TO THE EDITOR:
Myeloproliferative neoplasms (MPNs) are chronic bone marrow malignancies characterized by clonal proliferation of hematopoietic precursors and elevated cell counts in peripheral blood.1 Patients with MPN are at risk of progression to myelofibrosis or acute leukemia and experience a substantial burden of microvascular symptoms.2,3 However, thrombosis (both arterial and venous) represents the leading cause of morbidity and mortality for patients with polycythemia vera (PV) and essential thrombocythemia (ET).4-6
Translational studies have indicated that the platelet proteome influences pathways relating to immune response, inflammation, and malignancy.7,8 Thrombocytosis and platelet hyperactivity are hallmarks of MPN;9 however, platelet count in isolation is not predictive of clinical outcome, and conventional antiplatelet therapy does not fully mitigate thrombotic risk.10 A comprehensive picture of the MPN platelet molecular profile is lacking, and to date, no studies have evaluated the unbiased platelet proteome in a sizable clinical cohort of affected patients. Here, we performed untargeted quantitative profiling of the platelet proteome in a large (n = 140) cohort of patients with PV and ET.
Using standardized platelet isolation protocols (supplemental Methods), we prepared purified platelets from peripheral blood samples of patients with an established diagnosis of MPN (World Health Organization defined, n = 59 ET, n = 41 PV) and a cohort of healthy controls (n = 40) recruited across 2 sites: Hospital Papa Giovanni XXIII, Bergamo, Italy and Mater Misericordiae University Hospital, Dublin, Ireland. Pertinent clinical features are shown in Figure 1 (and listed in supplemental Table 1). Interpatient variability, including age, sex, and treatment, as well as experimental batch effects, were adjusted as confounding factors in downstream expression analyses (supplemental Methods). Focusing on the most prothrombotic subtypes of MPNs, we hypothesized that the platelet proteome differs in MPN, and its characterization would offer insights into the underlying pathobiology and possible mechanisms underlying the associated clinical complications.
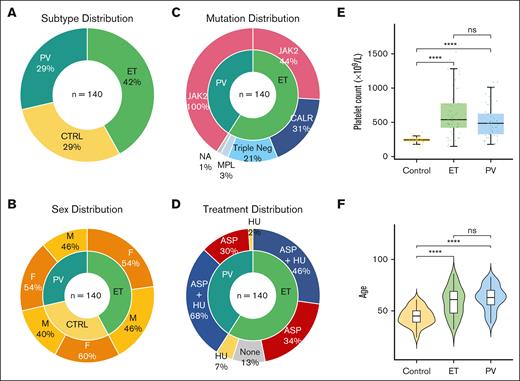
Clinical demographics of patients with MPN and healthy controls. (A) Similarity in distribution of MPN subtypes and controls, with slightly higher proportion ET (PV n = 41, ET n = 59, control n = 40). (B) Comparable and balanced distribution of sex across MPN subtypes and controls. Larger percentage of female healthy controls. (C) All patients with PV harbored the JAK2 V617F mutation, and in keeping with the general ET population JAK2 V617F was the most common driver mutation followed by CALR and MPL, with 12 patients with triple-negative ET included in this study. (D) MPN patient therapies reflecting current clinical practice. Most patients with PV and ET were prescribed aspirin (ASA), with hydroxyurea (HU) as a commonly used cytoreductive therapy. To control for any interpatient variability, all treatment, in addition to patient, sex and experimental batch are adjusted as confounding factors in downstream differential expression analyses. (E) Comparable distribution of age across MPN subtypes and controls. Violin plots of patient age from each MPN subtype reflect clinical expectation, with slightly higher median age noted for patients with ET and PV than that for controls. (F) Platelet counts, as box plots, measured at the same date and time as experimental platelet sampling. As expected, Mann-Whitney U tests marked by asterisks indicate a statistically significant difference between control and MPN groups (∗∗∗∗P ≤ 0.0001; ns, not significant).
Clinical demographics of patients with MPN and healthy controls. (A) Similarity in distribution of MPN subtypes and controls, with slightly higher proportion ET (PV n = 41, ET n = 59, control n = 40). (B) Comparable and balanced distribution of sex across MPN subtypes and controls. Larger percentage of female healthy controls. (C) All patients with PV harbored the JAK2 V617F mutation, and in keeping with the general ET population JAK2 V617F was the most common driver mutation followed by CALR and MPL, with 12 patients with triple-negative ET included in this study. (D) MPN patient therapies reflecting current clinical practice. Most patients with PV and ET were prescribed aspirin (ASA), with hydroxyurea (HU) as a commonly used cytoreductive therapy. To control for any interpatient variability, all treatment, in addition to patient, sex and experimental batch are adjusted as confounding factors in downstream differential expression analyses. (E) Comparable distribution of age across MPN subtypes and controls. Violin plots of patient age from each MPN subtype reflect clinical expectation, with slightly higher median age noted for patients with ET and PV than that for controls. (F) Platelet counts, as box plots, measured at the same date and time as experimental platelet sampling. As expected, Mann-Whitney U tests marked by asterisks indicate a statistically significant difference between control and MPN groups (∗∗∗∗P ≤ 0.0001; ns, not significant).
Using label-free quantification liquid chromatography mass spectrometry (supplemental Methods), we compared proteomic expression in PV and ET with that of healthy donors. Unsupervised principal component analysis of patients with MPN and controls (Figure 2A) confirmed that the collective variability from the first 2 principal components was MPN disease status (26% of total variance). A total of 1952 platelet proteins (listed in supplemental Tables 3 and 4) were quantified across MPN and control samples. Differential expression analysis of proteomic data (volcano plot, Figure 2B-C) resulted in highly significant expression signatures (false discovery rate <0.05) with 227 proteins differentially regulated in ET (113 increased and 114 decreased) and 166 in PV (122 increased and 44 decreased) compared with healthy donors (see supplemental Tables 5 and 6 for full list).
MPN platelet proteome distinguishes disease phenotype. (A) Unsupervised principal component analysis of normalized platelet protein expression adjusted for age, sex, treatment (antiplatelet and cytoreduction), and experimental batch. PC1 and PC2 colored by MPN subtype; and each contrasted with controls (n = 40, yellow): ET (n = 59, light green), PV (n = 41, blue). The first 2 principal components account for 26% of total variance in the data. (B-C) Volcano plots (2 panels of ET, PV) of differential protein expression showing log2 fold change vs statistical significance (negative log10 of P values) of each gene. Significant upregulated and downregulated genes are those with P values (false discovery rate [FDR]) ≤.05 and absolute value of fold changes ≥1.5. (D) Hierarchically clustered heat map of the top 10 differentially expressed proteins (FDR <0.01) from control vs MPN patient samples. Colored annotation is provided to indicate MPN subtype, mutation status, and sex. Rows indicate gradation in expression on a yellow (low) to orange (high) scale. Columns indicate sample type from controls, ET, and PV. (E) Pathway-enrichment analysis of proteins with MPN subtype–specific expression (color indicated; yellow ET, and light green PV). Each point represents a pathway; the x-axis gives the normalized enrichment score, which reflects the degree to which each pathway is overrepresented at the top of the ranked list of differentially expressed proteins, normalized to account for differences in gene set size and in correlations between gene sets and the expression data set. The y-axis lists the detail-level node of the most enriched pathways; solid lines mark gene set enrichment analysis–recommended11 Bonferroni-corrected statistical significance criterion of FDR <0.25 for exploratory analyses.
MPN platelet proteome distinguishes disease phenotype. (A) Unsupervised principal component analysis of normalized platelet protein expression adjusted for age, sex, treatment (antiplatelet and cytoreduction), and experimental batch. PC1 and PC2 colored by MPN subtype; and each contrasted with controls (n = 40, yellow): ET (n = 59, light green), PV (n = 41, blue). The first 2 principal components account for 26% of total variance in the data. (B-C) Volcano plots (2 panels of ET, PV) of differential protein expression showing log2 fold change vs statistical significance (negative log10 of P values) of each gene. Significant upregulated and downregulated genes are those with P values (false discovery rate [FDR]) ≤.05 and absolute value of fold changes ≥1.5. (D) Hierarchically clustered heat map of the top 10 differentially expressed proteins (FDR <0.01) from control vs MPN patient samples. Colored annotation is provided to indicate MPN subtype, mutation status, and sex. Rows indicate gradation in expression on a yellow (low) to orange (high) scale. Columns indicate sample type from controls, ET, and PV. (E) Pathway-enrichment analysis of proteins with MPN subtype–specific expression (color indicated; yellow ET, and light green PV). Each point represents a pathway; the x-axis gives the normalized enrichment score, which reflects the degree to which each pathway is overrepresented at the top of the ranked list of differentially expressed proteins, normalized to account for differences in gene set size and in correlations between gene sets and the expression data set. The y-axis lists the detail-level node of the most enriched pathways; solid lines mark gene set enrichment analysis–recommended11 Bonferroni-corrected statistical significance criterion of FDR <0.25 for exploratory analyses.
Differential markers in ET and PV highlight candidate proteins as potential mediators of the prothrombotic phenotype, including proteins associated with hypercoagulation (eg, MMP1, SERPINH1, FcγRIIA, and PDIA6) and inflammation (LGALS1, S100A6, SLC25A24, and CD63). Select candidates are highlighted in Figure 2B-C and discussed in detail in supplemental Table 2. Furthermore, although the profibrotic phenotype of myelofibrosis is well understood,12 candidate markers (LGALS1, S100A6) in the early, more indolent subtypes of chronic MPNs point to potential platelet mediators of fibrosis of relevance to myelofibrosis. Unsupervised hierarchical clustering further classified the top 10 differentially expressed platelet proteins for MPN, in contrast with those of healthy donors (Figure 2D). Although patients with MPN cluster into a group distinct from controls, we also note the overlay between PV and ET platelet signatures likely reflecting shared pathobiology.
PV and ET demonstrate overlapping clinical phenotypes; however, they remain recognized as distinct pathological entities with specific diagnostic and therapeutic considerations.13 Comparing platelet proteomic expression between patients with PV (n = 41) and those with JAK2-positive ET (n = 26) distinguished MPN subtypes and revealed subtype-specific signatures (supplemental Table 7); however, the biologic significance of these observations remains to be seen. The JAK2 V617F mutation is known to increase the risk of vascular events in ET, whereas the presence of the CALR mutation is associated with higher rates of bleeding on aspirin in low-risk patients.14,15 Upon subgroup analysis of our ET cohort, no difference was observed in platelet protein expression between JAK2- (n = 26) and CALR-positive patients (n = 18) (supplemental Table 8). In contrast, patients with triple-negative ET are recognized to have low rates of thrombosis and transformation.16 Although the number of patients with triple-negative ET included in this study was small (n = 12), 90 proteins were differentially expressed (false discovery rate <0.05; supplemental Table 9) relative to the mutation-positive ET cohort (n = 47). Intriguingly, proinflammatory proteins noted to be upregulated in the MPN platelet proteome (such as LGALS1, PDIA4) are decreased in our triple-negative population. Although this observation needs to be confirmed in larger studies, it does appear that the platelets of patients with triple-negative ET express lower levels of proinflammatory proteins.
To better decipher the functional significance of the observed proteomic changes, we performed enrichment analysis and identified biological pathways that are differentially activated between MPN and controls (Figure 2E; supplemental Tables 10 and 11). Gene set enrichment analysis (supplemental Methods) found that MPN (stratified by subtypes; ET and PV) primarily induces pathways associated with the unfolded protein response. Moreover, among the most enriched gene sets, MPN pathology induces activation of oxidative phosphorylation and mTORC1 signaling pathways. Proliferation pathways also reveal significant enrichment with overexpression of c-MYC target proteins among patients with PV. The MPN pathways exhibiting significant proteomic regulation by gene set enrichment analysis are consistent with our observations at the individual level for increased or decreased protein expression.
Here, we profile the MPN platelet proteome. Our findings validate our prior MPN platelet transcriptomic data from an independent cohort, confirming a strong possible role for platelet biology in MPN.17 Our data also expand on findings from other smaller or targeted studies,18-24 thus identifying the platelet proteome as a potential mediator of MPN proinflammatory, prothrombotic, and profibrotic processes. In keeping with the knowledge that vascular risk remains elevated among chronically treated patients,6 we demonstrate evidence of an altered platelet proteome in patients with ET and PV despite standard therapy. The observed platelet molecular profile supports our hypothesis that platelets contribute to remodeling of the circulatory microenvironment, which could lead to a self-reinforcing inflammatory milieu, possibly promoting disease progression and associated MPN vascular complications.
Using label-free, untargeted platelet proteomic profiling, we discover key potential mediators of immunothrombosis and proteostasis in patients with ET and PV. We identify significant differential platelet protein expression, revealing high-priority candidate markers for functional evaluation in MPN studies. We acknowledge that our healthy donor population was not screened for mutations associated with clonal hematopoiesis of indeterminate potential and recognize that future mechanistic studies are needed to substantiate our results. In addition, investigation into the circulatory microenvironment and the MPN megakaryocyte molecular signature may reveal further insights into the etiology of these observed proteomic differences. Finally, a longitudinal study following patients for the development of thrombotic outcomes would be critical to enabling the development of future risk prediction algorithms. Our ongoing efforts include proteomic analysis of platelets from prospectively recruited patients at the time of initial MPN diagnosis to identify candidates for mechanistic interrogation in treatment-naive patients. Our findings highlight that the circulating platelet molecular profile offers unique insights into MPN pathobiology, and we demonstrate the immense value of cross-institutional collaborations in advancing larger-scale translational studies, particularly for rare disease cohorts.
Acknowledgments: S.K. received a training fellowship from the International Society of Thrombosis Haemostasis and the International Network of VENous Thromboembolism Clinical Research Networks to complete this multisite study with A.K. as host mentor at Stanford University. S.K. is a fellow on the Irish Clinical Academic Training Programme, supported by the Wellcome Trust and the Health Research Board (grant 203930/B/16/Z), the Health Service Executive, National Doctors Training and Planning and the Health and Social Care, Research and Development Division, Northern Ireland. This work was also funded by US National Institutes of Health grants 1K08HG010061-01A1 (National Human Genome Research Institute) and 3UL1TR001085-04S1 (National Center for Advancing Translational Sciences), and the MPN Research Foundation (A.K.), and additional partial support from U01HG011762 (Genomics Research to Elucidate the Genetics of Rare Disease Consortium, principal investigator: Stephen B. Montgomery at Stanford University).
Contribution: S.K., B.K., and A.K. conceived of the overall study and secured funding; S.K. designed the experimental plan with input from P.M., F.N.A., B.K., and A.K.; S.K. and S.G. coordinated and performed the sample acquisition and processing according to established protocols; Z.S. and A.K. developed the code for computational analyses; A.K. and S.K. performed and interpreted the analyses and wrote and edited the manuscript; S. Madden and K.B. provided bioinformatic support; L.W. supported platelet protein preparation; C.S. operated mass spectrometry at the Proteomics Core Facility, Conway Institute, University College Dublin; A. Falanga, M.M., F.S., and S.G. provided clinical samples and annotation at the Hospital Papa Giovanni XXIII, Bergamo, Italy; A. Fortune, S. Maung, and M.F. provided clinical samples and annotation at the Mater Misericordiae University Hospital, Dublin, Ireland; and all authors critically reviewed and edited the draft manuscript and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anandi Krishnan, Department of Biomedical Engineering, Rutgers University, 599 Taylor Rd, Piscataway, NJ 08854; email: anandi.krishnan@rutgers.edu.
References
Author notes
S.K. conducted part of this research as a visiting training fellow with A.K. at Stanford University.
A.K. and B.K. are joint senior authors.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRoteomics IDEntifications Database (PRIDE) partner repository with the data set identifier PXD046324. RNA-sequencing data from this work (original FASQ files from paired-end sequencing of all 120 samples) is already deposited to the National Institutes of Health genomic database of Genotypes and Phenotype (dbGAP) under public accession number PHS-0021-21. v1.P1.
The full-text version of this article contains a data supplement.

![MPN platelet proteome distinguishes disease phenotype. (A) Unsupervised principal component analysis of normalized platelet protein expression adjusted for age, sex, treatment (antiplatelet and cytoreduction), and experimental batch. PC1 and PC2 colored by MPN subtype; and each contrasted with controls (n = 40, yellow): ET (n = 59, light green), PV (n = 41, blue). The first 2 principal components account for 26% of total variance in the data. (B-C) Volcano plots (2 panels of ET, PV) of differential protein expression showing log2 fold change vs statistical significance (negative log10 of P values) of each gene. Significant upregulated and downregulated genes are those with P values (false discovery rate [FDR]) ≤.05 and absolute value of fold changes ≥1.5. (D) Hierarchically clustered heat map of the top 10 differentially expressed proteins (FDR <0.01) from control vs MPN patient samples. Colored annotation is provided to indicate MPN subtype, mutation status, and sex. Rows indicate gradation in expression on a yellow (low) to orange (high) scale. Columns indicate sample type from controls, ET, and PV. (E) Pathway-enrichment analysis of proteins with MPN subtype–specific expression (color indicated; yellow ET, and light green PV). Each point represents a pathway; the x-axis gives the normalized enrichment score, which reflects the degree to which each pathway is overrepresented at the top of the ranked list of differentially expressed proteins, normalized to account for differences in gene set size and in correlations between gene sets and the expression data set. The y-axis lists the detail-level node of the most enriched pathways; solid lines mark gene set enrichment analysis–recommended11 Bonferroni-corrected statistical significance criterion of FDR <0.25 for exploratory analyses.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/16/10.1182_bloodadvances.2023012016/2/m_blooda_adv-2023-012016-gr2.jpeg?Expires=1769136742&Signature=l3iVOdyFR59wsUb0D6Z3Sn6zsU-JfDNQ4V3dPbXoAcC4D63vYrU4UxiD1bQFIipF9O3qFaqlZHeznWW32n-YTmc6CD4t9UwsykY2FqeiqmVPu7GIGSBFU6hxjpHTvHREowb1DTf0sbCecasR6OFpLfaCCrWbgFPpkAIVSY73ueA2Ve-AXag-Dc~198XMN6Y0frSqWMcq7j23NBB94AlAhHd4b4QhiVP6KGuJWep75V6~jDXFi8JGOzX~8VHX8-InKjIKFQD1Mye0Ui9SlEiLz3~r~hHZF8RgRUCWKZ22Donu3CgEb9Dp87XnMPdVVUUEgwwkQqtFpXAzzOcCi~Kcdg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)