Key Points
The inflammatory microenvironment that accompanies aging, and specifically TNF, favors TET2-mutant myeloid expansion.
Therapeutic blockage of TNF may be a therapeutic target to reduce TET2-mutant clonal hematopoiesis.
Visual Abstract
Somatic mutations in the TET2 gene occur more frequently with age, imparting an intrinsic hematopoietic stem cells (HSCs) advantage and contributing to a phenomenon termed clonal hematopoiesis of indeterminate potential (CHIP). Individuals with TET2-mutant CHIP have a higher risk of developing myeloid neoplasms and other aging-related conditions. Despite its role in unhealthy aging, the extrinsic mechanisms driving TET2-mutant CHIP clonal expansion remain unclear. We previously showed an environment containing tumor necrosis factor (TNF) favors TET2-mutant HSC expansion in vitro. We therefore postulated that age-related increases in TNF also provide an advantage to HSCs with TET2 mutations in vivo. To test this hypothesis, we generated mixed bone marrow chimeric mice of old wild-type (WT) and TNF–/– genotypes reconstituted with WT CD45.1+ and Tet2–/– CD45.2+ HSCs. We show that age-associated increases in TNF dramatically increased the expansion of Tet2–/– cells in old WT recipient mice, with strong skewing toward the myeloid lineage. This aberrant myelomonocytic advantage was mitigated in old TNF–/– recipient mice, suggesting that TNF signaling is essential for the expansion Tet2-mutant myeloid clones. Examination of human patients with rheumatoid arthritis with clonal hematopoiesis revealed that hematopoietic cells carrying certain mutations, including in TET2, may be sensitive to reduced TNF bioactivity following blockade with adalimumab. This suggests that targeting TNF may reduce the burden of some forms of CHIP. To our knowledge, this is the first evidence to demonstrate that TNF has a causal role in driving TET2-mutant CHIP in vivo. These findings highlight TNF as a candidate therapeutic target to control TET2-mutant CHIP.
Introduction
Clonal hematopoiesis of indeterminate potential (CHIP) has emerged as a significant age-related risk factor for multiple conditions including hematologic cancers, cardiovascular disease (CVD), ischemic stroke, respiratory infections,1 and all-cause mortality.2,3 CHIP occurs when a hematopoietic stem/progenitor cell (HSPC) acquires an advantageous somatic mutation, leading to clonal expansion, defined when the variant allele fraction reaches at least 2% in peripheral blood cells. Somatic mutations in >100 genes have been identified as candidate drivers of CHIP, with mutations in the epigenetic regulators DNA methyltransferase 3a (DNMT3A), ten-eleven translocation-2 (TET2), and additional sex combs-like 1 (ASXL1) being among the most common.4 These mutations, referred to as “DTA mutations” account for ∼80% of all CHIP cases and increase exponentially with age, occurring in at least 10% of older adults.5-7 Although DNMT3A and TET2 functionally differ, epigenetic dysregulation caused by mutations in either of these genes converge on progressive defects in hematopoiesis, including enhanced HSPC self-renewal and aberrant myeloid lineage expansion, culminating in inflammation.3,8-10 These phenotypes are ultimately thought to contribute to the development of CHIP-comorbid conditions. In support of this notion, it has been demonstrated that CHIP carriers of the hypomorphic interleukin-6 (IL-6) receptor gene variant (IL6R p.Asp358Ala),11 a genetic proxy of IL-6 inhibition, have reduced pneumonia1 and CVD risk among people with CHIP, especially driven by TET2- and other non-DNMT3A–mutant CHIP.12 Inhibition of IL-1β with canakinumab likewise decreased the risk of major cardiovascular events in carriers of acquired TET2-mutant CHIP.13 In addition, TET2 mutations are significantly associated with worse outcomes of cardiogenic shock patients, and those patients with TET2-mutant CHIP exhibit higher circulating levels of tumor necrosis factor α (TNF-α).14 These preliminary findings suggest that inflammatory mediators contribute to comorbidity and could serve as promising therapeutic targets in CHIP-related conditions.
Targeting inflammation may also have protective benefits in ameliorating the occurrence of CHIP. It has been proposed that DNMT3A or TET2 mutations may enhance the fitness of HSPCs to expand under adverse bone marrow (BM) environments.15 With age, there is an increase in the levels of proinflammatory cytokines such as TNF-α, IL-6, and IL-1β, a phenomenon that has been termed “inflammaging” (reviewed by Franceschi et al16). Inflammaging is a significant risk factor for morbidity and mortality in older adults,16 and it has been demonstrated that consistent exposure to proinflammatory cytokines is detrimental to the function of HSPCs. The HSPCs with DNMT3A or TET2 mutations, however, are refractory to these signals, providing them with a potential competitive advantage in an inflammatory environment.17 In support of this, we and others have demonstrated that Tet2–/– HSPCs show sustained survival in response to inflammation, with resistance to inflammation-induced damage and apoptosis in vitro.17,18 Similarly, older hematopoietic stem cells (HSCs) from IL-1R1 knockout (KO) mice have significantly mitigated inflammaging, and loss of IL-1 signaling rescues hematologic abnormalities associated with Tet2 deficiency at the HSPC level.19,20 Although these papers support the conjecture that TET2 mutations may enhance the fitness of HSPCs under inflammatory stress, investigations into the effects of age-associated inflammation have been lacking.
Here, we address this deficiency by determining what impact age-associated TNF has on mutant Tet2-HSPC fitness, lineage expansion, and function, using competitive transplantation studies in mice, and serial sequencing of TET2-mutant human participants during anti-TNF treatment.
Materials and methods
Animals
Specific pathogen–free female mice were maintained under a 12-hour light-dark cycle at 22 ± 2°C and 55 ± 5% air humidity at the McMaster Central Animal Facility (Hamilton, Ontario, Canada). To protect from age-related obesity, all mice were provided with an exercise wheel. Mice had ad libitum access to an irradiated aging chow diet (Envigo Teklad Diets 2914, Madison, WI) and autoclaved reverse osmosis water. Any mice that developed tumors during the entire period of observation were omitted from analyses. The C57Bl/6 wild-type (WT) and TNF-α KO (TNF–/–) mice on a C57Bl/6 background were obtained from The Jackson Laboratory (Bar Harbor, ME). The Tet2 KO (Vav-Cre–mediated, hematopoietic–specific disruption of Tet2 at exon 3; Tet2–/–) and floxed control mice (Tet2f/f) were generously provided by Michael Rauh (Queen’s University).
Human participants and CHIP analysis
Research participants diagnosed with rheumatoid arthritis (RA) were recruited from the Greater Hamilton Area (Ontario, Canada) from 2016 to 2018. All protocols were approved by the Hamilton Research Ethics Boards (number 2855). Venus blood was drawn in anticoagulant-free vacutainers for the isolation of serum, and in heparin-coated vacutainers for the experiments that required viable leukocytes.21 Blood draws occurred before any immunomodulatory treatment (baseline) and at 3 and 6 months after treatment with adalimumab (Humira), an anti-TNF agent. Patients who were treated with a different RA drug (eg, methotrexate, leflunomide, and naproxen) were included as controls. Participant demographic information (age, sex, and height) and health status (components of the Charlson comorbidity index, body mass index, medication history, vaccination history, and frailty scores) were provided at the time of sample collection. Only participants who had not required antibiotics within 2 weeks of sample collection were included in this analysis. CHIP status was determined by applying a successful 48-gene, targeted, Ion Torrent–based sequencing approach to isolated genomic DNA from peripheral blood mononuclear cells, as previously described.22 Confirmation of CHIP calls and increased sensitivity to detect clones with a variant allele frequency (VAF) <0.02 used an established single-molecule molecular inversion probe (smMIP)–targeted genomic capture technique and high-depth (47 500× average coverage) paired-end sequencing of sample duplicates, with the ability to detect clones down to 0.005 VAF.23 High-stringency filters for error suppression included the use of 2 control cord blood samples and binomial models to estimate background error rates at each allele, to find frequencies that are significantly higher in samples than controls, ensuring variants are found in each technical replicate and probe that covers the locus, with an average VAF of at least 0.005, having 1 single strand consensus sequence family size >4 for each read in the replicate, with a minor allele frequency <0.001 or not available, and not being intron or synonymous exon variants.
Establishment of BM chimeras
Young (6 months, n = 9 female) and old (18-22 months, n = 9 female) WT mice and old TNF–/– mice (n = 7 female) were subjected to nonirradiative myeloablation as previously described.24 Briefly, the chemotherapeutic drug busulfan (Busulfex; Otsuka Pharmaceuticals, Tokyo, Japan) was diluted using sterile phosphate-buffered saline to a concentration of 3 mg/mL. Mice were administered fractionated intraperitoneal doses of 20 mg/kg per day for a total of 80 mg/kg. Twenty-four hours after the final busulfan treatment, mice received retro-orbital injections of 8 × 106 cells per mL T-cell–depleted BM (catalog no. 18951; STEMCELL, Vancouver, BC, Canada), equally harvested from 4-month-old WT CD45.1 (n = 5) and Tet2–/– CD45.2 donor mice (n = 5). Flow cytometry was used to confirm that equal numbers of CD45.1+ and CD45.2+ cells were injected. Engrafted mice were euthanized, and spinal cords were harvested 8 weeks after BM transplant (BMT) for analysis of whole-blood and BM-derived cell accumulation. Female mice were used to accurately translate to the human samples that were predominately from female patients, in keeping with the increased reported incidence of RA in females.25
Immunophenotyping by flow cytometry
For whole-blood and BM immunophenotyping, 100 μL of heparinized blood and isolated BM were incubated with antibodies (supplemental Table 1) for 30 minutes at room temperature and then incubated in 1X Fix/Lyse buffer (eBioscience, Carlsbad, CA) for 10 minutes. Cells were washed and resuspended in fluorescence-activated cell sorting wash buffer (phosphate-buffered saline, 0.5% bovine serum albumin, and 2-mM EDTA) before analysis. For intracellular markers, samples were initially surface stained with antibodies, and then intracellular staining was performed after 30-minute permeabilization at room temperature with 1× Intracellular Staining Permeabilization Wash Buffer (BioLegend; catalog no. 421002) as previously described.26
Hierarchical gating strategies were used to acquire flow cytometry data for HSCs, monocyte, T-cell, B-cell, natural killer, and neutrophil populations (supplemental Figures 1-3). All fluorescence gates were set using appropriate isotype or fluorescence minus one controls (supplemental Table 1), and compensation of spectral overlap was performed for all fluorochromes. Flow cytometry was performed on a CytoFLEX (Beckman Coulter) and analyzed using FlowJo software (version 10.7.1; Becton Dickinson & Company). Total cell counts were determined with CountBright Absolute Counting Beads (Life Technologies; catalog no. C36950). Data are reported as percent positive or count for each cell subset.
Measurement of cytokine production
Serum IL-10, IL-1β, IL-6, monocyte chemoattractant protein (MCP)-1, and TNF were measured using a high-sensitivity enzyme-linked immunosorbent assay as per manufacturer’s recommendations (Meso Scale Discovery; catalog no. K15069L-1).
Statistical analysis
Statistical analyses were performed in GraphPad Prism V9.2 or R 4.1.2. Significance was calculated using a 2-way analysis of variance or Mann-Whitney test when appropriate. The results are expressed as the mean value with standard error of the mean, unless otherwise stated. Significance levels used are ∗P value <.05, ∗∗P value <.01, and ∗∗∗P value <.001.
The animal ethics have been approved by the McMaster Animal Research Ethics Board (number 21-04-13) and performed in accordance with the Canadian Council on Animal Care guidelines. All human protocols were approved by the Hamilton Research Ethics Board (numbers 1949 and 2855) and Queen’s University Health Sciences and Affiliated Teaching Hospitals Research Ethics Board (PATH-181-18). Informed consent was received before participation.
Results
TNF increases myeloid differentiation in BM of old mice after BMT
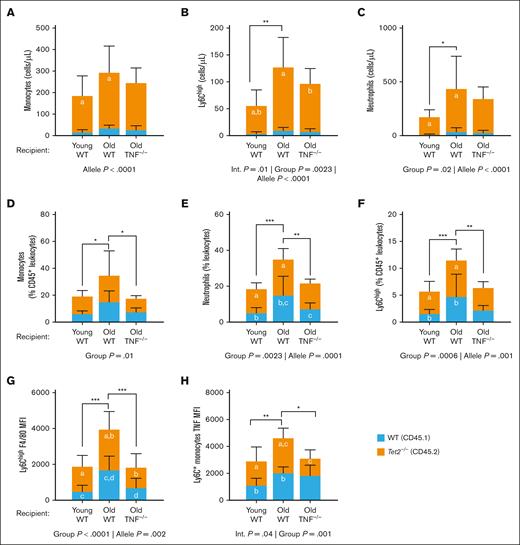
We have previously demonstrated that an in vitro environment containing TNF favors Tet2-mutant clonal hematopoiesis (CH).18 To determine whether these findings translated in vivo, we performed competitive transplantation studies using Tet2–/– (CD45.2+) and WT (CD45.1+) donor HSCs, injected into old WT and TNF–/– recipient mice (Figure 1A-B). Young WT recipient mice were also included to account for any age- but not TNF-driven changes in the BM environment. At 8 weeks after BMT, all recipient mice had similar absolute counts of WT (CD45.1+) leukocytes in the hematopoietic environment. However, the old TNF–/– recipient mice had fewer Tet2–/– (CD45.2+) leukocytes, resulting in a lower CD45.1:CD45.2 cell ratio, suggesting a role for TNF in Tet2–/– clonal expansion (Figure 1C). Within the BM compartment, chronic TNF exposures in old WT recipients expanded HSPCs (Figure 1D), giving rise to myeloid-biased multipotent progenitor cells. We found that the absolute number of common myeloid progenitors were increased in old WT recipient mice compared with old TNF–/– recipient mice (Figure 1E). Common myeloid progenitor cells differentiate into either granulocyte-monocyte progenitor cells or monocyte-dendritic progenitor cells. We show that the presence of age-associated TNF increased the numbers of both granulocyte-monocyte progenitor cells and monocyte-dendritic progenitor cells in old WT recipient mice (Figure 1F-G) compared with young WT and old TNF–/– recipient mice. Common monocyte progenitor cells, which are monocyte committed, were likewise increased in the old WT but not TNF–/– recipient mice (Figure 1H), suggesting a role for TNF in driving myelomonocytic differentiation in chimeric mice.
Age-associated TNF inflammation increases myeloid progenitor differentiation in female mice 8 weeks after BMT with WT and Tet2–/– HSPCs. (A) Experimental design and representative flow cytometric data showing CD45+ T-cell–depleted (TCD) BM donor cells transplanted in mice conditioned with busulfan. Young (6 months) and old (18-22 months) C57Bl/6-J WT and old TNF–/– animals received 80 mg/kg busulfan and received transplantation with 2 × 106 TCD-BM cells from young CD45.1+ WT and CD45.2+Tet2–/– mice. (B) Histogram showing percentage of CD45.1+ WT and CD45.2 Tet2–/– donor cells transplanted into recipient mice. (C) Ratio of CD45.2+Tet2–/– to CD45.1+ WT leukocytes in the BM of recipient mice 8 weeks after BMT. (D-I) Flow cytometric analyses showing absolute counts of HSPC (D), common myeloid progenitor (CMP) cells (E), monocyte-dendritic progenitor (MDP) cells (F), granulocyte-monocyte progenitor (GMP) cells (G), common monocyte progenitor (cMoP) cells (H), and mature monocytes (I). (J) Absolute count of Ly6Chigh inflammatory monocytes. (K) Geometric mean of CX3 chemokine receptor 1 (CX3CR1) in BM monocytes. (L) Absolute counts of circulatory Ly6Chigh monocytes. Statistical significance determined by 1-way analysis of variance (ANOVA). ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. ELISA, enzyme-linked immunosorbent assay; MFI, geometric mean fluorescence intensity.
Age-associated TNF inflammation increases myeloid progenitor differentiation in female mice 8 weeks after BMT with WT and Tet2–/– HSPCs. (A) Experimental design and representative flow cytometric data showing CD45+ T-cell–depleted (TCD) BM donor cells transplanted in mice conditioned with busulfan. Young (6 months) and old (18-22 months) C57Bl/6-J WT and old TNF–/– animals received 80 mg/kg busulfan and received transplantation with 2 × 106 TCD-BM cells from young CD45.1+ WT and CD45.2+Tet2–/– mice. (B) Histogram showing percentage of CD45.1+ WT and CD45.2 Tet2–/– donor cells transplanted into recipient mice. (C) Ratio of CD45.2+Tet2–/– to CD45.1+ WT leukocytes in the BM of recipient mice 8 weeks after BMT. (D-I) Flow cytometric analyses showing absolute counts of HSPC (D), common myeloid progenitor (CMP) cells (E), monocyte-dendritic progenitor (MDP) cells (F), granulocyte-monocyte progenitor (GMP) cells (G), common monocyte progenitor (cMoP) cells (H), and mature monocytes (I). (J) Absolute count of Ly6Chigh inflammatory monocytes. (K) Geometric mean of CX3 chemokine receptor 1 (CX3CR1) in BM monocytes. (L) Absolute counts of circulatory Ly6Chigh monocytes. Statistical significance determined by 1-way analysis of variance (ANOVA). ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. ELISA, enzyme-linked immunosorbent assay; MFI, geometric mean fluorescence intensity.
To determine whether the increase in committed progenitors cumulated in more mature monocytes within the BM niche, we examined the counts of monocyte subsets based on their expression of the surface marker Ly6C. As expected, old WT but not TNF–/– recipient mice had an increase in the number of BM monocytes (Figure 1I), including a significant number of Ly6Chigh monocytes (Figure 1J). Monocytes expressing high levels of Ly6C have proinflammatory functions and tend to express low levels of the CX3C chemokine receptor 1, which may promote monocyte trafficking and release into the bloodstream.27 Accordingly, we found that the expression of CX3 chemokine receptor 1 was decreased in old recipient mice (Figure 1K). This corresponded with a complementary increase of Ly6Chigh monocytes in the periphery (Figure 1L), especially in old WT recipient mice. These results recapitulate what is observed in WT, TNF–/–, and Tet2–/– mice at steady state (nonchimeric), as we have previously published1 (compare with supplemental Figure 4). Collectively, these data demonstrate that age-associated TNF contributes to the expansion of a monocyte–biased HSPC pool in competitive Tet2–/– mouse chimeras, increasing inflammatory monocyte subsets in the periphery.
TNF contributes to myeloid regeneration in chimeric mice after BMT
To gain insight into whether the microenvironment influenced the reconstitution potential of donor WT (CD45.1+) and Tet2–/– (CD45.2+) cells, we assessed leukocytes in the periphery using flow cytometry, with a focus on the myeloid lineage. Gating myeloid cells by CD45 allele type revealed that Tet2–/– cells had an advantage over WT cells in the circulation of all engrafted mice, shown by significant main effects for allele type. This Tet2–/– myeloid bias was exacerbated by TNF in the aging microenvironment. Although overall monocyte cell counts were similar among recipient mice, a 2-way analysis of variance accounting for allele type revealed that the absolute cell count of Tet2–/– monocytes was higher in old WT recipient mice than in young recipient mice after BMT (Figure 2A; Table 1). In accordance with what was observed in the BM, the absolute cell count of Ly6Chigh monocytes was increased in old WT recipients, and this corresponded with a significant increase in Tet2–/– cells (Figure 2B). Although the old TNF–/– recipient mice had an increase in the number of Tet2–/– Ly6Chigh monocytes, the effect was less pronounced, and total monocyte counts were comparable with that of young WT recipient mice (Figure 2A-B). Absolute cell count data of neutrophils similarly demonstrated higher Tet2–/– neutrophils in old WT but not old TNF–/– recipient mice (Figure 2C), cumulating in an overall higher neutrophil count than young WT mice. Together, these data show that increased myelomonopoiesis within the BM coincides with increased numbers of Tet2–/– monocytes and neutrophils in the blood.
Tet2 mutations increase circulating myeloid cells of old WT but not TNF–/– recipient mice. Circulating myeloid immune populations were compared in young WT, old WT, and old TNF–/– recipient mice. (A-C) Total counts of circulating monocytes (A), Ly6Chigh monocytes (B), and neutrophils (C). (D-F) Total circulating monocytes, neutrophils, and Ly6Chigh monocytes as a proportion of total leukocytes (CD45+) cells, respectively. (G) Surface expression of F4/80 on Ly6Chigh monocyte. (H) Intracellular expression of TNF in monocytes after stimulation with lipopolysaccharides. Data are shown as a stacked bar plot, in which orange represents gated CD45.2 Tet2–/– cells and blue represents CD45.1 WT cells. Statistical significance determined by 2-way ANOVA with Tukey multiple comparisons test. Letters in the orange and blue columns denote significant differences (P ≤ .05) in the group means of CD45.2 or CD45.1 alleles, respectively. For all variables with the same letter, the difference between the means is significantly different. If 2 variables have different letters or no letters, they are not significantly different. Black bars with asterisks denote group differences, inclusive of both CD45.1 and CD45.2 alleles. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001.
Tet2 mutations increase circulating myeloid cells of old WT but not TNF–/– recipient mice. Circulating myeloid immune populations were compared in young WT, old WT, and old TNF–/– recipient mice. (A-C) Total counts of circulating monocytes (A), Ly6Chigh monocytes (B), and neutrophils (C). (D-F) Total circulating monocytes, neutrophils, and Ly6Chigh monocytes as a proportion of total leukocytes (CD45+) cells, respectively. (G) Surface expression of F4/80 on Ly6Chigh monocyte. (H) Intracellular expression of TNF in monocytes after stimulation with lipopolysaccharides. Data are shown as a stacked bar plot, in which orange represents gated CD45.2 Tet2–/– cells and blue represents CD45.1 WT cells. Statistical significance determined by 2-way ANOVA with Tukey multiple comparisons test. Letters in the orange and blue columns denote significant differences (P ≤ .05) in the group means of CD45.2 or CD45.1 alleles, respectively. For all variables with the same letter, the difference between the means is significantly different. If 2 variables have different letters or no letters, they are not significantly different. Black bars with asterisks denote group differences, inclusive of both CD45.1 and CD45.2 alleles. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001.
Whole-blood leukocyte cell numbers by group and allele type
| Recipient . | Allele . | Young WT (n = 9F) . | Old WT (n = 9F) . | Old TNF–/– (n = 7F) . | |||
|---|---|---|---|---|---|---|---|
| Mean (SD) . | Median (IQR) . | Mean (SD) . | Median (IQR) . | Mean (SD) . | Median (IQR) . | ||
| Myeloid | |||||||
| Neutrophils | WT (CD45.1) | 11.3 (5.32) | 9.129 (6.78-15.58) | 38.16 (35.96) | 25.46 (15.08-51.17) | 28.29 (21.01) | 19.75 (17.69-43.87) |
| Tet2–/– (CD45.2) | 164.9 (64.24) | 164.1 (112-215.5) | 400.3 (299.4) | 305.7 (261.1-426.2) | 316.6 (108) | 332.8 (228.3-380.6) | |
| Monocytes | WT (CD45.1) | 15.68 (12.51) | 12.74 (8.048-18.16) | 34.95 (14.51) | 39.66 (19.17-45.71) | 27.71 (18.35) | 22.17 (16.6-35.3) |
| Tet2–/– (CD45.2) | 170.8 (91.24) | 150.3 (103.6-248.7) | 260.4 (120.6) | 193 (184.2-402.9) | 218.7 (68.06) | 224.3 (147.2-278.2) | |
| Ly6Chigh monocytes | WT (CD45.1) | 3.90 (3.50) | 3.038 (1.66-4.765) | 10.04 (5.69) | 11.03 (4.57-13.06) | 8.118 (5.10) | 6.862 (5.48-11.65) |
| Tet2–/– (CD45.2) | 52.44 (28.99) | 43.93 (27.43-80.43) | 118 (54.79) | 95.63 (75.98-179.2) | 89.26 (27.75) | 91.14 (60.79-117.3) | |
| Lymphoid | |||||||
| B cells | WT (CD45.1) | 156.5 (94.43) | 139.9 (92.75-185.6) | 108.6 (31.94) | 104.4 (82.2-136.1) | 176.5 (63.27) | 197.2 (118.9-222.7) |
| Tet2–/– (CD45.2) | 770.6 (434.8) | 626 (461.8-999.2) | 867.6 (328.2) | 833.6 (593.7-971) | 1109 (305.7) | 1154 (749.8-1304) | |
| T cells | WT (CD45.1) | 56.72 (36.5) | 46.57 (28.31-83.66) | 23.54 (8.336) | 25.3 (14.82-30.59) | 34.64 (25.39) | 33.09 (9.829-52.03) |
| Tet2–/– (CD45.2) | 183.8 (66.22) | 194.3 (137.5-213.8) | 62.78 (32.63) | 58.05 (40.19-84.66) | 88.38 (46.48) | 75.93 (50.38-103.7) | |
| CD8+ T cells | WT (CD45.1) | 21.35 (14.37) | 18.09 (10.44-28.78) | 7.969 (3.05) | 7.943 (5.092-10.67) | 10.28 (7.954) | 9.916 (2.583-14.87) |
| Tet2–/– (CD45.2) | 53.68 (20.76) | 52.32 (40.32-63.94) | 17.85 (10.88) | 17.6 (10.13-21.05) | 22.34 (13.16) | 18.31 (11.9-26.32) | |
| CD4+ T cells | WT (CD45.1) | 33.08 (22.01) | 27.7 (14.6-51.96) | 12.36 (5.414) | 13.78 (6.318-17.17) | 22.93 (16.63) | 21.41 (7.067-35.83) |
| Tet2–/– (CD45.2) | 124.1 (43.91) | 128.9 (89.41-146) | 42.1 (21.35) | 39.86 (25.33-59.83) | 63.15 (31.8) | 56.18 (36.86-73.87) | |
| Recipient . | Allele . | Young WT (n = 9F) . | Old WT (n = 9F) . | Old TNF–/– (n = 7F) . | |||
|---|---|---|---|---|---|---|---|
| Mean (SD) . | Median (IQR) . | Mean (SD) . | Median (IQR) . | Mean (SD) . | Median (IQR) . | ||
| Myeloid | |||||||
| Neutrophils | WT (CD45.1) | 11.3 (5.32) | 9.129 (6.78-15.58) | 38.16 (35.96) | 25.46 (15.08-51.17) | 28.29 (21.01) | 19.75 (17.69-43.87) |
| Tet2–/– (CD45.2) | 164.9 (64.24) | 164.1 (112-215.5) | 400.3 (299.4) | 305.7 (261.1-426.2) | 316.6 (108) | 332.8 (228.3-380.6) | |
| Monocytes | WT (CD45.1) | 15.68 (12.51) | 12.74 (8.048-18.16) | 34.95 (14.51) | 39.66 (19.17-45.71) | 27.71 (18.35) | 22.17 (16.6-35.3) |
| Tet2–/– (CD45.2) | 170.8 (91.24) | 150.3 (103.6-248.7) | 260.4 (120.6) | 193 (184.2-402.9) | 218.7 (68.06) | 224.3 (147.2-278.2) | |
| Ly6Chigh monocytes | WT (CD45.1) | 3.90 (3.50) | 3.038 (1.66-4.765) | 10.04 (5.69) | 11.03 (4.57-13.06) | 8.118 (5.10) | 6.862 (5.48-11.65) |
| Tet2–/– (CD45.2) | 52.44 (28.99) | 43.93 (27.43-80.43) | 118 (54.79) | 95.63 (75.98-179.2) | 89.26 (27.75) | 91.14 (60.79-117.3) | |
| Lymphoid | |||||||
| B cells | WT (CD45.1) | 156.5 (94.43) | 139.9 (92.75-185.6) | 108.6 (31.94) | 104.4 (82.2-136.1) | 176.5 (63.27) | 197.2 (118.9-222.7) |
| Tet2–/– (CD45.2) | 770.6 (434.8) | 626 (461.8-999.2) | 867.6 (328.2) | 833.6 (593.7-971) | 1109 (305.7) | 1154 (749.8-1304) | |
| T cells | WT (CD45.1) | 56.72 (36.5) | 46.57 (28.31-83.66) | 23.54 (8.336) | 25.3 (14.82-30.59) | 34.64 (25.39) | 33.09 (9.829-52.03) |
| Tet2–/– (CD45.2) | 183.8 (66.22) | 194.3 (137.5-213.8) | 62.78 (32.63) | 58.05 (40.19-84.66) | 88.38 (46.48) | 75.93 (50.38-103.7) | |
| CD8+ T cells | WT (CD45.1) | 21.35 (14.37) | 18.09 (10.44-28.78) | 7.969 (3.05) | 7.943 (5.092-10.67) | 10.28 (7.954) | 9.916 (2.583-14.87) |
| Tet2–/– (CD45.2) | 53.68 (20.76) | 52.32 (40.32-63.94) | 17.85 (10.88) | 17.6 (10.13-21.05) | 22.34 (13.16) | 18.31 (11.9-26.32) | |
| CD4+ T cells | WT (CD45.1) | 33.08 (22.01) | 27.7 (14.6-51.96) | 12.36 (5.414) | 13.78 (6.318-17.17) | 22.93 (16.63) | 21.41 (7.067-35.83) |
| Tet2–/– (CD45.2) | 124.1 (43.91) | 128.9 (89.41-146) | 42.1 (21.35) | 39.86 (25.33-59.83) | 63.15 (31.8) | 56.18 (36.86-73.87) | |
All cell numbers are cells per μL. Summary statistics of peripheral blood leukocyte cell numbers in young WT, old WT, and old TNF–/– recipient mice after engraftment of WT (CD45.1+) and Tet2–/– (CD45.2+) cells.
F, female; IQR, interquartile range; SD, standard deviation.
Despite dramatic differences in cell counts, we show that the frequencies of both WT and Tet2–/– myeloid subsets (as a proportion of total WT CD45.1+ and Tet2–/– CD45.2+ leukocytes, respectively) are not significantly different. The old WT recipient mice had a disproportionate increase in the prevalence of circulating monocytes (Figure 2D; Table 2) and neutrophils (Figure 2E), driven by changes in both WT and Tet2–/– cells. Surface expression of Ly6C (Figure 2F) and the maturity marker F4/80 (Figure 2G) were also increased in old WT but not old TNF–/– recipient mice. To determine whether the increase in inflammatory monocytes increased TNF-driven inflammation, we measured the intracellular expression of TNF after lipopolysaccharide challenge. As expected, the expression of TNF was significantly increased in Ly6C+ monocytes of old WT recipient mice, whereas the old TNF–/– recipients, whose donor cells can produce TNF, were similar to young WT mice (Figure 2H). The increase in TNF in WT recipients was confirmed using a multiplexed enzyme-linked immunosorbent assay, which showed no additional differences in cytokines (IL-10, IL-6, MCP-1, and IL-1β) among the recipient mice at steady state (supplemental Figure 5). Given that levels of other inflammatory cytokines were similar among recipient mice, these results associate the phenotypic changes observed with TNF. Taken together, these data demonstrate that Tet2–/– myeloid lineages have an advantageous regenerative capacity in peripheral blood after BMT, which is exacerbated by TNF inflammation in the aging microenvironment. These results also highlight that TNF in the aging microenvironment, rather than intrinsic changes in myeloid progenitors, drives changes in monocyte subsets.
Whole-blood leukocyte prevalence
| . | Allele . | Young WT (n = 9F) . | Old WT (n = 9F) . | Old TNF–/– (n = 7F) . | |||
|---|---|---|---|---|---|---|---|
| Mean (SD) . | Median (IQR) . | Mean (SD) . | Median (IQR) . | Mean (SD) . | Median (IQR) . | ||
| % CD45+leukocytes | |||||||
| Neutrophils | WT (CD45.1) | 5.28 (2.84) | 5.2 (3.07-6.24) | 15.13 (10.6) | 11.7 (7.035-24.25) | 7.38 (3.45) | 6.46 (4.83-8.94) |
| Tet2–/– (CD45.2) | 13.44 (3.23) | 12.9 (11.1-15.2) | 20.13 (5.88) | 17.8 (15.75-24.35) | 14.54 (2.21) | 14.7 (13.1-16.5) | |
| Monocytes | WT (CD45.1) | 6.27 (2.01) | 2.018 (6.27-0.672) | 15.15 (8.07) | 8.07 (15.15-2.69) | 7.54 (2.99) | 2.99 (7.54-1.13) |
| Tet2–/– (CD45.2) | 13.29 (4.01) | 4.016 (13.29-1.33) | 19.82 (18.12) | 18.12 (19.82-6.04) | 10.2 (1.93) | 1.93 (10.2-0.73) | |
| Ly6Chigh monocytes | WT (CD45.1) | 1.58 (0.79) | 1.42 (0.76-2.54) | 4.791 (4.15) | 3.76 (2.53-4.68) | 2.24 (0.86) | 1.97 (1.66-3.11) |
| Tet2–/– (CD45.2) | 4.19 (1.80) | 3.6 (2.92-5.175) | 6.75 (2.07) | 5.75 (5.32-8.63) | 4.21 (1.08) | 3.68 (3.41-4.96) | |
| B cells | WT (CD45.1) | 59.98 (8.96) | 61.6 (52.25-67.55) | 42.57 (20.34) | 51.2 (24.4-57.45) | 63.13 (20.55) | 70.3 (35.9-78.8) |
| Tet2–/– (CD45.2) | 52.6 (8.44) | 54.4 (46.5-59.7) | 47.12 (17.34) | 48.1 (45.95-59.05) | 65.84 (7.53) | 69.8 (57.5-70.9) | |
| T cells | WT (CD45.1) | 22.97 (8.52) | 23.3 (16.85-30.05) | 11.57 (4.35) | 11.65 (8.44-16) | 11.24 (4.19) | 10.5 (9.13-15.4) |
| Tet2–/– (CD45.2) | 14.21 (2.57) | 13.7 (11.7-16) | 4.028 (1.38) | 4.43 (2.96-5.19) | 5.14 (1.211) | 4.84 (4.32-6.71) | |
| CD8+ T cells | WT (CD45.1) | 8.66 (3.36) | 9.13 (5.08-12.1) | 3.89 (1.45) | 4.17 (2.70-4.82) | 3.28 (1.301) | 3.46 (2.51-4.54) |
| Tet2–/– (CD45.2) | 4.12 (0.73) | 4.29 (3.35-4.63) | 1.13 (0.42) | 1.24 (0.79-1.36) | 1.28 (0.36) | 1.14 (1.02-1.7) | |
| CD4+ T cells | WT (CD45.1) | 13.34 (5.60) | 12 (10.55-17.45) | 5.98 (2.54) | 5.97 (4.30-8.22) | 7.43 (2.787) | 6.78 (5.73-10) |
| Tet2–/– (CD45.2) | 9.63 (1.874) | 8.71 (8.17-11) | 2.70 (0.97) | 2.80 (2.00-3.64) | 3.69 (0.839) | 3.48 (3.13-4.85) | |
| % CD8+T cells | |||||||
| Naïve CD8+ | WT (CD45.1) | 36.46 (9.20) | 36.9 (29.9-42.05) | 16.19 (4.58) | 17.6 (14.35-19.7) | 30.34 (3.37) | 29.1 (29-33.1) |
| Tet2–/– (CD45.2) | 39.28 (5.80) | 37.1 (33.8-45.3) | 13.84 (3.89) | 13.45 (10.4-17.38) | 24.97 (6.36) | 26 (16.9-29.8) | |
| CD8+ TCM | WT (CD45.1) | 52.84 (5.73) | 53.4 (49.35-57.55) | 69.55 (7.42) | 66.75 (64.25-78.23) | 59.66 (3.35) | 60.4 (56.6-62.8) |
| Tet2–/– (CD45.2) | 52.1 (4.59) | 49.6 (48.1-56.6) | 63.9 (7.81) | 63.7 (56.73-70.45) | 49.57 (8.16) | 48.7 (43-55.9) | |
| CD8+ TEM | WT (CD45.1) | 9.911 (10.68) | 5.18 (3.97-13.65) | 13.8 (9.61) | 13.75 (5.695-17.18) | 8.836 (2.10) | 8.13 (7.64-9.95) |
| Tet2–/– (CD45.2) | 8.163 (4.08) | 7.12 (5.06-11) | 21.95 (7.43) | 20 (15.35-28.08) | 24.33 (11.6) | 21 (14-39.5) | |
| CD8+ TVM | WT (CD45.1) | 47.22 (5.81) | 48.2 (42.25-52.1) | 53.69 (7.24) | 54.95 (46.43-60.55) | 49.19 (5.55) | 48.7 (44.2-55.1) |
| Tet2–/– (CD45.2) | 46.34 (4.24) | 45.7 (42.4-50.15) | 50.96 (9.70) | 49.35 (43.68-59.7) | 42.74 (7.54) | 44.9 (38.4-50.1) | |
| % CD4+T cells | |||||||
| Naïve CD4+ | WT (CD45.1) | 30.64 (5.64) | 31.7 (25.25-34.6) | 17.58 (3.44) | 18.45 (15.9-19.3) | 21.49 (3.36) | 19.8 (18.9-24) |
| Tet2–/– (CD45.2) | 23.58 (1.69) | 23.7 (23.05-24.35) | 17.24 (2.56) | 18.05 (14.68-19.13) | 17.59 (2.46) | 16.7 (15.4-20.5) | |
| CD4+ TCM | WT (CD45.1) | 56.97 (4.83) | 56.4 (53.05-62.65) | 54.23 (7.29) | 55.2 (46.95-59.7) | 47.49 (8.97) | 48.5 (38-55.7) |
| Tet2–/– (CD45.2) | 62.51 (3.81) | 61.7 (59.55-65.85) | 51.7 (6.15) | 51.65 (45.05-58.23) | 47.36 (7.36) | 49.7 (41.5-52.9) | |
| CD4+ TEM | WT (CD45.1) | 11.51 (3.84) | 13.1 (8.58-14.05) | 27.45 (8.04) | 25.1 (20.48-35.45) | 27.89 (9.64) | 27.3 (18.1-34.7) |
| Tet2–/– (CD45.2) | 13.17 (3.96) | 13.9 (9.09-17.1) | 30.19 (5.99) | 33.1 (23.7-34.8) | 33.06 (7.45) | 30 (28-38.5) | |
| CD4+ TVM | WT (CD45.1) | 27.2 (4.00) | 28.6 (23.7-30.25) | 49.69 (5.58) | 49.9 (45.65-54.53) | 33.94 (5.53) | 30.8 (30.1-39.6) |
| Tet2–/– (CD45.2) | 24.28 (3.70) | 26.3 (21.2-27.05) | 37.11 (4.37) | 36.75 (34.03-40.4) | 27.89 (4.66) | 25.8 (24.6-31.4) | |
| . | Allele . | Young WT (n = 9F) . | Old WT (n = 9F) . | Old TNF–/– (n = 7F) . | |||
|---|---|---|---|---|---|---|---|
| Mean (SD) . | Median (IQR) . | Mean (SD) . | Median (IQR) . | Mean (SD) . | Median (IQR) . | ||
| % CD45+leukocytes | |||||||
| Neutrophils | WT (CD45.1) | 5.28 (2.84) | 5.2 (3.07-6.24) | 15.13 (10.6) | 11.7 (7.035-24.25) | 7.38 (3.45) | 6.46 (4.83-8.94) |
| Tet2–/– (CD45.2) | 13.44 (3.23) | 12.9 (11.1-15.2) | 20.13 (5.88) | 17.8 (15.75-24.35) | 14.54 (2.21) | 14.7 (13.1-16.5) | |
| Monocytes | WT (CD45.1) | 6.27 (2.01) | 2.018 (6.27-0.672) | 15.15 (8.07) | 8.07 (15.15-2.69) | 7.54 (2.99) | 2.99 (7.54-1.13) |
| Tet2–/– (CD45.2) | 13.29 (4.01) | 4.016 (13.29-1.33) | 19.82 (18.12) | 18.12 (19.82-6.04) | 10.2 (1.93) | 1.93 (10.2-0.73) | |
| Ly6Chigh monocytes | WT (CD45.1) | 1.58 (0.79) | 1.42 (0.76-2.54) | 4.791 (4.15) | 3.76 (2.53-4.68) | 2.24 (0.86) | 1.97 (1.66-3.11) |
| Tet2–/– (CD45.2) | 4.19 (1.80) | 3.6 (2.92-5.175) | 6.75 (2.07) | 5.75 (5.32-8.63) | 4.21 (1.08) | 3.68 (3.41-4.96) | |
| B cells | WT (CD45.1) | 59.98 (8.96) | 61.6 (52.25-67.55) | 42.57 (20.34) | 51.2 (24.4-57.45) | 63.13 (20.55) | 70.3 (35.9-78.8) |
| Tet2–/– (CD45.2) | 52.6 (8.44) | 54.4 (46.5-59.7) | 47.12 (17.34) | 48.1 (45.95-59.05) | 65.84 (7.53) | 69.8 (57.5-70.9) | |
| T cells | WT (CD45.1) | 22.97 (8.52) | 23.3 (16.85-30.05) | 11.57 (4.35) | 11.65 (8.44-16) | 11.24 (4.19) | 10.5 (9.13-15.4) |
| Tet2–/– (CD45.2) | 14.21 (2.57) | 13.7 (11.7-16) | 4.028 (1.38) | 4.43 (2.96-5.19) | 5.14 (1.211) | 4.84 (4.32-6.71) | |
| CD8+ T cells | WT (CD45.1) | 8.66 (3.36) | 9.13 (5.08-12.1) | 3.89 (1.45) | 4.17 (2.70-4.82) | 3.28 (1.301) | 3.46 (2.51-4.54) |
| Tet2–/– (CD45.2) | 4.12 (0.73) | 4.29 (3.35-4.63) | 1.13 (0.42) | 1.24 (0.79-1.36) | 1.28 (0.36) | 1.14 (1.02-1.7) | |
| CD4+ T cells | WT (CD45.1) | 13.34 (5.60) | 12 (10.55-17.45) | 5.98 (2.54) | 5.97 (4.30-8.22) | 7.43 (2.787) | 6.78 (5.73-10) |
| Tet2–/– (CD45.2) | 9.63 (1.874) | 8.71 (8.17-11) | 2.70 (0.97) | 2.80 (2.00-3.64) | 3.69 (0.839) | 3.48 (3.13-4.85) | |
| % CD8+T cells | |||||||
| Naïve CD8+ | WT (CD45.1) | 36.46 (9.20) | 36.9 (29.9-42.05) | 16.19 (4.58) | 17.6 (14.35-19.7) | 30.34 (3.37) | 29.1 (29-33.1) |
| Tet2–/– (CD45.2) | 39.28 (5.80) | 37.1 (33.8-45.3) | 13.84 (3.89) | 13.45 (10.4-17.38) | 24.97 (6.36) | 26 (16.9-29.8) | |
| CD8+ TCM | WT (CD45.1) | 52.84 (5.73) | 53.4 (49.35-57.55) | 69.55 (7.42) | 66.75 (64.25-78.23) | 59.66 (3.35) | 60.4 (56.6-62.8) |
| Tet2–/– (CD45.2) | 52.1 (4.59) | 49.6 (48.1-56.6) | 63.9 (7.81) | 63.7 (56.73-70.45) | 49.57 (8.16) | 48.7 (43-55.9) | |
| CD8+ TEM | WT (CD45.1) | 9.911 (10.68) | 5.18 (3.97-13.65) | 13.8 (9.61) | 13.75 (5.695-17.18) | 8.836 (2.10) | 8.13 (7.64-9.95) |
| Tet2–/– (CD45.2) | 8.163 (4.08) | 7.12 (5.06-11) | 21.95 (7.43) | 20 (15.35-28.08) | 24.33 (11.6) | 21 (14-39.5) | |
| CD8+ TVM | WT (CD45.1) | 47.22 (5.81) | 48.2 (42.25-52.1) | 53.69 (7.24) | 54.95 (46.43-60.55) | 49.19 (5.55) | 48.7 (44.2-55.1) |
| Tet2–/– (CD45.2) | 46.34 (4.24) | 45.7 (42.4-50.15) | 50.96 (9.70) | 49.35 (43.68-59.7) | 42.74 (7.54) | 44.9 (38.4-50.1) | |
| % CD4+T cells | |||||||
| Naïve CD4+ | WT (CD45.1) | 30.64 (5.64) | 31.7 (25.25-34.6) | 17.58 (3.44) | 18.45 (15.9-19.3) | 21.49 (3.36) | 19.8 (18.9-24) |
| Tet2–/– (CD45.2) | 23.58 (1.69) | 23.7 (23.05-24.35) | 17.24 (2.56) | 18.05 (14.68-19.13) | 17.59 (2.46) | 16.7 (15.4-20.5) | |
| CD4+ TCM | WT (CD45.1) | 56.97 (4.83) | 56.4 (53.05-62.65) | 54.23 (7.29) | 55.2 (46.95-59.7) | 47.49 (8.97) | 48.5 (38-55.7) |
| Tet2–/– (CD45.2) | 62.51 (3.81) | 61.7 (59.55-65.85) | 51.7 (6.15) | 51.65 (45.05-58.23) | 47.36 (7.36) | 49.7 (41.5-52.9) | |
| CD4+ TEM | WT (CD45.1) | 11.51 (3.84) | 13.1 (8.58-14.05) | 27.45 (8.04) | 25.1 (20.48-35.45) | 27.89 (9.64) | 27.3 (18.1-34.7) |
| Tet2–/– (CD45.2) | 13.17 (3.96) | 13.9 (9.09-17.1) | 30.19 (5.99) | 33.1 (23.7-34.8) | 33.06 (7.45) | 30 (28-38.5) | |
| CD4+ TVM | WT (CD45.1) | 27.2 (4.00) | 28.6 (23.7-30.25) | 49.69 (5.58) | 49.9 (45.65-54.53) | 33.94 (5.53) | 30.8 (30.1-39.6) |
| Tet2–/– (CD45.2) | 24.28 (3.70) | 26.3 (21.2-27.05) | 37.11 (4.37) | 36.75 (34.03-40.4) | 27.89 (4.66) | 25.8 (24.6-31.4) | |
Summary statistics of peripheral blood leukocyte proportions in young WT, old WT, and old TNF–/– recipient mice after engraftment of WT (CD45.1+) and Tet2–/– (CD45.2+) cells.
Abbreviations are explained in Table 1.
The aging microenvironment promotes T-cell remodeling
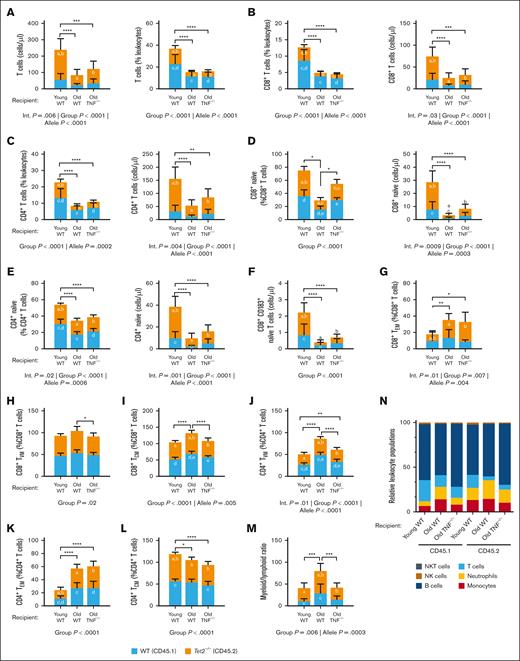
To determine the role of TNF in mutant-Tet2 lymphocyte expansion, we assessed T cells in whole blood of the chimeric mice. Much similar to what was observed in myelomonocytic cells, the absolute counts of Tet2–/– (CD45.2+) cells were higher than WT (CD45.1+) cells in all T-cell subsets; however, the bias toward Tet2–/– cell counts was less pronounced. We found that the proportion and absolute counts of T cells were lower in aged recipient mice (Figure 3A), as were CD8+ and CD4+ T cells (Figure 3B-C). This was primarily driven by a reduction in Tet2–/– (CD45.2+) cells, suggesting that Tet2–/– lymphoid cells have less of a selective advantage in old mice. Beyond the aging-dependent loss of T-cell populations, the chimeras revealed a role for TNF in the differentiation of T-cell subsets. The relative proportion of naïve T cells are known to decrease with age28; however, we show that old TNF–/– recipient mice maintain similar proportions of CD8+ naïve T cells to young recipient mice (Figure 3D). In contrast, the old WT recipients had a significant reduction in both the proportions and absolute counts of CD8+ naïve T cells compared with the old TNF–/– recipients. A similar trend was observed in circulating CD4+ naïve T cells (Figure 3E). Thus, naïve T cells decrease with age, and this reduction is exacerbated in mutant-Tet2 cell subsets, particularly in CD8+ naïve T cells within a TNF-rich environment. One prominent T-cell change that occurred was the loss of CD183 expression on circulating CD8+ naïve cells, important for T-cell trafficking and function29 (Figure 3F). Counter to naïve cells, memory cells increase with aging. Results from the chimeric mice show that both old WT and old TNF–/– recipient mice have an increase in CD8+ effector memory T cells (TEM; Figure 3G). However, only the old WT chimeras had an increase in CD8+ virtual memory T cells (TVM) and central memory T cells (TCM; Figure 3H-I), whereas the TNF–/– recipients were protected from this aging phenomenon. In old WT recipient mice, the proportion of CD4+ TVM cells was likewise higher than young WT and old TNF–/– recipients (Figure 3J). In contrast, old WT recipient CD4+ TEM and CD4+ TCM were similar to that of old TNF–/– recipients, indicating no apparent impact on the aging inflammatory environment on the generation of these cells (Figure 3K-L). The changes in lymphoid cell counts, alongside changes in myeloid cells, resulted in an increased myeloid-to-lymphoid ratio in old WT recipient mice compared with young WT and old TNF–/– recipients (Figure 3M). A summary of leukocyte populations among the chimeric mice is shown in Figure 3N. Collectively, these show a role for Tet2 in driving features of T-cell senescence and immune aging in old mice. The effect of age on lymphocyte populations is worsened in the presence of TNF.
TNF exacerbates Tet2–mutant T-cell remodeling with age. Peripheral whole blood of recipient mice was collected 8 weeks after BMT and analyzed by flow cytometry for the absolute and relative counts of circulating lymphocytes. (A-C) Absolute and relative (as a percentage of total CD45+ leukocytes) counts of total T cells (A), CD8+ T cells (B), and CD4+ T cells (C). (D-E) Absolute and relative counts of CD8+ (D) and CD4+ naïve T cells (E) as a percentage of total CD8+ and CD4+ T cells, respectively. (F) Absolute counts of circulating CD8+ naïve T cells expressing CD183. (G-I) Relative count of CD8+ TEM (G), TVM (H), and TCM cells (I). (J-M) Relative count of CD4+ TVM (J), TEM (K), and TCM cells (L). (M) Myeloid-to-lymphoid ratio. (N) Leukocyte summary. Significance was assessed by a 2-way ANOVA with Tukey multiple comparisons test. Letters in the orange and blue columns denote significant differences (P ≤ .05) in the group means of CD45.2 or CD45.1 alleles, respectively. For all variables with the same letter, the difference between the means is significantly different. If 2 variables have different letters or no letters, they are not significantly different. Black bars with asterisks denote group differences, inclusive of both CD45.1 and CD45.2 alleles. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗P ≤.0001. Abbreviations: TEM, effector memory T cells; TVM, virtual memory T cells; TCM, central memory T cells; NK, natural killer; NKT, natural killer T cells.
TNF exacerbates Tet2–mutant T-cell remodeling with age. Peripheral whole blood of recipient mice was collected 8 weeks after BMT and analyzed by flow cytometry for the absolute and relative counts of circulating lymphocytes. (A-C) Absolute and relative (as a percentage of total CD45+ leukocytes) counts of total T cells (A), CD8+ T cells (B), and CD4+ T cells (C). (D-E) Absolute and relative counts of CD8+ (D) and CD4+ naïve T cells (E) as a percentage of total CD8+ and CD4+ T cells, respectively. (F) Absolute counts of circulating CD8+ naïve T cells expressing CD183. (G-I) Relative count of CD8+ TEM (G), TVM (H), and TCM cells (I). (J-M) Relative count of CD4+ TVM (J), TEM (K), and TCM cells (L). (M) Myeloid-to-lymphoid ratio. (N) Leukocyte summary. Significance was assessed by a 2-way ANOVA with Tukey multiple comparisons test. Letters in the orange and blue columns denote significant differences (P ≤ .05) in the group means of CD45.2 or CD45.1 alleles, respectively. For all variables with the same letter, the difference between the means is significantly different. If 2 variables have different letters or no letters, they are not significantly different. Black bars with asterisks denote group differences, inclusive of both CD45.1 and CD45.2 alleles. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗P ≤.0001. Abbreviations: TEM, effector memory T cells; TVM, virtual memory T cells; TCM, central memory T cells; NK, natural killer; NKT, natural killer T cells.
TNF blockade affects TET2-mutant clonal detection in humans
To determine whether TNF gives an advantage to mutant-TET2 leukocytes in humans, we analyzed the blood of 12 patients with RA (average age, 53 year [range, 33-74]; 11 female and 1 male) before and after TNF blockade using antibodies (HUMIRA [adalimumab]) and 4 patients with RA who were treated with other therapeutics. We first used our successful targeted Ion Torrent–based sequencing approach to isolated genomic DNA from peripheral blood mononuclear cells.20 However, only 1 patient was identified with detectable CHIP (ie, variant present with VAF ≥2%). Because only 1 of 12 patients (8%) manifested with CHIP, as would be expected given the average age of the RA cohort, we sought to increase the sensitivity of our approach to detect smaller CH clones. Variants below the 2% VAF threshold required for CHIP are reportedly more common in adults.30 Therefore, we applied a more sensitive, higher-depth, error-suppression approach to finding CH clones near or below the CHIP VAF threshold using our established smMIP toolset and Illumina sequencing.23 For the patient with detectable CHIP, duplicate sequencing using the smMIP identified the presence of CBL and PPM1D variants, in agreement with the initial Ion Torrent–based sequencing approach. Two additional variants were detected at baseline in the CHIP patient, namely TP53 R196∗ (VAF = 1.34%) and ASXL1 P689L (VAF = 3.13%), but these were not detected at 3 and 6 months after treatment. Four additional patients had detectible CH clones before intervention (Figure 4A-B), with 1 patient having CH clones at 3 months after treatment only. In 1 patient, we detected a STAG2 R1033Q variant at baseline (average VAF = 1.23%) but not after treatment. Similarly, in another patient, a TET2 baseline E798K variant (average VAF = 1.71%) was detectable but not during anti-TNF treatment. The third patient had 2 TP53 variants (R273H and R248Q [average VAF = 2.0%]), which were not detectable at the 3- and 6-month follow-up assessments. The fourth patient had a TET2 baseline variant (average VAF = 1.18%), which was not detectable after treatment; however, a PHF6 R226Q variant (average VAF = 1.11%) was detected at 3 months after treatment. No CH clones were detected at 6 months after treatment. Finally, 1 patient had a PPM1D R458Q variant detected at 3 months after treatment, only (average VAF = 1.4%). Of the patients with RA who were not treated with adalimumab, 1 patient had a detectible CH clone before treatment (TP53 R248Q, 10.7%). Intriguingly, this variant was likewise abrogated after treatment with naproxen, leflunomide, and methotrexate. We questioned whether there were changes in inflammation that may explain the reduction in CH clones and found that over the course of the study the patient’s erythrocyte sedimentation rate was reduced from 44 mm/hour to 2 mm/hour, and C-reactive protein was reduced from 150 mg/L to 1.5 mg/L, indicating that the severe levels of inflammation in this individual at baseline were resolved by the treatment. These findings are the first, to our knowledge, to examine the impact of TNF blockade on CH dynamics in humans and suggest promise for reducing clonal burden. Larger, prospective studies are required to confirm these findings and to determine whether inflammatory cytokine blockade or effective inflammation control with other agents may improve CHIP-comorbid conditions and myeloid cancer risk.
CH clones, including mutant TET2, become undetectable in the peripheral blood of older adults administered the TNF blocker, HUMIRA (adalimumab). Mutant-TET2, ASXL1, TP53, CBL, PPM1D, PHF6, and STAG2 clones identified in the peripheral whole blood of patients with RA before any immunomodulatory treatment (baseline) and at 3 and 6 months after treatment with the adalimumab (V1 and V2, respectively), show a significant reduction in CH clones after treatment. (A-B) Mean ± standard error of the mean (A) and detected mutant clones (B) shown. nd, none detected; V1, visit 1; V2, visit 2.
CH clones, including mutant TET2, become undetectable in the peripheral blood of older adults administered the TNF blocker, HUMIRA (adalimumab). Mutant-TET2, ASXL1, TP53, CBL, PPM1D, PHF6, and STAG2 clones identified in the peripheral whole blood of patients with RA before any immunomodulatory treatment (baseline) and at 3 and 6 months after treatment with the adalimumab (V1 and V2, respectively), show a significant reduction in CH clones after treatment. (A-B) Mean ± standard error of the mean (A) and detected mutant clones (B) shown. nd, none detected; V1, visit 1; V2, visit 2.
Discussion
Here, we provide, to our knowledge, the first in vivo evidence to demonstrate that the clonal dominance of Tet2 mutations is exacerbated by TNF in the aging microenvironment. Somatic mutations in epigenetic regulators, TET2 and DNMT3A, are recurrently detected in people with CHIP.6,31-33 These mutations are thought to contribute a competitive advantage to HSPCs, resulting in clonal expansion of mutant hematopoietic cells.18 In this study, we confirmed that Tet2 mutations cause a competitive advantage of HSPCs after transplantation.34 However, we show that the acquired growth advantage is mitigated in the absence of TNF. Our results provide insights into potential therapeutic targets for CHIP, whereby TET2-mutant and other mutant hematopoietic cells may be reduced after TNF blockade.
The WT/Tet2–/– competitive transplantation experiments enabled us to determine whether the reported competitive advantage of mutant-Tet2 HSPCs is an exclusive cell-intrinsic phenomenon.35 There was a strong myelomonocytic advantage in the BM of old WT recipient mice. This advantage was mitigated in young WT and aged TNF–/– recipient mice, suggesting that age-associated TNF signaling in the BM is essential for the expansion myeloid clones in the chimeric mice. Despite being unable to delineate by CD45 allele type, we have shown that there were no age or genotypic differences in progenitor cells obtained from WT and TNF–/– mice without transplantations. In contrast, there are genotypic differences in the progenitor cells obtained from WT and Tet2–/– mice without transplantation,1 which recapitulate what was observed in the chimeric mice. These results suggest that the increase in myeloid lineage progenitor cells in old WT recipient mice was likely driven by the reconstituted mutant-Tet2 cells. Distinguishing between leukocyte allele types was prioritized in the peripheral blood of chimeric mice because CHIP is defined by the frequency of mutations in circulation. We show that old WT recipient mice have an increase in CD45.2 Tet2–/– inflammatory monocytes and neutrophils in the peripheral blood as compared with young WT recipient mice. The old TNF–/– recipient mice were protected from this phenotype, suggesting that TNF-dependent mechanisms are integrated to coordinate aberrant Tet2–/– myeloid regeneration.
We considered whether mutations in Tet2 provide a differential advantage to myeloid cell subsets. When myeloid cell populations were analyzed as a proportion of their parent population (eg, CD45.2+ Ly6Chigh monocytes as a percentage of CD45.2+ leukocytes), we found that both WT and Tet2–/– myeloid cell populations were dictated by the inflammatory environment and not intrinsic cell mechanisms. Our results show that TNF inflammation precipitates an increase in the proportion of Ly6Chigh inflammatory monocytes that contribute to TNF inflammation in a positive feedback loop. This inflammation, in turn, leads to the expansion of Tet2–/– myeloid cell numbers. Given these results, we surmised that targeting age-associated TNF should reduce inflammatory myeloid subsets as well as the number of circulating mutant-Tet2 myeloid clones.
Within the lymphoid compartment, we report a diminished advantage of mutant-Tet2 leukocytes in old recipient mice, contributing to immunosenescence. With age, there is a known change in the number and composition of lymphocytes in circulation. T-cell subsets shift from naïve T cells to TCM and TEM, accompanied by an increase in antigen naïve but semidifferentiated TVM.36 These alterations, together with an increase in the myeloid-to-lymphoid ratio, are key features of immunosenescence.37 We found that the absolute counts of T cells were lower in aged recipient mice, as were the CD8+ and CD4+ T cells. This was driven by a marked reduction in the number of Tet2–/– (CD45.2+) cells, whereas WT (CD45.1+) cells were not different among the groups. The reduction in mutant-Tet2 lymphocytes together with the increase in myeloid cells culminated in a pronounced myeloid-to-lymphoid ratio in the old WT recipient mice, whereas the old TNF–/– recipient mice were protected from this aging hallmark. In agreement with published literature,36 we show that the distribution of T-cell subpopulations change with aging. The WT (CD45.1+) naïve T-cell counts decreased in old WT recipient mice when compared with young WT recipients. This was accompanied by an increase in CD8+ TCM and CD4+ TVM, as well as TEM. We show that the old TNF–/– recipients were better able to maintain the number of WT (CD45.1+) naïve T cells as well as CD8+ TCM, suggesting that age-associated TNF drives features of immunosenescence in normal cells. Analyses of the Tet2–/– (CD45.2+) lymphocyte subpopulations revealed a more pronounced decrease in naïve T cells with aging in both WT and TNF–/– recipient mice. The remaining changes in Tet2–/– (CD45.2+) memory T-cell subsets strongly resembled the changes in WT cells, with the old TNF–/– recipient mice maintaining a better T-cell subset distribution. In prior studies of mutant-Tet2 T-cell differentiation, it was reported that the loss of Tet2 promotes CD8+ T-cell memory differentiation.38 We speculate that this phenotype is a product of mutant-Tet2–driven TNF inflammation, rather than a cell-intrinsic event.
The present results suggest that age-associated changes in TNF underlie many observed features of mutant-Tet2 fitness and lineage expansion in mice, and therefore, therapeutic interventions aimed at improving TNF inflammation may reduce mutant-TET2 and perhaps other forms of CHIP. For instance, it has been demonstrated that aging-induced TNF signaling through TNF receptor 1 (TNFR1) promotes the selective advantage of DNMT3A-mutant CH,39 suggesting similar underpinning mechanisms of multiple forms of CHIP. Herein, we show a reduction in TET2- and other mutant clones in older patients with RA after TNF blockade with adalimumab. We hypothesize that a reduction in mutant clones might be responsible for reducing the risk of CVD in patients with RA on TNF inhibitors40; however, further study in a larger cohort is needed to evaluate this relationship. In addition, the presented data were acquired from female mice and predominately female patients with RA. We encourage further research on the impact of biological sex, because aging phenotypes vary. We also acknowledge that we did not have young TNF–/– recipient mice, which is a limitation in this study. Finally, although the results highlight TNF blockade as a therapeutic target, we also found a reduction in a CH clone in a patient with RA who was not on adalimumab. This patient had a significant reduction in inflammatory markers, and we hypothesize that targeting other inflammatory mediators with overlapping biological properties, (eg, TNF, IL-1, and IL-6) may also reduce CH clones. In support of this, it has been shown that IL-1 signaling drives leukogenesis induced by Tet2 loss,20 and blockage of IL-1 signaling in old WT mice reverses myeloid-biased output of their HSC population.19 Genetic deletion or pharmacologic inhibition of IL-1 signaling impairs Tet2+/– clonal expansion in aging mice, identifying the IL-1 pathway as another relevant and therapeutically targetable driver of Tet2+/– CHIP progression.41 Taken together, our murine model and human data provide evidence that targeting inflammatory mediators such as TNF may be a viable means to reduce TET2-mutant and other genetic drivers of CHIP.
Acknowledgments
D.M.E.B. was funded through the Canadian Research Chairs program and by project grants from the Canadian Institutes of Health Research (grant numbers PJT-156291 and PJT-163047). C.Q. was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship Award. Additional funding was provided to S.A. and M.J.R. from the Ontario Institute for Cancer Research (project number CPTRG-056).
Authorship
Contribution: D.M.E.B. and M.J.R. conceived and funded the experiments; D.M.E.B., M.J.R., and C.Q. designed the experiments; C.Q. and E.N.D. performed flow cytometry and tissue collection; A.J.M.M. bred and genotyped all mice; C.Q. performed all analysis and critically evaluated the data; M.J.L. diagnosed, treated, and collected blood from patients with rheumatoid arthritis; M.M.B., S.B., and S.A. performed clonal hematopoiesis of indeterminate potential analysis; and C.Q., M.J.R., and D.M.E.B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dawn M. E. Bowdish, Department of Medicine, McMaster University, 1280 Main St W West, Hamilton, ON, L8S 4K1, Canada; email: bowdish@mcmaster.ca.
References
Author notes
Detailed protocols and data sets supporting the results of this article are available in the Open Science Framework database repository at https://osf.io/jy7c5/.
The full-text version of this article contains a data supplement.