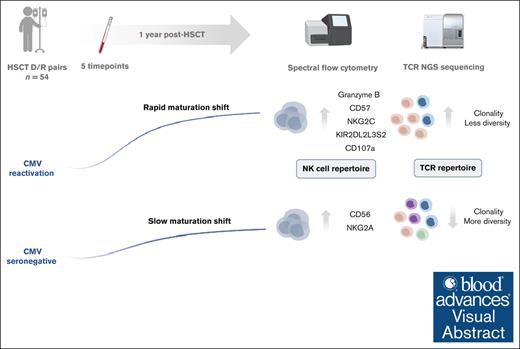
Key Points
The TCR and NK-cell repertoire establish within the first 90 days after transplantation.
CMV reactivation is associated with enhanced TCR clonality and a shift toward enhanced maturation and functionality of NK cells.
Visual Abstract
Besides genetic influences, nongenetic factors such as graft-versus-host disease and viral infections have been shown to be important shapers of the immune reconstitution and diversification processes after hematopoietic stem cell transplantation (HSCT). However, differential susceptibility to immune modulation by nongenetic factors is not fully understood. We determined to follow the reconstitution of the T-cell receptor (TCR) repertoire through immune sequencing of natural killer (NK) cells using a 35-marker spectral flow cytometry panel and in relation to clinical events. A longitudinal investigation was performed on samples derived from 54 HSCT recipients during the first year after HSCT. We confirmed a significant contraction in TCR repertoire diversity, with remarkable stability over time. Cytomegalovirus (CMV) reactivation had the ability to significantly change TCR repertoire clonality and composition, with a long-lasting imprint. Our data further revealed skewing of NK-cell reconstitution in CMV reactivated recipients, with an increased frequency of KIR2DL2L3S2+ adaptive, cytolytic, and functional CD107a+ NK cells, concomitant with a reduced pool of NKG2A+ NK cells. We provided support that CMV might act as an important driver of peripheral homeostatic proliferation of circulating specific T and NK cells, which can be viewed as a compensatory mechanism to establish a new peripheral repertoire.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a well-established immune therapy for patients with hematological malignancies such as leukemia or primary immunodeficiencies.1 Although long-term survival has strongly improved over the last decade, it remains a high-risk treatment, and its success is hampered by the occurrence of infections, immunological complications, and disease relapse.2 Its potential curative effect relies on the recreation of new competent hematopoietic and immune cells, combined with the eradication of residual tumor cells through the graft-versus-leukemia effect. Restoration of the state of the immune system thereafter is thought to be the contribution of the first wave of mature donor-derived immune cells, followed by a second wave of de novo stem cell–derived cells from the innate immune system, followed by the adaptive immune system.3,4 Alongside this process, the developing immune system is exposed to a wide variety of extrinsic factors, forcing this system to respond and adapt. Among these factors, we and others have demonstrated that cytomegalovirus (CMV) is an important immune state modulator that imprints the T-cell receptor (TCR) repertoire at 1 year after transplantation and persists for >5 years.5,6 Natural killer (NK)-cell repertoire reconstitution has been less studied but similar observations regarding the altering role of CMV have been published.7-9 These shifts are specifically driven by the massive clonal expansion of antigen-specific terminally differentiated CD8+ T cells and NKG2C+ CD57+ adaptive NK cells, combined with skewed expression of self-specific inhibitory killer cell immunoglobulin-like receptors (KIRs).10,11 Recent findings hint toward a similar inflationary memory response, with a lack of adaptive memory–like NK-cell contraction, as previously shown for CD8+ T cells during viral latency.11,12
In light of such findings and with regard to the limited data available on their combined reconstitution pattern, this report aims to dissect the interdependent temporal NK-cell repertoire and TCR repertoire restoration in a unique human allo-HSCT case model and further trace relations to clinical events. Comprehensive immune profiling was performed by leveraging a high-dimensional single-cell proteomic technique with next-generation sequencing in 54 HSCT recipients monitored longitudinally over the time course of 1 year after transplantation.
Materials and methods
Study design
All patients having undergone an allo-HSCT at the Geneva University Hospital between December 2020 and April 2022 were enrolled in this study, encompassing a total of 54 donor/recipient (D/R) pairs. Detailed demographic and clinical characteristics of the cohort are summarized in Table 1. Medical complications, such as infectious events (viral or bacterial), acute graft-versus-host disease (GVHD) and chronic GVHD episodes, relapse of the initial disease, transplant-related or nonrelated deaths, and CMV serologic status/reactivation (defined as CMV DNA in plasma above the limit of detection, ie, 2.1E + 1 UI/mL, in patients with or without clinical symptoms) were recorded (Table 2; supplemental Figure 1A-B). All recipients with a positive CMV serology received letermovir CMV prophylaxis (480 mg per day with tacrolimus-based immunosuppression or 240 mg per day with ciclosporin-based immunosuppression) from the day of transplantation until 100 days after transplantation. For all recipients with negative CMV serology, no prophylaxis was given, and CMV viremia was monitored on a regular basis after transplantation. All patients were treated for CMV reactivation with ganciclovir, which was stopped once viremia reached an undetectable level (<21 copies).
Demographic and transplant-related characteristics of the study cohort
| Parameter . | All (n = 54) . |
|---|---|
| Recipient age at HSCT, y (median, IQR) | 57 (62) |
| Recipient genre (M:F) | 37:17 |
| Donor age, y (median, IQR) | 41 (21.5) |
| Donor genre (M:F) | 39:15 |
| Underlying diagnosis, n (%) | |
| AML | 27 (50) |
| ALL | 4 (7.4) |
| AL nonspecific | 2 (3.7) |
| CML/CLL | 2 (3.7) |
| Lymphoma | 4 (7.5) |
| Myeloma | 1 (1.9) |
| MDPS/MDS/MPS | 12 (22.2) |
| Hemoglobinopathy | 2 (3.7) |
| Donor type, n (%) | |
| MUD | 22 (40.7) |
| MMUD | 2 (3.7) |
| MRD | 9 (16.7) |
| Haploidentical | 21 (38.9) |
| First transplantation, n (%) | 49 (90.7) |
| Conditioning with ATG, n (%) | 39 (72.2) |
| PTCy, n (%) | 22 (40.7) |
| No T-cell depletion, n (%) | 53 (98.1) |
| Cryopreservation, n (%) | 43 (79.6) |
| Stem cell source, n (%) | |
| BM | 3 (5.5) |
| PBSC | 51 (94.4) |
| CMV serostatus, n (%) | |
| D+/R+ | 22 (40.7) |
| D–/R+ | 12 (22.2) |
| D+/R– | 7 (13) |
| D–/R– | 13 (24.1) |
| Disease risk index, n (%) | |
| Low | 4 (7.4) |
| Intermediate | 41 (75.9) |
| High/very high | 7 (13) |
| NA | 2 (3.7) |
| Parameter . | All (n = 54) . |
|---|---|
| Recipient age at HSCT, y (median, IQR) | 57 (62) |
| Recipient genre (M:F) | 37:17 |
| Donor age, y (median, IQR) | 41 (21.5) |
| Donor genre (M:F) | 39:15 |
| Underlying diagnosis, n (%) | |
| AML | 27 (50) |
| ALL | 4 (7.4) |
| AL nonspecific | 2 (3.7) |
| CML/CLL | 2 (3.7) |
| Lymphoma | 4 (7.5) |
| Myeloma | 1 (1.9) |
| MDPS/MDS/MPS | 12 (22.2) |
| Hemoglobinopathy | 2 (3.7) |
| Donor type, n (%) | |
| MUD | 22 (40.7) |
| MMUD | 2 (3.7) |
| MRD | 9 (16.7) |
| Haploidentical | 21 (38.9) |
| First transplantation, n (%) | 49 (90.7) |
| Conditioning with ATG, n (%) | 39 (72.2) |
| PTCy, n (%) | 22 (40.7) |
| No T-cell depletion, n (%) | 53 (98.1) |
| Cryopreservation, n (%) | 43 (79.6) |
| Stem cell source, n (%) | |
| BM | 3 (5.5) |
| PBSC | 51 (94.4) |
| CMV serostatus, n (%) | |
| D+/R+ | 22 (40.7) |
| D–/R+ | 12 (22.2) |
| D+/R– | 7 (13) |
| D–/R– | 13 (24.1) |
| Disease risk index, n (%) | |
| Low | 4 (7.4) |
| Intermediate | 41 (75.9) |
| High/very high | 7 (13) |
| NA | 2 (3.7) |
AL, acute leukemia; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ATG, antithymocyte globulin; BM, bone marrow; CLL, chronic lymphoblastic leukemia; CML, chronic myeloid leukemia; MDPS, myelodysplastic/myeloproliferative syndrome; MDS, myelodysplastic syndrome; MMUD, mismatched unrelated donor; MPS, myeloproliferative syndrome; MRD, matched related donor; MUD, matched unrelated donor; PBSC, peripheral blood stem cells; PTCy, posttransplant cyclophosphamide.
Posttransplantation clinical events and transplant outcomes of the study cohort
| Parameter . | All (n = 54) . |
|---|---|
| Immunogenic posttransplantation complications | |
| aGVHD, n (%)∗ | 40 (74.1) |
| >1 episode within 1 year after HSCT, n (%) | 8 (14.8) |
| Severe aGVHD, grade ≥3, n (%) | 6 (11.1) |
| cGVHD, n (%) | 13 (24.1) |
| >1 episode within 1 year after HSCT, n (%) | 2 (3.7) |
| Moderate-to-severe cGVHD, n (%) | 9 (16.7) |
| Infectious posttransplantation complications | |
| Chronic viral infection, n (%) | |
| CMV | 13 (24) |
| EBV | 11 (20.4) |
| HHV-6 | 6 (11.1) |
| HHV-8 | 1 (1.9) |
| HSV | 3 (5.6) |
| BKV | 5 (9.3) |
| Respiratory viral infections, n (%) | |
| Influenza | 5 (9.3) |
| SARS-CoV-2 | 16 (29.6) |
| RSV | 4 (7.4) |
| Rhinovirus | 5 (9.3) |
| Bacterial infections, n (%) | 26 (48.2) |
| Transplantation outcomes, n (%) | |
| Disease relapse | 11 (20.4) |
| Overall survival at 1 year after HSCT | 50 (92.6) |
| Parameter . | All (n = 54) . |
|---|---|
| Immunogenic posttransplantation complications | |
| aGVHD, n (%)∗ | 40 (74.1) |
| >1 episode within 1 year after HSCT, n (%) | 8 (14.8) |
| Severe aGVHD, grade ≥3, n (%) | 6 (11.1) |
| cGVHD, n (%) | 13 (24.1) |
| >1 episode within 1 year after HSCT, n (%) | 2 (3.7) |
| Moderate-to-severe cGVHD, n (%) | 9 (16.7) |
| Infectious posttransplantation complications | |
| Chronic viral infection, n (%) | |
| CMV | 13 (24) |
| EBV | 11 (20.4) |
| HHV-6 | 6 (11.1) |
| HHV-8 | 1 (1.9) |
| HSV | 3 (5.6) |
| BKV | 5 (9.3) |
| Respiratory viral infections, n (%) | |
| Influenza | 5 (9.3) |
| SARS-CoV-2 | 16 (29.6) |
| RSV | 4 (7.4) |
| Rhinovirus | 5 (9.3) |
| Bacterial infections, n (%) | 26 (48.2) |
| Transplantation outcomes, n (%) | |
| Disease relapse | 11 (20.4) |
| Overall survival at 1 year after HSCT | 50 (92.6) |
aGVHD, acute graft-versus-host disease; BKV, polyomavirus BK; cGVHD, chronic graft-versus-host disease; EBV, Epstein-Barr virus; HHV-6, human herpesvirus 6; HHV-8, human herpesvirus 8; HSV, herpes simplex virus; RSV, respiratory syncytial virus.
Classification of aGVHD grading according to the National Institutes of Health criteria in which aGVHD can occur beyond 3 months based on clinical manifestations.
This study was approved by the ethical committee of the institution Geneva University Hospital (CER 06-208 and 08-208R).
Sample collection and processing
Whole blood samples were serially collected from HSCT recipients 3, 6, 9, and 12 months after HSCT and from the donor before transplantation. Peripheral blood mononuclear cell isolation was performed using Ficoll density gradient centrifugation and DNA extraction (supplemental Material).
High-resolution human leucocyte antigen typing
Human leucocyte antigen (HLA) typing was performed using reverse polymerase chain reaction sequence-specific oligonucleotide microbead arrays and high-throughput sequencing (One Lambda, Canoga Park, CA) or polymerase chain reaction sequence-specific primers (GenoVision, Milan Analytika AG, Switzerland).
Bulk TCR CDR3β sequencing
High-throughput TCR complementarity-determining region 3 beta (CDR3β) sequencing at survey resolution was performed using the immunoSEQ Assay (Adaptive Biotechnologies, Seattle, WA), according to the manufacturer’s instructions (supplemental Material).
High-resolution KIR genotyping
All donors were genotyped at a high resolution for all KIR loci. To this end, a DNA probe–based capture method was used as described here13 (supplemental Material).
Spectral flow cytometry immunophenotyping
The antibody panel for immunophenotyping comprised 35 markers. The antibody clones, concentrations, corresponding fluorochromes, and suppliers are summarized in supplemental Table 1. Surface and intracellular staining and sample acquisition were performed according to a standard protocol (supplemental Material).
CD107a mobilization assay
NK-cell cytotoxicity potential assessment was based on the CD107a mobilization assay, as previously described.14 Shortly, after overnight resting, 100 μL of the previously thawed peripheral blood mononuclear cells (2 × 106 to 4 × 106) were transferred from each sample to a 5-mL polypropylene tube and cocultured with 100 μL of K562 cells at an effector:tumor cell ratio of 5:1 or without a target for unstimulated controls. CD107a-BUV396 (BD Biosciences, San Diego, CA) antibody was added to each tube before the stimulation, including all unstimulated samples to assess any spontaneous degranulation. After 1 hour of stimulation, brefeldin A and monensin were added to the coculture and the cells were stimulated for another 4 hours at 37°C and 5% CO2. After 5 hours of stimulation, samples were washed for subsequent flow cytometry staining, as described in the section below. The internal control of stimulated cells without CD107a staining was added to assess the positive signals.
Computational and statistical analysis
Pushing Immunogenetics to the Next Generation bioinformatic pipeline
Pushing Immunogenetics to the Next Generation pipeline was applied for sequencing filtering, alignment, gene content, and allelic genotype determination derived from next-generation sequencing fastq files.13,15 High-resolution KIR genotyping, copy number, and alleles were determined for all KIR genes (KIR2DS1, 2DS2, 2DS3, 2DS4, 2DS3/S5, 3DL1/3DS1, 2DL1, 2DL2/L3/, 2DL4, 2DL5A/B, 3DL2, and 3DL3) and 2 pseudogenes (KIR2DP1 and 3DP1).
TCR data processing
Raw data processing and sequencing analyses were performed using the immunoSEQ Analyzer 4.0 online platform and in the R environment (version 4.1.3, R Core Team, R foundation for statistical computing) (supplemental Material). TCR clonality, overlap analysis, and identification of public and private TCR clones are detailed in the supplemental Material.
Prediction and inference of antigenic specificity
For the assessment of pathology specificity, several public databases were downloaded (VDJd,16McPAS-TCR,17 and ImmuneCODE18) and merged together, and individual CDR3 amino acid (aa) sequences were filtered based on the following categories: viral and bacterial disease, autoimmunity, allergenic, cancerous diseases, and GVHD. Subsequently, individual CDR3aa sequences from the cohort were queried against cleaned databases. HLA restriction was available for all CMV-specific clonotypes and is listed in supplemental Table 4.
Spectral flow cytometry data preprocessing and gating
FCS3.0 files were extracted from SpectroFlo Software v3.1.0 after spectral unmixing and compensation adjustments and analyzed by manual gating (supplemental Material).
Statistical analysis
Continuous data are reported as medians with interquartile ranges (IQRs). The normality distribution of the data was tested using the Shapiro-Wilk test. Two-group comparisons were analyzed using a 2-tailed unpaired Wilcoxon rank-sum test and a matched–paired Wilcoxon signed-rank test. A false discovery rate correction was applied for multiple tests. Linear regression using the glm function in R was used to assess the contribution of CMV–specific TCR-clone frequency to TCR clonality, assuming nonparametric data. All statistical analyses were performed in the R environment (version 4.1.3, R Core Team, R foundation for statistical computing) with a P value of .05 set as the threshold for significance.
Results
In this study, we characterized the coordinated changes involved in the immune reconstitution of NK- and T-cell compartments after allo-HSCT and interrogated the effect of nongenetic factors on immune state restoration. To this end, 54 recipients and their respective donors were enrolled in a prospective manner at Geneva University Hospital between December 2020 and April 2022. We monitored TCR repertoire and NK/T-cell compartment restoration in longitudinal blood samples drawn at 3 (t3), 6 (t6), 9 (t9), and 12 (t12) months after HSCT from recipients and from their respective donors before HSCT (t0). We performed targeted TCR CDR3β chain high-throughput sequencing on 254 samples and analyzed overall 2 710 450 productive TCR rearrangements after correcting for nonproductive CDR3aa sequences. To interrogate temporal changes in the NK- and T-cell compartments after HSCT, we designed and applied a high-dimensional antibody panel targeting 35 surface and intracellular markers on 228 samples derived from 47 recipients (supplemental Figure 1A). In a subset of recipients (n = 24), we additionally performed CD107a in vitro assay to interrogate NK-cell functional competence.
Recipient’s TCR repertoire is subject to long-lasting restricted diversity after transplantation
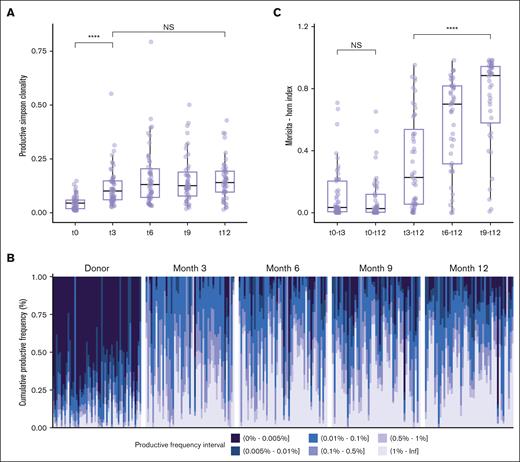
In the first place, we aimed to establish a comprehensive view of TCR repertoire reconstitution dynamics during the first year after transplantation with respect to its diversity, clone organization, and composition. The recipient’s TCR repertoires showed a significant increase in clonality (ie, reduced diversity) over the first 3 months, which remained stable at subsequent time points (P = 1.157e-08 between t0 and t3; P = .076 between t3 and t12; Figure 1A). Despite the clonality being stable, the different clone size categories stratified according to their productive frequency were subject to changes after transplantation, although the repertoire during the first 3 months was almost exclusively dominated by expanded clones, a progressive significant increase in nonexpanded clones (1-2 template) was apparent upon 6 months and lasted until 12 months (P = 1.5e-06) (Figure 1B). To better understand the evolution of TCR composition with time after HSCT, we computed the TCR overlap between any 2 time points before and after transplantation on an intraindividual level, considering abundance and clone identity. Donor-recipient overlap indices were low and did not increase during the entire follow-up time until 1 year after transplantation (P = .566) suggesting no further major donor-derived clone expansion (Figure 1C). The overlap between 3 and 12 months after transplantation was low (median, 0.228; IQR, 0.0556-0.537) and significantly increased from 6 months onward, with the highest overlap being between 9 and 12 months (median, 0.885; IQR, 0.579-0.943; P = 2.986e-06 between t3-t12 and t9-t12), indicating that the major clone size hierarchy was set up at an early time after transplantation, whereas the TCR composition turnover and qualitative reconstitution stabilized over time (Figure 1C).
TCR diversity restricted during the first year after HSCT. (A) Evolution of TCR-productive Simpson clonality before allo-HSCT (t0, n = 53) in donors and at serial time points t3 (n = 54), t6 (n = 52), t9 (n = 47), and t12 (n = 48) after allo-HSCT in recipients. (B) Fractal clonal size organization defined by the productive frequency of clones at the indicated time points (t0, n = 53; t3, n = 54; t6, n = 52; t9, n = 47; and t12, n = 48). Each bar represents a single individual. The color-coded legend bar represents stratification according to the productive frequency of the individual clone. (C) TCR repertoire overlaps between donor and recipient and between post-HSCT time points in recipients calculated using the Morisita-Horn index. Morisita-Horn indices vary between 0 (no overlap) and 1 (complete overlap) and are represented along the y-axis. Box plots display medians and IQRs, with whiskers representing 1.5× IQR. Wilcoxon rank-sum test with false discovery rate correction in panel A. Kruskal-Wallis test and post hoc Dunn test in panel C. All P values were 2-sided. Statistical thresholds: ∗∗∗∗P < .0001. NS, not significant.
TCR diversity restricted during the first year after HSCT. (A) Evolution of TCR-productive Simpson clonality before allo-HSCT (t0, n = 53) in donors and at serial time points t3 (n = 54), t6 (n = 52), t9 (n = 47), and t12 (n = 48) after allo-HSCT in recipients. (B) Fractal clonal size organization defined by the productive frequency of clones at the indicated time points (t0, n = 53; t3, n = 54; t6, n = 52; t9, n = 47; and t12, n = 48). Each bar represents a single individual. The color-coded legend bar represents stratification according to the productive frequency of the individual clone. (C) TCR repertoire overlaps between donor and recipient and between post-HSCT time points in recipients calculated using the Morisita-Horn index. Morisita-Horn indices vary between 0 (no overlap) and 1 (complete overlap) and are represented along the y-axis. Box plots display medians and IQRs, with whiskers representing 1.5× IQR. Wilcoxon rank-sum test with false discovery rate correction in panel A. Kruskal-Wallis test and post hoc Dunn test in panel C. All P values were 2-sided. Statistical thresholds: ∗∗∗∗P < .0001. NS, not significant.
CMV reactivation drives a strong TCR immune response
Although genetics partly predicts the TCR repertoire state after transplantation, extrinsic factors play an important role in shaping immune states.19 To interrogate their effects, we stratified our cohort into 3 distinct groups with respect to the CMV serostatus of the recipient20: CMV seronegative (D–/R– and D+/R–, n = 20), CMV seropositive but no documented reactivation (D–/R+ and D+/R+, n = 21), and whether they were subject to a CMV reactivation (D–/R+ and D+/R+, n = 13) (supplemental Tables 2 and 3). Only recipients who were positive for CMV reactivated after transplantation and displayed CMV reactivation up to 9 months after transplantation (supplemental Figure 1B).
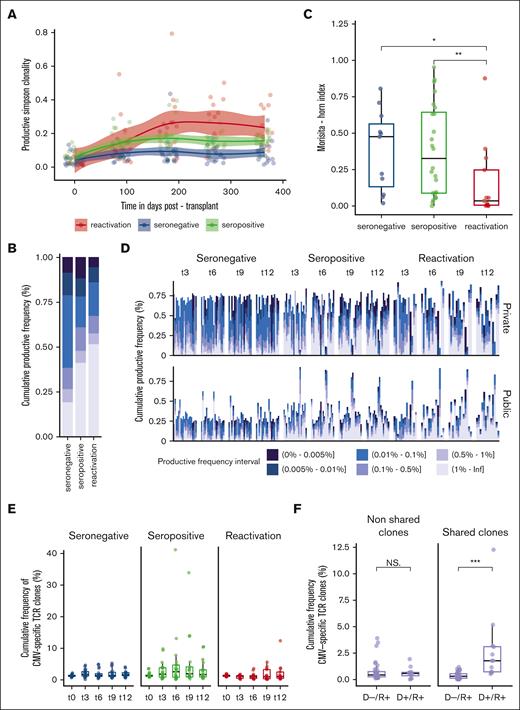
CMV seronegative and seropositive recipients displayed a posttransplantation repertoire stable in terms of diversity (P = .87 and .65, respectively), as compared with CMV seropositive recipients subject to a CMV reactivation episode who experienced a significant increase in clonality (P = 4.07e-02) (Figure 2A; supplemental Figure 7). In CMV-reactivated recipients at 12 months, this shift in TCR repertoire clonality was dominated by a large-scale clone expansion of >1% (Figure 2B). Representative reconstitution trajectories of the selected recipients are provided in supplemental Figure 8. The overlap between the 3 and 12 months was the lowest in recipients with CMV reactivation (median, 0.035; IQR, 0.005-0.247; P = .013 between CMV seronegative and reactivated), likely reflecting a rapid turnover of the TCR repertoire with major changes in the clone composition and expansion of competitive T-cell clones (Figure 2C). In light of such differential reconstitution trajectories, we thought to interrogate the origin and the presence of distinct molecular features associated with the presence of CMV. CMV-reactivated recipients showed an incremental and significant increase in the cumulative frequency of hyperexpanded (>1%) public clones (P = .046) between the 3 and 12 months as compared with CMV seronegative and seropositive recipients (P = .85 and .945, respectively) (Figure 2D). To assess immune response specificity, we queried our data set against public databases encompassing 20 018 experimentally annotated CMV-specific TCR clones. All recipients had a good capacity to present with CMV peptides according to their HLA class I restriction. We did not observe any significant enrichment in the cumulative frequency of CMV-specific clones after CMV reactivation compared with CMV seronegative or seropositive recipients (P = .687 at t12) (Figure 2E). We further extended the query to all pathogen-associated clones; however, no significant increase in the cumulative frequency of potential cross-reactive clones was noted (supplemental Figure 3). However, under the assumption of transferred immunity, we observed a significant enrichment in the cumulative frequency of CMV-specific TCR clones shared between the donor and recipient after transplantation in D+/R+ (n = 4) compared with D–/R+ CMV-reactivated recipients (n = 9) (P = 2.039e-06) (Figure 2F; supplemental Figure 4A). The cumulative frequency of shared anti-CMV clones in CMV-mismatched nonreactivated recipients was comparable to that in CMV-matched nonreactivated recipients (P > .05) (supplemental Figure 4B). In line with this, we found that there was a significant contribution of the CMV-specific TCR clone’s productive frequency on the clonality in D+/R+ reactivated recipients (P = 6.89e-07, linear regression), whereas the effect was nonsignificant on the clonality in D–/R+ CMV reactivated recipients (P = .443, linear regression).
TCR repertoire restoration in relation to CMV serostatus and infection/reactivation. Stratification according to the following groups: seronegative (n = 20), seropositive (n = 21), and reactivated (n = 13) CMV recipients applies to plots (A-E). (A) Evolution of TCR-productive Simpson clonality at serial time points after HSCT fitted using LOESS regression with a 95% confidence interval. (B) Productive frequency distribution at 12 months after HSCT with respect to groups. (C) TCR repertoire overlap computed using the Morisita-Horn index between 3 and 12 months after HSCT. Morisita-Horn indices vary between 0 (no overlap) and 1 (complete overlap) and are represented along the y-axis. (D) Private (upper panel) (ie, observed in only 1 D/R pair) and public (lower panel) (ie, observed in ≥2 D/R pairs or matching clonotypes present in public databases with antigen-specific validated TCRs) fractal clonal size organization at serial time points after HSCT. Each bar represents a single individual. The color-coded legend bar represents the stratification according to the individual clone’s productive frequency. (E) Cumulative productive frequency of CMV–specific TCR clones identified by in silico matching with public databases at serial time points after HSCT. (F) Cumulative productive frequency of donor-recipient nonshared and shared (ie, between each D/R pair) CMV-specific TCR clones in D–/R+ (n = 9) and D+/R+ (n = 4) CMV reactivated recipients at all time points combined. The cumulative frequencies at each time point are detailed in supplemental Figure 4A. Box plots display medians and IQRs, with whiskers representing 1.5× IQR. Wilcoxon-rank sum test for pairwise comparisons in panels B,F. All P values were 2-sided. Statistical thresholds: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
TCR repertoire restoration in relation to CMV serostatus and infection/reactivation. Stratification according to the following groups: seronegative (n = 20), seropositive (n = 21), and reactivated (n = 13) CMV recipients applies to plots (A-E). (A) Evolution of TCR-productive Simpson clonality at serial time points after HSCT fitted using LOESS regression with a 95% confidence interval. (B) Productive frequency distribution at 12 months after HSCT with respect to groups. (C) TCR repertoire overlap computed using the Morisita-Horn index between 3 and 12 months after HSCT. Morisita-Horn indices vary between 0 (no overlap) and 1 (complete overlap) and are represented along the y-axis. (D) Private (upper panel) (ie, observed in only 1 D/R pair) and public (lower panel) (ie, observed in ≥2 D/R pairs or matching clonotypes present in public databases with antigen-specific validated TCRs) fractal clonal size organization at serial time points after HSCT. Each bar represents a single individual. The color-coded legend bar represents the stratification according to the individual clone’s productive frequency. (E) Cumulative productive frequency of CMV–specific TCR clones identified by in silico matching with public databases at serial time points after HSCT. (F) Cumulative productive frequency of donor-recipient nonshared and shared (ie, between each D/R pair) CMV-specific TCR clones in D–/R+ (n = 9) and D+/R+ (n = 4) CMV reactivated recipients at all time points combined. The cumulative frequencies at each time point are detailed in supplemental Figure 4A. Box plots display medians and IQRs, with whiskers representing 1.5× IQR. Wilcoxon-rank sum test for pairwise comparisons in panels B,F. All P values were 2-sided. Statistical thresholds: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
We analyzed the impact on TCR repertoire reconstitution according to the presence of posttransplant cyclophosphamide (PTCy), GVHD, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with minor noticeable changes (supplemental Table 5).
Dynamic immune cell regeneration during the first year after transplantation
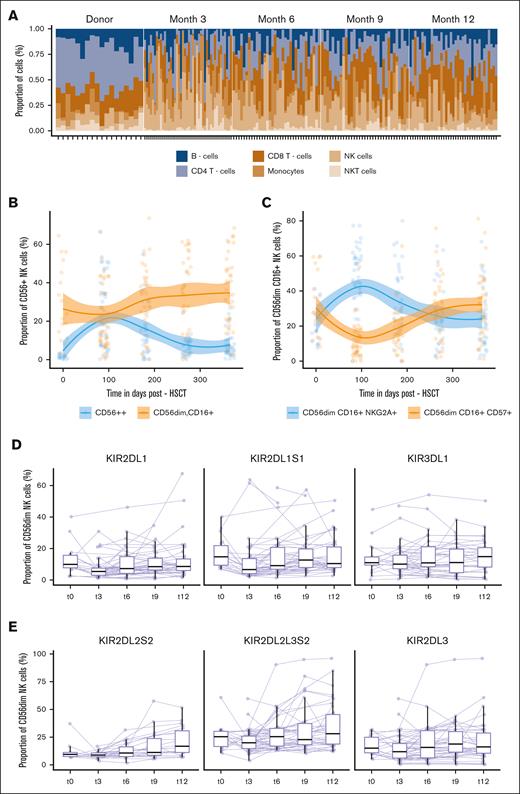
Alongside TCR repertoire assessment, we systematically assessed NK- and T-cell compartments and their recreation at the phenotypic level based on a 35-color antibody panel. This global view showed differential recovery patterns for the regeneration of the major canonical cell subsets: at 3 months, the immune compartment was dominated by NK cells and monocytes, with a descendant kinetic up to 12 months. NK cells were significantly enriched compared with their donor counterparts (P = .018; supplemental Table 6). CD4+ and CD8+ T cells were significantly reduced early after transplantation (P = 7.5e-06; P = .024) and slowly recovered up to 12 months (P = 4.4e-05; P = 8.1e-05) (Figure 3A; supplemental Table 6). The frequency of circulating B cells was subject to a gradual increase from 3 months up to 12 months, without reaching statistical significance (P = .11) (Figure 3A). CD56bright NK cells expanded transiently after transplantation, significantly outnumbering donor values (P = 7.1e-07), and were subject to a sharp significant decrease from 3 months (P = 8.4e-06), with a progressive conversion toward NK-cell maturation, reflected by a significant increase in CD56dim CD16+ NK cells (P = .0094) (Figure 3B).
Immune-cell composition during the first year after HSCT. (A) Relative proportions of 6 major cell populations: CD8+ and CD4+ T cells, NK T cell, NK cell, monocytes, and B cells assessed at the indicated time points after HSCT: t0 (n = 18), t3 (n = 44), t6 (n = 35), t9 (n = 32), and t12 (n = 37). Each stacked bar represents an individual. (B) Evolution of the frequency of CD56bright, CD56dim, CD16+ NK cells at indicated time points after HSCT: t0 (n = 18), t3 (n = 44), t6 (n = 35), t9 (n = 32), and t12 (n = 37) fitted using LOESS regression with a 95% confidence interval. Day 0 indicates a pre-HSCT state of the donor. Color-coded lines represent linear regression according to cell subset. (C) Evolution of the frequency of CD56dim, CD16+, NKG2A+, CD57neg and CD56dim, CD16+, NKG2Aneg, and CD57+ NK cells at the indicated time points after HSCT: t0 (n = 18), t3 (n = 44), t6 (n = 35), t9 (n = 32), and t12 (n = 37) fitted using LOESS regression with a 95% confidence interval. Day 0 indicates a pre-HSCT state of the donor. Color-coded lines represent linear regression according to the cell subset. (D, E) Proportion of KIR+ CD56dim NK cell at the indicated time points after HSCT: t0 (n = 18), t3 (n = 44), t6 (n = 35), t9 (n = 32), and t12 (n = 37). The lines connect paired samples. Box plots display medians and IQRs, with whiskers representing 1.5× IQR.
Immune-cell composition during the first year after HSCT. (A) Relative proportions of 6 major cell populations: CD8+ and CD4+ T cells, NK T cell, NK cell, monocytes, and B cells assessed at the indicated time points after HSCT: t0 (n = 18), t3 (n = 44), t6 (n = 35), t9 (n = 32), and t12 (n = 37). Each stacked bar represents an individual. (B) Evolution of the frequency of CD56bright, CD56dim, CD16+ NK cells at indicated time points after HSCT: t0 (n = 18), t3 (n = 44), t6 (n = 35), t9 (n = 32), and t12 (n = 37) fitted using LOESS regression with a 95% confidence interval. Day 0 indicates a pre-HSCT state of the donor. Color-coded lines represent linear regression according to cell subset. (C) Evolution of the frequency of CD56dim, CD16+, NKG2A+, CD57neg and CD56dim, CD16+, NKG2Aneg, and CD57+ NK cells at the indicated time points after HSCT: t0 (n = 18), t3 (n = 44), t6 (n = 35), t9 (n = 32), and t12 (n = 37) fitted using LOESS regression with a 95% confidence interval. Day 0 indicates a pre-HSCT state of the donor. Color-coded lines represent linear regression according to the cell subset. (D, E) Proportion of KIR+ CD56dim NK cell at the indicated time points after HSCT: t0 (n = 18), t3 (n = 44), t6 (n = 35), t9 (n = 32), and t12 (n = 37). The lines connect paired samples. Box plots display medians and IQRs, with whiskers representing 1.5× IQR.
We then focused on the NK-cell repertoire using manual serial gating to profile the expression level and frequency of each functional marker in total CD56+, CD56dim, and CD56bright NK cells. There was significant enrichment of CD56dim CD16+ CD57+ (P = 2.0e-07) NK cells at the expense of CD56dim CD16+ NKG2A+ NK cells (P = 1.1e-05) (Figure 3C). This equilibrium stabilized at 9 months, with no further significant changes in either subset (P = .705; P = .755) (Figure 3C). No significant changes were noted in terms of the frequency of NKp30, NKp46, NKp44, TIGIT, PD-1, and NKG2D expressing CD56dim NK cells between 3 and 12 months (supplemental Figure 5A). The maturation and functional competence of NK cells are tightly regulated by the interaction between inhibitory KIRs and their HLA-I ligand, dictating their cytotoxic strength.21 Given the interindividual differences in the KIR gene content,22 we performed high-resolution KIR genotyping to accurately assess which KIRs could be expressed. Although HLA-B–specific KIRs showed stable behavior in the posttransplantation course (P = .97), HLA-C–specific inhibitory KIRs displayed differential reconstitution kinetics: KIR2DL1+ subsets were significantly downregulated compared with the donors (P = .018) and displayed a progressive reconstitution up to 12 months (P = .017) (Figure 3D). KIR2DL2L3S2+ subsets showed a similar frequency to the donors early after transplantation and showed a significant increase during the time course of 1 year after transplantation (P = .022), specifically the KIR2DL2+S2+ subsets (P = .0018) (Figure 3E). The proportion of KIR2DL5+, KIR2DS4+, KIR2DS1+, and KIR3DS1+ cells did not change across all the observed time points (P > .05) (supplemental Figure 5B).
CMV reactivation induces an accelerated maturation of NK-cell subsets
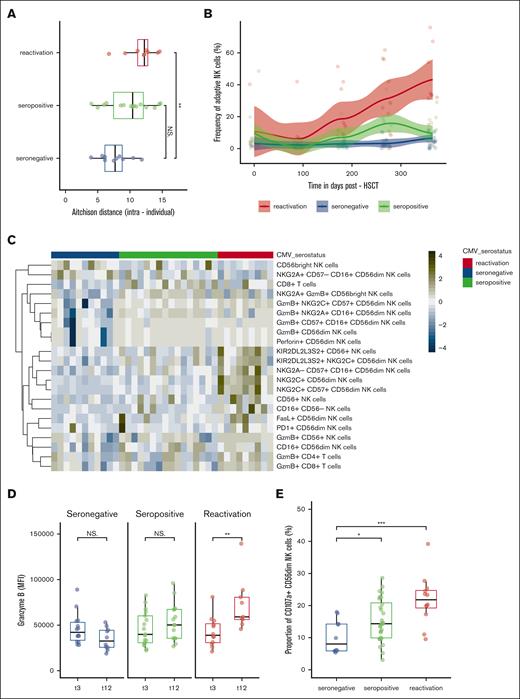
The presence of CMV has been shown to be also a decisive regulator of the NK-cell immune state.10 To assess whether these groups were subject to a divergent reconstitution trajectory, we first calculated Aitchison distance, measuring the difference in the cell composition based on 62 manually gated cell subsets. Intraindividual distances between 3 and 12 months were significantly higher in CMV reactivation recipients (P = .006) reflecting major changes in the NK- and T-cell immune state composition of CMV-reactivated recipients (Figure 4A). This observation was firstly driven by a significant increase in adaptive NK cells coexpressing NKG2C and CD57, which were associated with the presence of CMV (P = 1.97e-05) (Figure 4B). CMV seropositivity had a higher frequency of total CD8+ T cells than CMV-reactivated recipients at 3 months (P = .021). At 12 months, there was a significant enrichment in CMV reactivated recipients in circulating CD56dim NK cells expressing mature and cytolytic markers, such as CD16+, CD57+, granzyme B, perforin and FasL, CD56neg CD16pos unconventional NK cells and granzyme B expressing CD8+ and CD4+ T cells. The data also revealed a higher rate of CD56dim expression of the checkpoint inhibitory marker PD-1 in CMV-reactivated recipients (P = 4.60e-02) (Figure 4C). Finally, the frequency of KIR2DL2L3S2+ CD56dim NKG2C+ NK cells was significantly tuned upwards (P = .01) in CMV-reactivated recipients in contrast to KIR2DL1+ and KIR3DL1+ CD56dim NKG2C+ NK cells and showed an incremental increase in granzyme B content over time (P = .0033) (Figure 4D). In comparison, the NK-cell compartment in CMV seronegative recipients was dominated by immature CD56bright NK cells and NKG2A+-expressing CD56dim and CD56bright NK cells (Figure 4C). There were no differences in the frequency of NKp30, NKp44, NKp46, TIGIT, NKG2D, and TRAIL-expressing CD56dim NK cells (P > .05) (data not shown). Representative reconstitution trajectories of selected recipients are provided in supplemental Figure 8.
NK-cell compartment restoration in relation to CMV serostatus and infection/reactivation. Stratification according to the following groups: seronegative (n = 17), seropositive (n = 18), and reactivated (n = 12) CMV recipients applies to plots (A-E). (A) Intraindividual distance (Aitchison distance) between 3 and 12 months after HSCT among stratified groups based on 62 manually gated cell frequencies. (B) Evolution of the CD57+, NKG2C+ CD56dim adaptive NK cells at 1-year follow-up after HSCT stratified among the groups fitted using LOESS regression with a 95% confidence interval. Color-coded lines represent linear regression according to the cell subset. (C) Heat map of the frequency of cell subsets at 12 months that were statistically different between CMV seronegative (green) and CMV reactivated (blue) recipients. Heat map rows display cell subsets and are colored by the z score normalized per row. Each column represents the recipient. (D) Granzyme B expression (MFI) in KIR2DL2L3S2+ NKG2C+ CD56dim NK cells among stratified groups at 3 and 12 months after HSCT. (E) Frequency of CD107a+ CD56dim NK cells in stratified groups: seronegative (n = 17), seropositive (n = 18), and reactivated (n = 12). Box plots display medians and IQRs, with whiskers representing 1.5× IQR. Wilcoxon rank-sum test in panels A,C-E. All P values were 2-sided. Statistical thresholds: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. MFI, mean flurorescence intensity.
NK-cell compartment restoration in relation to CMV serostatus and infection/reactivation. Stratification according to the following groups: seronegative (n = 17), seropositive (n = 18), and reactivated (n = 12) CMV recipients applies to plots (A-E). (A) Intraindividual distance (Aitchison distance) between 3 and 12 months after HSCT among stratified groups based on 62 manually gated cell frequencies. (B) Evolution of the CD57+, NKG2C+ CD56dim adaptive NK cells at 1-year follow-up after HSCT stratified among the groups fitted using LOESS regression with a 95% confidence interval. Color-coded lines represent linear regression according to the cell subset. (C) Heat map of the frequency of cell subsets at 12 months that were statistically different between CMV seronegative (green) and CMV reactivated (blue) recipients. Heat map rows display cell subsets and are colored by the z score normalized per row. Each column represents the recipient. (D) Granzyme B expression (MFI) in KIR2DL2L3S2+ NKG2C+ CD56dim NK cells among stratified groups at 3 and 12 months after HSCT. (E) Frequency of CD107a+ CD56dim NK cells in stratified groups: seronegative (n = 17), seropositive (n = 18), and reactivated (n = 12). Box plots display medians and IQRs, with whiskers representing 1.5× IQR. Wilcoxon rank-sum test in panels A,C-E. All P values were 2-sided. Statistical thresholds: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. MFI, mean flurorescence intensity.
We finally interrogated the functional competence of NK cells using a stimulation assay, coculturing NK cells against HLA-I deficient cancer cell lines (K562). Among 62 stimulated samples, we assessed the CD107a degranulation response in manually gated CD56dim NK cells (supplemental Figure 6A-B). CMV seropositive and reactivated recipients produced a higher frequency of CD107a+ CD56dim NK cells after stimulation compared with seronegative recipients after transplantation. These differences were statistically significant (P = .045 and 6e-04, respectively) (Figure 4E).
We analyzed the impact on NK-cell repertoire reconstitution according to the presence of PTCy, GVHD, and SARS-CoV-2 with minor changes (supplemental Table 5).
Discussion
Allo-HSCT represents a unique case study to understand immune ontogeny and its behavior under external nongenetic influences. In line with our previous findings and those of others, we confirmed a significant contraction in TCR repertoire diversity after transplantation.5,6,23-25 Our data further revealed remarkable stability in clonality over a 1-year follow-up, suggesting that the clone size hierarchy is established at a very early stage after transplantation and does not fluctuate significantly over distinct immunomodulatory stages. Supportive data from mathematical modeling in healthy individuals profiled that adaptation of these systems early in life involves a rapid and long-lasting expansion of TCR clones as a direct response to the lymphopenic environment with reduced clonal competition.26 The reappearance of nonexpanded clones at later stages after transplantation in our data likely suggests a progressive increase in peripheral clonal competition and stabilization in quantitative homeostatic turnover.
Although studies have demonstrated the influential effect of many factors, such as the use of PTCy,23,27 we believe based on our data that CMV reactivation has a dominant immunomodulatory effect. This is reflected by the ability to enhance TCR clonality and induce compositional changes with a long-lasting effect.5,28 Unexpectedly, we were unable to assess an enhanced frequency of CMV-specific or other antigen-experienced TCR clones in CMV-reactivated recipients based on in silico matching. Although undeniably these public databases lack completeness, inducing a bioinformatic bias in detecting antigen-specific TCR clones, several biological explanations might account for this observation: there are supportive data demonstrating a reduced thymic activity after CMV infection as a consequence of damaging CD8+ T-cell infiltration.29,30 The observation is further supported by the reduced presence of circulating T-cell receptor expansion circles after CMV reactivation.31 Thus, we hypothesized that a reduced thymic output associated with or as a consequence of CMV reactivation might lead to an even greater peripheral homeostatic proliferation of circulating clones as a compensatory mechanism. The absence of CMV-specific T cells could be explained by the copresence of GVHD, which has been shown to impair major histocompatibility complex-I and major histocompatibility complex-II processes by dendritic cells, thus corrupting efficient CMV-specific T-cell priming.32 Alternatively, the proliferation of nonspecific T cells could be induced as a bystander effect after cytokine release or based on the fitness of nonspecific, unexperienced T-cell clones.30 Less likely, we might attribute this effect to a broader feature of the adaptive immune system, as clonal unspecific expansion has been reported in other settings, that is, the B-cell receptor repertoire after a SARS-CoV-2 infection.33
Similar to the TCR repertoire, the NK-cell repertoire showed a substantially high level of reconstitution at 3 months after HSCT. Our observation of the differential reconstitution trajectory of KIRs along the posttransplantation time course mirrors previous findings.34-36 The expansion of adaptive NK cells has been observed in recipients with CMV reactivation.8,9,37 Our data further point toward long-lasting immune reconstitution skewing in terms of highly mature, cytolytic, and functional NK- and T-cell subsets in CMV-reactivated recipients, reflecting the highly activated immune environment after a CMV. Specifically, we first showed a bias toward KIR-mediated reactivity with the increase in KIR2DL2L3S2+ adaptive NK cells concomitant with a reduced pool of NKG2A-expressing NK cells. Horowitz et al showed a concomitant increase in HLA-C expression on monocytes and myeloid lineages alongside CMV reactivation, likely related to the upregulation of immunomodulatory cytokines, such as interferon gamma and tumor necrosis factor α.38 The developing nature of the NK-cell compartment early after HSCT could be particularly sensitive to changes in the HLA environment and substantiate questions about the contribution of HLA-C ligands to accelerated and enhanced NK-cell licensing, especially given the fact that dendritic cells have been shown to be important mediators of NK-cell education.38 Supportive data show that donor and recipient immunogenetic backgrounds play a role: the magnitude of single KIR2DL2L3+ NKG2C cell expansion has been shown to be rendered in donors and recipients bearing the C1/C1 and C1C1Bw4 HLA genotypes.39 A recent genetic study further demonstrated the protective effect of KIR cenB02 motifs encompassing KIR2DL2 and KIR2DS2 on relapse risk.40 In contrast, data on the impact of CMV infection in healthy individuals have shown an upward tuning of KIR2DL1 on adaptive NK cells, which we could not confirm in our cohort and might question the implication of KIR/HLA mismatch with regard to the KIR2DL1 locus.11
Several groups have reported that skewing NK-cell repertoire into a maturation status promotes the graft-versus-leukemia effect, with CD57+ NKG2C+ NK cells being acknowledged as the main cell contributor to this observation independently of the cell source origin.41-44 Based on our findings showing that CMV–induced immune changes were still present at least 3 months after CMV reactivation, it remains to be examined whether these cells undergo changes in their functional competence and phenotype, as we observed an increase in NK cells expressing exhaustion markers, such as PD-1. Indeed, single-cell proteomics has similarly highlighted an enrichment in the expression of PD-1 and LAG-3 in adaptive NK cells, suggesting progressive exhaustion and dysfunction over time.45 In this regard, we might interrogate the presence of a certain immunological time window in which the graft-versus-leukemia effect might reach its maximum and subsequently vanish.
Finally, although our study questions the impact of clinical events on immune system reconfiguration, it remains a difficult task to fully disentangle the cause and consequence, which we cannot fully distinguish in our study. Our data revealed an enhanced expansion of shared CMV–specific T cells in CMV–matched reactivated recipients in contrast to CMV–mismatched reactivated recipients. Thus, from a clinical point of view, consideration of CMV serostatus would be key in predicting immune response, as intrinsic donor’s graft immunity could be “transferred” to the recipient with CMV–specific immune cells undergoing peripheral homeostatic proliferation and contributing to protective CMV-specific immunity. In addition, most CMV seropositive recipients without reactivation showed no enhanced frequency of adaptive memory–like NK cells, suggesting that memory CMV–specific T-cell clones are sufficient for first-line protection against further viral replication, whereas adaptive NK cells are required to unleash NK-cell antiviral activity in acute manifested CMV replication.46 These observations could also indicate that patients at high risk for primary infection might benefit from the infusion of third-party CMV–specific CD8+ T cells or in case of refractory severe CMV infection, as it is performed in several centers.47,48
This study has several limitations. The limited number of patients does not give us sufficient power to rule out the effect of other codependent nongenetic influences, such as immunosuppressive therapy and specific infections, in a small subgroup of patients. The restricted number of identified specific antigenic TCR epitopes limited our in silico matching and prevented us from ruling out the possibility that the proliferation of some T-cell clones that are considered bystanders could have specificity. Finally, despite high-resolution KIR genotyping, KIR phenotyping cannot represent the real repertoire because of the limited number of antibodies that cover KIR proteins at the NK-cell surface.
In summary, our data indicated that the T- and NK-cell repertoire reconstitution after allo-HSCT is majorly established during the first 3 months and seems to be driven to a large part by the CMV serostatus and CMV reactivation and is persistent for at least 1 year.
Acknowledgments
The authors are grateful to the technicians of the National Reference Laboratory for Histocompatibility for their most efficient support for HLA genotyping.
This study was supported by the Academic Society of Geneva, International Research Group on unrelated Hematopoietic stem cell Transplantation, Henri Dubois-Ferrière Dinu Lippatti Foundation, and Philanthropy Settlement. A.S. was supported by a grant from the Fondation de Reuter. P.J.N. was supported by National Institutes of Health, University of Colorado School of Medicine (Aurora, CO) grant U01AI090905.
Authorship
Contribution: J.V. designed the study; A.S., M.-P.H., and Z.C.S. performed the experiments; A.S. acquired and analyzed the data and performed the statistical analysis; A.S. and J.V. wrote and drafted the manuscript; and all authors contributed to manuscript revision and read and approved the submitted version.
Conflict-of-interest disclosure: Y.C. reports consulting fees from Merck Sharp & Dohme (MSD), Novartis, Incyte, Bristol Myers Squibb, Pfizer, AbbVie, Roche, Jazz, Gilead, Amgen, AstraZeneca, and Servier; and travel support from MSD, Roche, Gilead, Amgen, Incyte, AbbVie, Janssen, AstraZeneca, Jazz, and Sanofi, all via the institution. The remaining authors declare no competing financial interests.
Correspondence: Jean Villard, Transplantation Immunology Unit and National Reference Laboratory for Histocompatibility, Geneva University Hospitals, Gabrielle-Perret-Gentil 4, 1205 Geneva, Switzerland; email: jean.villard@hcuge.ch.
References
Author notes
The data sets used and/or analyzed during the study are available on reasonable request from the corresponding author, Jean Villard (jean.villard@hcuge.ch).
The data set for the T-cell receptor sequencing generated during and/or analyzed during the current study is available in the Yareta repository at https://doi.org/10.26037/yareta:uluquhaq7zf4tb2lxbxbkvvzse.
The full-text version of this article contains a data supplement.