | Thrombocytopenia | Anemia | Pneumonia with resistance to antibiotics and hematological abnormalities | Pancytopenia | Gum bleeding, lightheadedness, shortness of breath, nausea, and fatigue | Thrombocytopenia |
| Developmental abnormalities | Hearing loss | Left thumb aplasia/ hypoplasia, axillary freckling, and café-au-lait spots | Left rib fusion and café-au-lait spots | None | Bilateral bifid thumb, and clinodactyly | None |
| Age (y) at onset of hematologic disease | 19 | 12 | 14 | 26 | 33 | 48 |
| Age (y) at BMT | 25 | 21 | No transplantation | 43 | 34 | No transplantation |
| BMT type | No records | Unrelated (7-of-8 matched antigens) | No transplantation | Nonmyeloablative allogeneic (sibling) | Unrelated (8-of-8 matched antigens) | No transplantation |
| Fertile‡ | Yes | Yes | Yes | Yes | Yes | No |
| Cancer history | None | Cervical dysplasia | None | None | Skin squamous cell carcinoma | Melanoma, MDS |
| Vital status | Deceased | Alive | Deceased | Deceased | Alive | Deceased |
| COD | Pulmonary aspergillosis sepsis s/p BMT with failure to engraft | N/A | Official COD unknown; based on predeath complications, COD most likely multiple bacterial blood infections/sepsis | Post-BMT complications (intraparenchymal reperfusion hemorrhage) | N/A | MDS complications |
| Age (y) at last follow-up | 25 | 33 | 26 | 43 | 43 | 66 |
| FANCA variant (p.Ser1208=) . | Individual 1 . | Individual 2 . | Individual 3 . | Individual 4 . | Individual 5 . | Individual 6 . |
|---|---|---|---|---|---|---|
| c.3624C>T . | c.3624C>T . | c.3624C>T . | c.3624C>T . | c.3624C>T . | c.3624C>T . | |
| Second FANCA variant | c.1073_1074delTG (p.Tyr357Profs∗49) | Large deletion Exons 31-43∗ | c.1027C>T (p.Gln343∗) | c.2021C>A (p.Ser674∗) | c.1115_1118delTTGG (p.Val372Alafs∗42) | c.2534T>C (p.Leu845Pro) |
| Sex | M | F | F | F | M | M |
| Age (y) at FA diagnosis | 23 | 11 | 18 | 42 | 33 | 63 |
| Chromosome breaks per metaphase at baseline | 0.2 | 0.14 | 0.24 | 0.2 | 0.2 | 0 |
| Chromosome breaks per metaphase after DEB/MMC | 3.6 | 2.2 | 2.1 | 2.3 | 1.06 | 0.1† |
| Clinical presentation | Thrombocytopenia | Anemia | Pneumonia with resistance to antibiotics and hematological abnormalities | Pancytopenia | Gum bleeding, lightheadedness, shortness of breath, nausea, and fatigue | Thrombocytopenia |
| Developmental abnormalities | Hearing loss | Left thumb aplasia/ hypoplasia, axillary freckling, and café-au-lait spots | Left rib fusion and café-au-lait spots | None | Bilateral bifid thumb, and clinodactyly | None |
| Age (y) at onset of hematologic disease | 19 | 12 | 14 | 26 | 33 | 48 |
| Age (y) at BMT | 25 | 21 | No transplantation | 43 | 34 | No transplantation |
| BMT type | No records | Unrelated (7-of-8 matched antigens) | No transplantation | Nonmyeloablative allogeneic (sibling) | Unrelated (8-of-8 matched antigens) | No transplantation |
| Fertile‡ | Yes | Yes | Yes | Yes | Yes | No |
| Cancer history | None | Cervical dysplasia | None | None | Skin squamous cell carcinoma | Melanoma, MDS |
| Vital status | Deceased | Alive | Deceased | Deceased | Alive | Deceased |
| COD | Pulmonary aspergillosis sepsis s/p BMT with failure to engraft | N/A | Official COD unknown; based on predeath complications, COD most likely multiple bacterial blood infections/sepsis | Post-BMT complications (intraparenchymal reperfusion hemorrhage) | N/A | MDS complications |
| Age (y) at last follow-up | 25 | 33 | 26 | 43 | 43 | 66 |
COD, cause of death; DEB, diepoxybutane; F, female; M, male; MMC, mitomycin C; N/A, not applicable; s/p, status post.
cDNA: c.2982-?_4368+?del, genomic (hg19): chr16:89,803,856-89,824,374.
Clinical.
Fertility confirmed through gamete analysis or biological offspring.
FANCA variant c.3624C>T generates a cryptic splice site. (A) Schematic of FANCA alleles in fibroblasts from individual 2. A large deletion extending from exons 31 to 43 is present on 1 allele. The other allele is c.3624C>T (top). Traces from Sanger sequencing of the reverse transcription PCR product amplified from the region around FANCA exon 36 using RNA derived from the RA2349 cell line. The c.3624C>T variant appears to create an aberrant splice donor site that is 4 bp upstream of the canonical exon 36 donor site, predicted to lead to a frameshift (bottom). (B) Schematic showing that the sequence around the c.3624C>T variant, predicted to be synonymous, affects RNA splicing. (C) Classification of the c.3624C>T variant according to American College of Medical Genetics and Genomics ACMG criteria.
FANCA variant c.3624C>T generates a cryptic splice site. (A) Schematic of FANCA alleles in fibroblasts from individual 2. A large deletion extending from exons 31 to 43 is present on 1 allele. The other allele is c.3624C>T (top). Traces from Sanger sequencing of the reverse transcription PCR product amplified from the region around FANCA exon 36 using RNA derived from the RA2349 cell line. The c.3624C>T variant appears to create an aberrant splice donor site that is 4 bp upstream of the canonical exon 36 donor site, predicted to lead to a frameshift (bottom). (B) Schematic showing that the sequence around the c.3624C>T variant, predicted to be synonymous, affects RNA splicing. (C) Classification of the c.3624C>T variant according to American College of Medical Genetics and Genomics ACMG criteria.
Clinical presentation of 6 individuals carrying the FANCA c.3624C>T variant
Table 1 lists the clinical presentation of 6 individuals with FANCA c.3624C>T variant. All individuals exhibited a delayed onset of hematologic disease that corresponded with a longer time to diagnosis of FA. The median age at diagnosis was 28 years (range, 11-63 years), whereas a typical FA diagnosis occurs at a median age of 10 years.28,29 The cross-link–induced breaks per metaphase in 4 individuals were <3, and individual 6 displayed only 0.1 breaks per metaphase, even below the number required for FA diagnosis. Individual 6 was diagnosed at age 63 years, after developing MDS. The median age at onset of hematological disease among the 6 individuals was 22.5 years (range, 12-48 years), which is threefold later than the general FA population (7 years).6 Four individuals underwent allogeneic hematopoietic cell transplantation at a median age of 29.5 years (range, 21-43 years). Two individuals died from complications after BMT at age 25 and 43 years whereas 2 individuals are alive at age 33 and 43 years after undergoing BMT at age 21 and 34 years, respectively. Individuals 3 and 6, who never underwent BMT, died at age 26 and 66 years, respectively. The official cause of death for individual 3 is unknown, but likely sepsis, and individual 6 died from MDS complications.
Reduced fertility in females and sterility in most males is 1 of the common features in adults with FA. Interestingly, 5 of these individuals (2 of 3 males, and 3 of 3 females) were confirmed to be fertile either through gamete analysis or the presence of biological offspring. Only 2 individuals had history of cancer: individual 5 had squamous cell carcinoma of the skin, and individual 6 had melanoma at age 58 years and MDS at age 62 years. Individual 2 had a precancerous condition of cervical dysplasia.
Functional analysis reveals that the c.3624C>T variant is hypomorphic
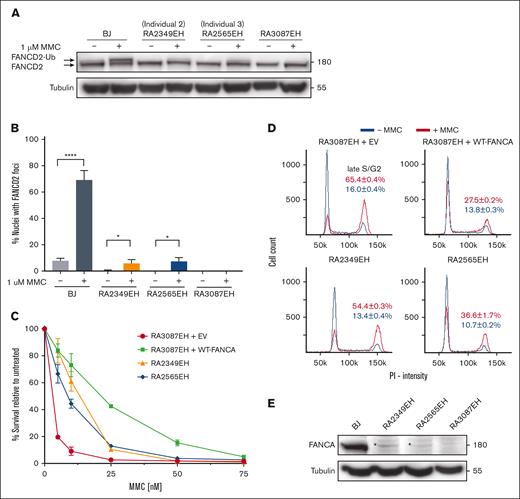
We performed functional analysis using patient-derived cells to determine the extent of FA pathway deficiency associated with the c.3624C>T variant. Specifically, the immortalized fibroblast cells from individuals 2 (RA2349EH) and 3 (RA2565EH) were subjected to functional evaluation after treatment with MMC using these assays: FANCD2 monoubiquitination, FANCD2 foci formation, cell survival, and cell cycle analysis. The phenotypes of cells from patients with the c.3624C>T variant were compared with FANCA-null cells (RA3087EH). FANCD2 monoubiquitination and foci formation assays revealed residual FA pathway activity in cells from both individuals (Figure 2A-B; supplemental Figure 1), suggesting retention of some FANCA function. Additionally, an intermediate level of cell survival and mild late S/G2 accumulation (Figure 2C-D) further supported presence of the residual FANCA function in these cells. Consistent with this, we observed a faint FANCA-size protein band in western blot analysis (Figure 2E). Overall, the observed residual FA pathway activity and presence of a faint band that corresponds to the presence of FANCA protein in both the patient-derived cell lines suggest that the c.3624C>T is a hypomorphic variant.
Functional evaluation of patient fibroblast cell lines shows hypomorphic nature of the c.3624C>T variant. (A) Western blot of the indicated cell lines with and without MMC treatment. Weak monoubiquitination of FANCD2 is observed in cells RA2349EH (from individual 2), and RA2565EH (from individual 3) in response to MMC. RA3087EH cells (individual with FA with biallelic deletions in FANCA) cells were used as negative controls. BJ cells were used as positive control. (B) Quantification of FANCD2 foci in the indicated cell lines. Proficiency of FANCD2 localization to sites of damage is suppressed in RA2349EH and RA2565EH in response to MMC but is higher than in FANCA-null cells (RA3087EH). Unpaired t test was used to test for significance between untreated and MMC-treated fibroblasts. Also see supplemental Figure 1. (C) Intermediate level of survival is observed for RA2349EH and RA2565EH cells in response to increasing concentrations of MMC. Cell survival was calculated relative to untreated cells. Error bars represent standard deviation (SD) of an experiment done in triplicate. Data from a representative experiment are shown with the SD showing the range of technical replicates. RA3087EH complemented with empty vector (EV) were used as a negative controls. RA3087EH complemented with wildtype FANCA (WT-FANCA) were used as a positive control. Statistical analysis was performed using data from 2 independent experiments, each with 3 technical replicates. Adjusted P values from multiple comparison 2-way analysis of variance analysis of differences in survival between RA3087EH+EV (FANCA null) and the other cell lines were P < .0001 at 5 and 10 nM of MMC for all 3 cell lines. P < .0001, P < .0057, and P < .0002 for WT, RA2349, and RA2565, respectively, at 25 nM MMC. At 50 nM MMC, the P value was significant (P < .0001) for the WT cell line only. (D) Cell cycle analysis without MMC treatment (blue line) or after exposure to 25 nM MMC (red line) in the indicated cell lines. Three independent experiments were performed and the average percentages of cells in the late S/G2 cell cycle stage are indicated. (E) Western blot analysis of FANCA in the indicated cell lines. RA2349EH and RA2565EH cells showed expression of residual endogenous FANCA protein (indicated by an asterisk “∗”).
Functional evaluation of patient fibroblast cell lines shows hypomorphic nature of the c.3624C>T variant. (A) Western blot of the indicated cell lines with and without MMC treatment. Weak monoubiquitination of FANCD2 is observed in cells RA2349EH (from individual 2), and RA2565EH (from individual 3) in response to MMC. RA3087EH cells (individual with FA with biallelic deletions in FANCA) cells were used as negative controls. BJ cells were used as positive control. (B) Quantification of FANCD2 foci in the indicated cell lines. Proficiency of FANCD2 localization to sites of damage is suppressed in RA2349EH and RA2565EH in response to MMC but is higher than in FANCA-null cells (RA3087EH). Unpaired t test was used to test for significance between untreated and MMC-treated fibroblasts. Also see supplemental Figure 1. (C) Intermediate level of survival is observed for RA2349EH and RA2565EH cells in response to increasing concentrations of MMC. Cell survival was calculated relative to untreated cells. Error bars represent standard deviation (SD) of an experiment done in triplicate. Data from a representative experiment are shown with the SD showing the range of technical replicates. RA3087EH complemented with empty vector (EV) were used as a negative controls. RA3087EH complemented with wildtype FANCA (WT-FANCA) were used as a positive control. Statistical analysis was performed using data from 2 independent experiments, each with 3 technical replicates. Adjusted P values from multiple comparison 2-way analysis of variance analysis of differences in survival between RA3087EH+EV (FANCA null) and the other cell lines were P < .0001 at 5 and 10 nM of MMC for all 3 cell lines. P < .0001, P < .0057, and P < .0002 for WT, RA2349, and RA2565, respectively, at 25 nM MMC. At 50 nM MMC, the P value was significant (P < .0001) for the WT cell line only. (D) Cell cycle analysis without MMC treatment (blue line) or after exposure to 25 nM MMC (red line) in the indicated cell lines. Three independent experiments were performed and the average percentages of cells in the late S/G2 cell cycle stage are indicated. (E) Western blot analysis of FANCA in the indicated cell lines. RA2349EH and RA2565EH cells showed expression of residual endogenous FANCA protein (indicated by an asterisk “∗”).
Quantitative transcript analysis reveals that the c.3624C>T variant generates 6% to 10% of WT FANCA transcript
Next, we explored the molecular events that may contribute to the hypomorphic nature of the c.3624C>T variant. We hypothesized that a fraction of transcripts with the c.3624C>T variant is either undergoing canonical splicing to generate low levels of WT transcript/protein or generating an additional aberrant splice product that goes back in-frame and has residual activity. To resolve these possibilities, we performed quantitative evaluation of FANCA transcripts by 3 independent methods: cloning and sequencing; fragment length analysis; and deep sequencing. For these assays, we used multiple cell lines from individual 2 who lacked expression of the second allele. For cloning, we used the reverse transcription PCR product generated using RNA from the RA2349 cell line, and sequenced 64 clones. In total, 62 exhibited the expected 4-bp deletion from the end of exon 36, and the other 2 clones were full-length transcripts with a c.3624C>T variant (Figure 3A). For qfPCR and deep sequencing, we used RNA from 2 lymphoblastoid cell lines cell lines (RA2219 and RA2140, both from individual 2) in addition to the fibroblast cell line. The qfPCR revealed the extent of the full-length WT transcript and the 4-bp deletion variant transcript (Figure 3B-C). Among the 3 patient-derived cell lines, we found ∼6% to 10% of canonically spliced WT transcript. Deep sequencing to a read depth of >200 k also showed a similar levels of canonical splice product (Figure 3D-E). Overall, all 3 methods clearly demonstrated that a small percentage of transcripts underwent canonical splicing with the c.3624C>T; p.Ser1208= variant, leading to a functional FANCA protein. The results confirm the hypomorphic nature of the variant and explain the relatively milder phenotype of these individuals.
Transcript analysis identifies low levels of canonical splice products associated with the c.3624C>T variant. (A) Traces of Sanger sequencing from 64 cloned reverse transcription PCR products amplified from region around FANCA exon 36 using RNA derived from RA2349 fibroblasts. Two of 64 clones exhibited normal spliced product at the splice donor site of exon 36 with the synonymous variant present. The remaining 62 clones showed a 4-bp deletion. (B) Schematic of the qf-PCR method used for relative quantification of splice products at exon 36-37 junction. (C) qf-PCR peak profiles showing that 6% to 10% of products had normal splicing. The average peak amplitude from 3 independent experiments was used to calculate relative percentage of product(s). (D) Integrative Genomics Viewer showing MiSeq deep sequencing of the c.3624C>T variant position in RA2349 cell line. Low-level expression of the normally spliced product. (E) Number of reads and percentages at each indicated position from MiSeq analysis of 3 cell lines including fibroblast RA2349 and 2 lymphoblastoid cell lines, RA2219 and RA2140, all derived from individual 2.
Transcript analysis identifies low levels of canonical splice products associated with the c.3624C>T variant. (A) Traces of Sanger sequencing from 64 cloned reverse transcription PCR products amplified from region around FANCA exon 36 using RNA derived from RA2349 fibroblasts. Two of 64 clones exhibited normal spliced product at the splice donor site of exon 36 with the synonymous variant present. The remaining 62 clones showed a 4-bp deletion. (B) Schematic of the qf-PCR method used for relative quantification of splice products at exon 36-37 junction. (C) qf-PCR peak profiles showing that 6% to 10% of products had normal splicing. The average peak amplitude from 3 independent experiments was used to calculate relative percentage of product(s). (D) Integrative Genomics Viewer showing MiSeq deep sequencing of the c.3624C>T variant position in RA2349 cell line. Low-level expression of the normally spliced product. (E) Number of reads and percentages at each indicated position from MiSeq analysis of 3 cell lines including fibroblast RA2349 and 2 lymphoblastoid cell lines, RA2219 and RA2140, all derived from individual 2.
Discussion
Here, we report 6 individuals with FA, each compound heterozygous for the FANCA variant c.3624C>T on 1 allele, and a distinct second pathogenic FANCA variant on the other allele. We demonstrate that the c.3624C>T variant is pathogenic because it induces aberrant splicing but is hypomorphic by enabling low-level expression of the correct splice product resulting in a small amount of WT FANCA protein. The second allele in 5 of 6 individuals were null, allowing us to ascribe the hypomorphic effect and the retention of residual FA pathway repair function observed in the individual-derived cell lines entirely to the c.3624C>T variant. Our study revealed that this residual function is sufficient to reduce the extent of DNA damage, explaining a delay in the onset of bone marrow failure, and improved germ cell development in a subset of patients that leads to longer survival when compared with typical patients with FANCA mutations. In addition to our 6 individuals reported here, 3 additional individuals compound heterozygous for the FANCA c.3624C>T variant have been reported30-32; however, their clinical presentations were not included in the reports.
Apart from the known founder FANCA variants in Afrikaners from South Africa,33 Spanish Gypsies,34 and Tunisian 35 populations, and 2 common variants across all populations,36 most patients with FA in the FA-A complementation group carry a distinct combination of 2 disease-causing FANCA variants,23 making it difficult to identify genotype–phenotype correlations. However, an earlier study had recognized that patients with FANCA with biallelic null variants exhibited earlier onset of hematological disease and increased incidence of MDS and AML compared with those with a FANCA missense variant, seemingly because of some level of residual functional protein.7 Recently, detailed evaluations have demonstrated that certain missense variants are hypomorphic based on residual FANCA protein function and associated retention of a fraction of the FA pathway function. A study of 11 individuals, who were homozygous or compound heterozygous for His913Pro and Arg951Gln/Trp FANCA missense variants, revealed that the variants retained some FA pathway function and that the individuals harboring these variants presented with milder disease.37
As mentioned earlier, 5 of 6 individuals in this study carried loss-of-function variants, but the second variant in the sixth individual was a missense variant, p.Leu845Pro, which was determined to be pathogenic/likely pathogenic in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/variation/556016/), supported by a detailed experimental functional evaluation of the variant.26 The clinical presentation of this individual was even milder than that of the other 5 individuals. Thrombocytopenia was the first hematological disease manifestation, which appeared at age 48 years, which is later than the median age at onset in the FA population. Notably, the chromosomal breakage test with cross-linking agent for this individual exhibited an unusually low number of breaks per metaphase, 0.1, which was not in the diagnostic range for FA. Not surprisingly, this individual was not diagnosed with FA until age 63 years and after the onset of MDS at the age of 62 years. The fact that the clinical presentation of individual 6 is milder than that of the other 5 individuals suggests that the p.Leu845Pro variant, despite being demonstrated to be pathogenic, may also provide some residual FA pathway function.
We previously reported 2 siblings in a family whose FA was not diagnosed until the individuals developed squamous cell carcinoma of esophagus at age 52 years, exacerbated by subsequent chemotherapy. Functional evaluation revealed that 1 of the FANCA variants they carried, a missense variant (c.4199G>A; p.Arg1400His), was hypomorphic. The siblings had very subtle thumb-related anomalies with otherwise unremarkable physical findings, and neither sibling exhibited any characteristic hematological findings associated with FA.38 Both affected male siblings had biological children.38 It is interesting to note that in the present study, all 3 females and 2 of 3 males were fertile, indicating that fertility may not be compromised for patients with FA carrying a subset of hypomorphic variants, which retain, to varying degrees, a functional FA pathway.
This study reveals the important role of transcript analysis in determining the precise molecular consequence leading to pathogenicity. Although predicted to be synonymous, the c.3624C>T causes aberrant splicing. We previously observed aberrant splicing as the pathogenic event for a subset of variants predicted to encode synonymous, nonsynonymous, or stop-gain changes.22,23,25,39 Similarly, this study and our previous work also illustrate that sensitive sequencing technologies need to be used for quantification and reliable evaluation of the functional consequences of potential hypomorphic variants to accurately determine genotype–phenotype correlations. Even certain splice junction variants may allow normal splicing and milder disease, as illustrated by those with FANCC variant c.165+1G>T.40
Correction of pre-messenger RNA splicing defects is an actively pursued therapeutic approach with some clear success treating genetic diseases such as Duchenne muscular dystrophy and spinal muscular atrophy.41 It will be important to explore such novel therapeutical interventions for those with hypomorphic variants due to splicing defects. This would not only potentially improve the function of the bone marrow but may also further delay or prevent cancer formation, which is a difficult challenge for patients with FA.
Acknowledgments
The authors thank the individuals enrolled in the International Fanconi Anemia Registry for participation in this study.
This study was supported by the National Institutes of Health National Heart, Lung, and Blood Institute (R01 HL120922; A.S.), the National Cancer Institute (R01 CA204127; A.S.), and the National Center for Advancing Translational Sciences (UL1 TR001866; R.M. and A.S.). A.S. was a Howard Hughes Faculty Scholar. This work was supported by the Intramural research program of the National Human Genome Research Institute, National Institutes of Health (R.R.-B., F.X.D., and S.C.C.)
Authorship
Contribution: R.R.-B., R.T., A.S., and S.C.C. designed the study; R.R.-B. and F.P.L. performed the experiments; R.R.-B., F.P.L., F.X.D., A.S., and S.C.C. analyzed the data; R.T., J.E.W., M.L.M., A.D.A., R.M., A.R., F.X.D., and A.S. analyzed the clinical presentations; S.C.C. and A.S. supervised the study; R.R.-B., F.X.D., and S.C.C. wrote the manuscript; and all authors discussed and revised the manuscript.
Conflict-of-interest disclosure: A.R. obtained salary support from Rocket Pharmaceuticals. A.S. obtained partial salary support from, and is an advisor to, Rocket Pharmaceuticals. J.E.W. is an advisor to Rocket Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Agata Smogorzewska, Laboratory of Genome Maintenance, The Rockefeller University, 1230 York Ave, New York, NY 10065-6399; email: asmogorzewska@rockefeller.edu; and Settara C. Chandrasekharappa, National Institutes of Health, National Human Genome Research Institute, 49 Convent Dr, Building 49, Room 4A68, Bethesda, MD 20892-8004; email: chandra@mail.nih.gov.
References
Author notes
R.R.-B. and R.T. contributed equally to this study.
Data and reagents are available on request from the corresponding authors, Settara C. Chandrasekharappa (chandra@mail.nih.gov) and Agata Smogorzewska (asmogorzewska@rockefeller.edu).
The full-text version of this article contains a data supplement.